Abstract
Staphylococcus sciuri is an emerging human pathogen widely found in dairy industries. In this study, we have isolated methicillin resistant Staphylococcus sp. from biofilm formed on utensil used in the dairy society situated at Raia, Goa and was designated as NN14. The isolate NN14 was identified through 16S rRNA sequencing as S. sciuri (GenBank accession number MF621976). This report reveals that the S. sciuri strain NN14 responds positively to the, acyl-homoserine lactone (AHL) having 6-carbon long acyl chain i.e. N-hexanoyl-homoserine lactone molecule (C6-HSL) with gradual rise in their biofilm establishing potential as the concentration of AHL was increased from 250 nM, 500 nM to 1 µM when compared to control (without C6-HSL) by performing crystal violet assay using 48 well microtiter plate. Also, exopolysaccharide (EPS) production was found to increase with gradual increase in C6-HSL concentration from 250 nM, 500 nM to 1 µM proving potential role of EPS in biofilm formation. These results were further proved by scanning electron microscopy where increased in biofilm and EPS production with increase in C6-HSL concentration was observed. The biofilm forming capability of S. sciuri strain NN14 was found to decreased significantly when it was subjected to 10 µg/ml of (R)-2-(2-hydroxynaphthalen-1-yl)-thiazolidine-4-carboxylic acid, however with the addition of 250 and 500 nM, C6-HSL in presence of the antimicrobial compound (R)-2-(2-hydroxynaphthalen-1-yl)-thiazolidine-4-carboxylic acid, the biofilm development in bacterial strain NN14 was increased when compared with control. Our results demonstrated that the C6-HSL molecule neutralize the effect of antibacterial compound and enhances EPS production and biofilm development in S. sciuri.
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3123-0) contains supplementary material, which is available to authorized users.
Keywords: Staphylococcus sciuri, Biofilm, Acyl homoserine lactone, Antimicrobial compound, EPS
Introduction
Staphylococcus sciuri is coagulase negative Gram-positive cocci and generally isolated from body of various farm animals, cow milk, pets, wild animals, humans and from various food products of animal origin (Becker et al. 2012; Piessens et al. 2012). Although S. sciuri are primarily associated with animals they are also reportedly isolated from various human clinical samples (Stepanovic et al. 2003, 2005). They are well known emerging human pathogens and found associated with serious infections like Pelvic inflammatory disease, septic shock, urinary tract infection, peritonitis, endocarditis, endopthalmitis and wound infections (Hedin and Widerstrom 1998; Wallet et al. 2000; Horii et al. 2001; Stepanovic et al. 2002, 2003). It is a temporary coloniser of human body entry portals (nasal passage, ear, eye, skin etc.). But any skin abrasions or person with compromised immune system gives opportunity for this bacterium to cause infection (Stepanovic et al. 2003). S. sciuri is an opportunistic pathogen found in cow milk and dairy utensils of controversial clinical significance and found to possess virulence factors like clumping factor, enterotoxin, capacity to stimulate nitric oxide production, hemolysin, production of spreading factor, methicillin resistance encoded by mecA gene and biofilm forming potential (Stepanovic et al. 2002, 2003; Piessens et al. 2012; Mohantana et al. 2015). Transmission of S. sciuri normally occurs from infected udder to uninfected udder during milking processes and utensils used during milking, improper handling of milk and hygiene can result into transmission of S. sciuri to humans.
Quorum sensing (QS) is a cell to cell communication mechanism between bacteria which helps in regulating biofilm development, plasmid transfer, swarming, bioluminescence etc. (Irie and Parsek 2008; LaSarre and Federle 2013; Naik et al. 2017). It also helps in provoking the expression of virulence genes like exotoxin, siderophores, enzymes, adhesion molecules (Novick and Muir 1999; Gonzalez and Keshavan 2006; LaSarre and Federle 2013). Both Gram positive and Gram-negative bacteria possess different types of quorum sensing systems (QS). Gram positive bacteria use peptides based QS and Gram-negative bacteria use acyl homoserine lactone (AHL) molecules based QS (LaSarre and Federle 2013; Naik et al. 2017). There is only one report demonstrating AHL as quorum sensing system in a marine Gram-positive bacteria Exiguobacterium sp. (Biswa and Doble 2013), but till date there is no report on production of AHL by Staphylococcus spp. There is report saying that Staphylococcus aureus responds negatively to exogenous AHL by down regulation of virulence genes such as exotoxin, cell wall fibronectin binding proteins and agr QS system (Qazi et al. 2006), whereas Listeria monocytogenes strain BN3 reported to respond positively to AHL molecules by increased biofilm development (Naik et al. 2017). But till date there is no report on response of Staphylococcus spp. to AHL molecules in respect to biofilm formation. Gram negative prokaryotic bacteria viz. Citrobacter freundii, Pseudomonas aeruginosa, Proteus mirabilis and Enterobacter agglomerans, reported to produce cyclic dipeptides which are capable of activating AHL biosensor in concentration dependent manner (Holden et al. 1999). Holden et al. (1999), confirmed that Gram negative bacteria apart from AHL production are also capable of producing cyclic peptide molecules for quorum sensing cross talk. Salmonella enterititis is a Gram-negative bacterium but never reported to produce AHL, however recently de Almeida et al. 2017, reported exogenous acyl homoserine lactone stimulates biofilm formation in Salmonella enteritidis. Therefore, it is imperative to study biofilm forming emerging pathogens like S. sciuri in dairy industry and response of S. sciuri to exogenous AHL to understand the reason behind persistence of S. sciuri in dairy industry where Gram-negative and Gram-positive bacteria share the same econiche.
In this investigation we have isolated S. sciuri from biofilm formed on milk collecting utensils of the local dairy society at Raia, Goa, which supply milk to Goa Dairy, Goa. We have also tried to investigate response of emerging pathogen S. sciuri to exogenous AHL molecule i.e. C6-HSL in EPS production and biofilm formation.
Materials and methods
Isolation of Staphylococcus sp. capable of forming biofilm from local dairy
Swabbing were taken using sterile swabs moisten with sterile saline (0.85%) from surfaces of dairy utensil (steel) which are used for collecting milk in local dairy society at Raia, Goa and transferred in 5 mL of 0.85% sterile saline in 50 mL centrifuge tube and then vortexed. This sample was processed within 24 h for enrichment and isolation of Staphylococcus sp.
Enrichment of sample
Sample (5 mL) was inoculated in sterile Peptone water broth (50 mL) in Erlenmeyer flask (100 mL capacity) and flask was incubated at 37 °C for 48 h with continuous shaking at 150 rpm for enrichment of Staphylococcus spp.
Isolation of Staphylococcus sp. on Mannitol Salt Agar
A loop full of enriched peptone water broth was streaked on selective and differential Mannitol salt agar (MSA) plates and incubated for 24–48 h at 37 °C. After incubation the typical yellow coloured colony surrounded by yellow zone due to mannitol fermentation was picked and purified on fresh MSA plates (Ateba et al. 2010; Dharmik and Gomashe 2011). The bacterial isolate was preserved at 4 °C on Brain heart infusion agar (BHI) slants and was designated as NN14.
Growth on Baird Parker agar
The bacterial isolate NN14 was further streaked on Baird Parker agar and incubated for 24–48 h at 37 °C. After incubation, shiny greyish black colony was observed due to tellurite reduction and opaque zones of precipitation around colonies was seen which is due to lecithinase production (Thaker et al. 2013). This colony was presumed to be Staphylococcus sp.
Biochemical tests for identification of bacterial isolate NN14
Staphylococcus isolate on Baird Parker agar showing lecithinase production as opaque zones of precipitation around colony was further identified using biochemical tests. Biochemical tests like sugar fermentation, Gram staining, methyl red, nitrate reduction and motility at 37 °C etc. were tested and identified by following Bergey’s manual of systematic bacteriology (Sneath et al. 1986).
16S rRNA sequencing of bacterial isolate NN14 for identification
Eubacterial primers: 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) were used for PCR amplification of 16S rRNA gene using genomic DNA of Staphylococcus sciuri as template. Sequencing was performed at Yaaz Xenomics, Madurai, Tamil Nadu, India. Using NCBI BLAST search (https://www.ncbi.nlm.nih.gov/Genbank), 16S rRNA sequence was compared against GenBank database and analysed.
Antibiotic sensitivity test
Antibiotic sensitivity test for S. sciuri strain NN14 was performed using Hexa G-plus disc HiMedia containing VANCOMYCIN (30 µg/disc), STREPTOMYCIN (10 µg/disc), LINEZOLID (30 µg/disc), METHICILLIN (10 µg/disc), GENTAMICIN (10 µg/disc) and PENICILLIN-G (10 units/disc) on Muller- Hinton agar plates (Bauer et al. 1966).
Siderophore and blood hemolysin production by S. sciuri strain NN14
Staphylococcus sciuri strain NN14 was spot inoculated on Chrome Azurol S (CAS) agar plates and incubated for 24–48 h at 37 °C to test siderophore production potential (Schwyn and Neilands 1987). The S. sciuri strain NN14 was further tested for the hemolysin producing ability by streaking on 5% sheep blood agar to check its virulence potential.
Bioreporter Chromobacterium violaceum MCC2216 bacteria for AHL detection assay
In the present investigation Chromobacterium violaceum strain MCC2216 was procured from Microbial Culture Collection (MCC) situated at National Centre for Cell Science (NCCS), Pune, which is a bioreporter strain used for AHL detection. Luria–Bertani agar (LB) plate was prepared and bioreporter C. violaceum MCC2216 was streaked on it. AHL molecules production by S. sciuri was checked by cross-streaking S. sciuri strain NN14 against C. violaceum MCC2216 on LB agar and incubating for 24–48 h at 37 °C. Wild type C. violaceum synthesize violet coloured pigment on LB agar, whereas C. violaceum MCC2216 does not produce the violet pigment in the absence of external AHL molecules because it is a mutated strain of C. violaceum. Therefore, if S. sciuri produces AHL molecules, then C. violaceum MCC2216 will produce violet pigment at the streak of S. sciuri strain NN14, demonstrating AHL production (Naik et al. 2017). For AHL production assay, bioluminescent marine bacterium Vibrio harveyi, which is well known to produce AHL molecules was used as positive control. Bioluminescent V. harveyi is a symbiont of marine squid and was isolated on bioluminescent agar.
Response to C6-HSL by S. sciuri strain NN14
AHL molecule N-hexanoyl-homoserine lactone (C6-HSL) was obtained from Sigma Aldrich, USA. AHL stock solution (1 M) was prepared, filtered through 0.22 µM size nitrocellulose filter and stored at 4 °C. Crystal violet assay was performed to check the biofilm developing ability of S. sciuri strain NN14 using 48 well microtiter plate. Here 500 µL (0.5 mL) of 16 h old S. sciuri strain NN14 grown in BHI broth was added to 500 μL sterile BHI broth in sterile polystyrene microtiter plate. Plain BHI broth (1 mL) without NN14 inoculation was kept as negative control and then microtiter plate was incubated at 37 o C for 48 h. Culture broth in microtiter plate was drained after 48 h incubation and the microtiter plate was gently washed with phosphate buffered saline (PBS) followed by washing twice with 1 mL sterile 0.85% saline. The plate was dried by inverting it on blotting paper for 25 min. Bacterial biofilm formed on walls of microtiter well (polystyrene surface) was then stained with crystal violet by adding 1 mL of 0.1% crystal violet to microtiter plates and kept for 40 min at room temperature. Excess crystal violet was then drained and washed off by rinsing microtiter plate gently with 1 mL sterile distilled water for 2-3 times. Microtiter plate was then air dried and crystal violet was solubilised by adding 1 mL of 30% acetic acid and agitated for 5 min and OD was taken at 595 nm using UV–Vis spectrophotometer (Shimadzu, Model UV 2450, Japan) by keeping blank as 30% acetic acid (Merritt et al. 2011; Naik et al. 2017).
Likewise, response of S. sciuri strain NN14 to different concentration of C6-HSL was tested by performing CV assay in sterile polystyrene microtiter plate (48 well) using BHI broth. Effect of exogenous AHL molecules on biofilm forming ability of S. sciuri strain NN14 was checked in presence of varying concentration (250 nM, 500 nM and 1 μM) of C6-HSL molecule and also in presence of 10 μg/mL of A(4)2015 i.e. (R)-2-(2-hydroxynaphthalen-1-yl)-thiazolidine-4-carboxylic acid) an antibacterial compound by using CV assay. Biofilm forming ability of S. sciuri strain NN14 was also tested in presence of thiozolidine derivative and C6-HSL together i.e. A(4)2015 + 250 nM C6-HSL and A(4)2015 + 500 nM C6-HSL. For each concentration of C6-HSL or A(4)2015 three replicates were maintained. BHI broth with S. sciuri strain NN14 without C6-HSL and A(4)2015 was considered as control whereas plain BHI broth was kept as negative control. Different thiazolidine derivatives are well known for their antibacterial activities against Gram positive bacteria (Jain et al. 2012; Naik et al. 2017). Antimicrobial compound A(4)2015 used in the present study was synthesized and provided by Dr. Chinmay Bhat (Online resourse 1).
Scanning electron microscopy (SEM) was also used to examine biofilm developing potential of S. sciuri strain NN14 in the presence of C6-HSL and antibacterial compound A(4)2015. Sterile thin cover glass pieces (0.5 cm2) were put in microtiter wells containing varying concentrations of C6-HSL molecule and 10 μg/mL of A(4)2015 and also in negative and positive control microtiter wells to examine biofilm formation. Three replicates were kept for each concentration. Cover glass pieces were removed using sterile forceps after incubation for 48 h at 37 °C and then BHI broth from covers glass was removed by washing gently with sterile saline. Cover glass pieces were then fixed for 8 h with 3% glutaraldehyde in 50 mM PBS at 4 °C. After this step gently wash cover glass thrice with PBS (pH 7) and bacterial cells dehydrated in progressively increasing concentrations of acetone i.e. 10, 20, 50, 70, 80, 90 and 100% for 15-20 min each (Naik et al. 2017). After dehydrating with 100% acetone cover glass pieces were air dried and kept in desiccator prior to SEM analysis to prevent spoilage of sample. The airdried cover glass pieces (size 0.5 cm2) were then coated with thin film of gold and examined under SEM (Zeiss EVO18). Within 2 days of sample preparation SEM analysis was carried out.
EPS production potential of bacterial isolate S. sciuri strain NN14 in response to C6-HSL
Exopolysaccharide (EPS) production in S. sciuri strain NN14 in presence of different concentration (250 nM, 500 nM and 1 μM) of C6-HSL molecules was studied in 100 mL sterile BHI broth in Erlenmeyer flask (250 mL) and then incubated at 37 °C for 48 h. BHI broth without added C6-HSL was kept as control. Culture broth after 48 h was centrifuged at 8000 rpm and resultant culture supernatant was filtered through sterile 0.22 µM nitrocellulose filter. Precipitation of EPS from final culture filtrate was done by using ice cold ethanol precipitation method (Naik et al. 2012). EPS was then lyophilised and dry weight of all 4 samples (in triplicate) was recorded on analytical electronic balance and compared.
Results and discussion
Isolation of Staphylococcus sp. capable of forming biofilm from local dairy
After enrichment of Staphylococcus spp. from swabbing taken from surface of dairy utensils in peptone broth for 48 h, a loop full from enriched peptone water broth was streaked on MSA agar plate. After incubation for 24-48 h at 37 °C, one yellow colour isolated colony on MSA and changing surrounding media from red colour to yellow due to mannitol fermentation was picked and further purified on MSA agar (Online resource 2), and tentatively identified as Staphylococcus sp. NN14. The isolate NN14 was then streaked on Baird Parker agar. After incubation at 37 °C for 48 h greyish black colonies with lecithinase activity (white little opaque circular ring around the colonies) was seen on Baird parker agar which confirmed Staphylococcus sp. (Online resource 3).
Biochemical tests and 16S rRNA sequencing of bacterial isolate NN14 for identification
Bacterial isolate NN14 was Gram positive cocci and non-motile. Bacterium showed ability to utilize MANNITOL but was not able to ferment RAFFINOSE sugar. After comparing all the biochemical results with Bergey’s manual of systematic bacteriology, the bacterial strain NN14 was identified as S. sciuri. Furthermore, through 16S rRNA, sequence analysis and comparing with GenBank database by means of NCBI BLAST search, bacterial isolate NN14 was identified with confirmation as S. sciuri strain NN14 having GenBank accession number MF621976 (https://www.ncbi.nlm.nih.gov/Genbank).
Antibiotic sensitivity test
It was observed that the dairy isolate S. sciuri strain NN14 showed resistant towards PENICILLIN-G and METHICILLIN but was susceptible to LINEZOLID, STREPTOMYCIN, VANCOMYCIN and GENTAMYCIN antibiotics. The treatment of infections caused by multi-antibiotic resistant strains of emerging pathogen S. sciuri may become a global issue in upcoming years.
Siderophore and blood hemolysin production by S. sciuri strain NN14
Orange coloured zone around the colony of S. sciuri on CAS agar plate confirmed siderophore synthesis by S. sciuri strain NN14 (Online resource 4). S. sciuri strain NN14 synthesis siderophores to survive in iron (Fe) starved situations and also chelate Fe from human during infection to weaken their immune system which is important for survival of pathogens and thus establish itself in host. β-hemolysis was seen on sheep blood agar further confirmed pathogenic potential of emerging coagulase negative pathogen S. sciuri strain NN14 (Online resource 5).
Bioreporter Chromobacterium violaceum MCC2216 bacteria for AHL detection assay
Bioreporter C. violaceum MCC2216 didn’t produce violet colour pigment when S. sciuri strain NN14 was cross-streaked with bioreporter C. violaceum MCC2216. However, when bioluminescent V. harveyi isolated from marine squid cross-streaked with bioreporter C. violaceum MCC2216, it was seen that bioreporter C. violaceum MCC2216 synthesized the violet colour pigment where V. harveyi was cross streaked (Online Resource 6). This proved that S. sciuri strain NN14 do not produce AHL molecules. Till date, there is no report on S. sciuri synthesizing AHL molecules and our results also support earlier reports saying only agr QS system is present in Gram positive bacteria (Novick and Muir 1999; Gray et al. 2013; LaSarre and Federle 2013; Naik et al. 2017).
Response of S. sciuri strain NN14 to C6-HSL molecule
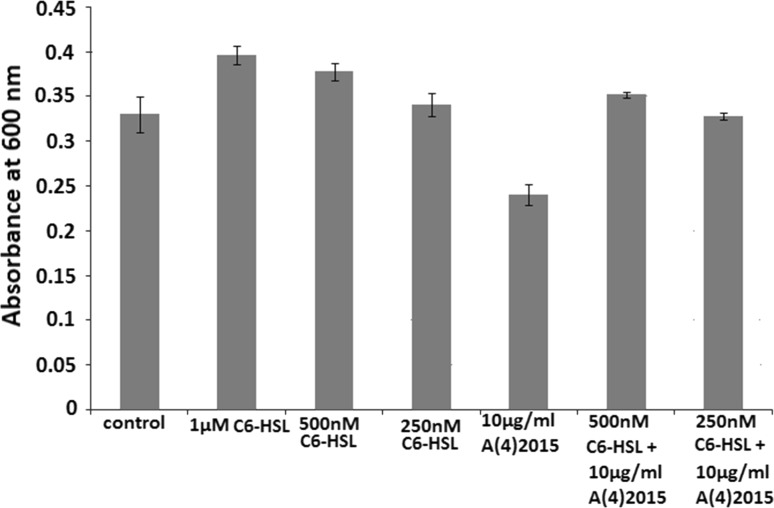
Gram positive S. sciuri was never reported to produce AHL molecules and till date there are no reports on S. sciuri responds to external AHL molecules. In current investigation by performing crystal violet assay using microtiter plate we have seen that there is 6.8% increase in biofilm forming potential of S. sciuri strain NN14 when 250 nM, C6-HSL was added in BHI broth (Fig. 1; Online resource 7). Similarly, when 500 nM C6-HSL was added to microtiter wells there was 18.49% enhancement in biofilm formation as compared to control and furthermore there was 24.76% rise in biofilm development in presence of 1 µM C6-HSL (Fig. 1). When biofilm formation in control microtitre wells was statistically compared with biofilm formation when 500 nM C6-HSL was added, the difference was found to be statistically significant (p value = 0.019; p value < 0.05). Biofilm formation in S. sciuri strain NN14 was significantly increased in presence of 500 nM C6-HSL as compared to control. Also, when biofilm formation in control microtitre wells was statistically compared with biofilm formation when 1 µM C6-HSL was added, the difference was found to be statistically significant (p value = 0.0062; p value < 0.05). Here biofilm formation in S. sciuri strain NN14 was significantly increased in presence of 1 µM C6-HSL as compared to control. These results revealed that S. sciuri strain NN14 responds positively to C6-HSL by enhanced biofilm formation in concentration dependent manner when compared to control. Therefore, it is concluded that AHL plays a crucial role in biofilm development in S. sciuri strain NN14, although it doesn’t produce AHL molecules.
Fig. 1.
Crystal violet assay to study response of S. sciuri strain NN14 to different concentrations of exogenous 6 carbon long acyl chain acyl homoserine lactone (AHL) molecules i.e. N-hexanoyl-l-homoserine lactone (C6-HSL) in biofilm development
When antibacterial compound A(4)2015 (thiazolidine derivative) was added to microtiter well containing BHI broth inoculated with strain NN14 broth at 10 μg/mL concentration there was 20.68% decrease in biofilm formation revealed by Crystal violet assay which proved that this antimicrobial compound has antimicrobial effect on S. sciuri strain NN14 and biofilm formation. When biofilm formation in control microtitre wells was statistically compared with biofilm formation when 10 μg/mL A(4)2015 was added, the difference was found to be statistically significant (p value = 0.02; p value < 0.05). Biofilm formation in S. sciuri strain NN14 was significantly decreased in presence of 10 μg/mL A(4)2015 as compared to control. But when we added 250 nM of C6-HSL molecules in presence of antimicrobial compound A(4)2015 (10 μg/mL) we had observed that there was only 2.8% decrease in biofilm development when compared with control i.e. when 250 nM C6-HSL added it nullify overall 17.88% effect of A(4)2015. Moreover when 500 nM, C6-HSL is added in the presence of 10 μg/mL antibacterial compound A(4)2015, there was overall 7.2% increase in biofilm forming potential of S. sciuri strain NN14 as compared to control (Fig. 1) which proves that the C6-HSL molecules cancel out the effect of antibacterial compound A(4)2015. When biofilm formation in microtitre wells containing only 10 μg/mL A(4)2015 was statistically compared with biofilm formation when 500 nM, C6-HSL was added to microtitre plate containing 10 μg/mL A(4)2015, the difference was found to be statistically significant (p value = 0.01; p value < 0.05). Biofilm formation in S. sciuri strain NN14 was significantly increased when 500 nM C6-HSL was added to microtitre plate containing 10 μg/mL A(4)2015 as compared to biofilm formation in presence of only 10 μg/mL A(4)2015 (Fig. 1) which confirmed that 500 nM C6-HSL nullify antibacterial effect of A(4)2015.
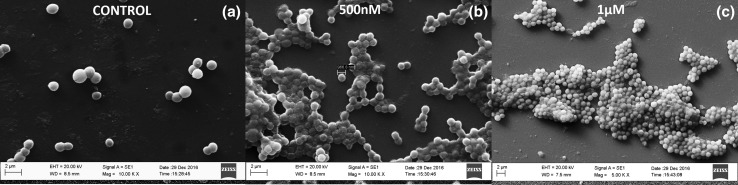
Results obtain using crystal violet assay in microtitre plate revealing positive response of S. sciuri strain NN14 to C6-HSL by enhanced biofilm formation in concentration dependent manner was further supported by Scanning electron microscopy data. Here we observed that as there was increase in concentration of C6-HSL (500 nM and 1 µM) there was significant increment in biofilm development in concentration dependent manner as compared to control (Fig. 2a–c). The SEM image of control showed isolated cells of S. sciuri strain NN14 but when the C6-HSL concentration was rise to 500 nM, cells started forming aggregates (biofilm) and significant amount of EPS production was seen, and when C6-HSL concentration was further increased to 1 µM there was a dense multilayer biofilm developed and EPS production was further increased approximately 2.5 times compared to control, which revealed the positive effect of C6-HSL QS molecule on biofilm development and EPS production (Fig. 2a–c). But when we exposed S. sciuri strain NN14 to 10 µg/mL concentration of A(4)2015 we observed that there was significant reduction in biofilm development as compared to control (Fig. 3a). However, when S. sciuri strain NN14 was treated with antibacterial compound A(4)2015 with 10 μg/mL concentration in the presence of 500 nM C6-HSL molecule, we observed significant increment in biofilm development, which suggests the involvement of AHL molecule in biofilm development and the biofilm developed results in resistance of S. sciuri strain NN14 to A(4)2015 (Fig. 3b, c). The increase in biofilm formation in Gram negative bacteria in response to C6-HSL is well studies (Irie and Parsek 2008; Shrout et al. 2011; LaSarre and Federle 2013) but present study is a primary report on Gram Positive S. sciuri strain NN14 respond to C6-HSL by increase in biofilm development.
Fig. 2.
Biofilm development ability of S. sciuri strain NN14 in the presence of a 0 nM C6-HSL (control), b 500 nM C6-HSL and c) 1 µM C6-HSL in BHI broth in a microtiter plate. A sterile and clean cover glass (0.5 cm2 size) was inserted into each microtiter well and incubated for 48 h and then observed for biofilm development on cover glass using SEM
Fig. 3.
Biofilm development ability of S. sciuri strain NN14 in the presence of a 10 μg/mL A(4)2015; b, c 10 μg/mL A(4)2015 + 500 nM C6-HSL in BHI broth on microtiter plate. A sterile and clean cover glass (0.5 cm2 size) was inserted into each microtiter well and incubated for 48 h and then observed for biofilm development on a thin cover glass using SEM
EPS production potential of bacterial isolate of S. sciuri strain NN14 in response to C6-HSL
Exopolysaccharide (EPS) production in S. sciuri strain NN14 in presence of different concentrations (250 nM, 500 nM and 1 μM) of C6-HSL molecules was studied. It was observed that in control flask EPS production was found to be 30 ± 1.5 mg/L but when S. sciuri strain NN14 was exposed to 250 nM C6-HSL there was increase in EPS production to 44.5 ± 2.6 mg/L. Similarly, when 500 nM C6-HSL was added to BHI broth there was significant increase in EPS production noted i.e. 67 ± 3.1 mg/L. Moreover, enhancement of EPS production by S. sciuri strain NN14 to 79.8 ± 3.7 mg/L was observed when 1 µM C6-HSL was added in the media. These results along with SEM images confirm that there is positive enhancement of EPS production in S. sciuri strain NN14 in response to increase in concentration of C6-HSL in concentration dependent manner. Therefore, these results confirmed positive relationship between EPS production and biofilm formation since increase in biofilm formation was observed with corresponding increase in EPS production. C6-HSL was found to positively regulate EPS production and biofilm formation, because gradual increase in C6-HSL concentration resulted in gradual increase in EPS production and biofilm formation. It is well known that EPS production increases in Gram negative bacteria in response to C6-HSL (LaSarre and Federle 2013) but present investigation is a first report on response of Gram-positive bacteria S. sciuri to C6-HSL by increased EPS production which helps in biofilm development.
The capacity to develop biofilm is the most significant virulence factor present in coagulase negative Staphylococci (CoNS) like S. sciuri, which facilitates its adherence and colonisation (Soumya et al. 2017). Coagulase negative Staphylococci are emerging as multiple drug resistant pathogens (e.g. methicillin and vancomycin) over the years and also there is increased incidence of biofilm formation on catheter and catheter associated infections (Soumya et al. 2017). In the present study it was observed that methicillin resistant S. sciuri strain NN14 isolated from local dairy society was found positive for virulence factors hemolysin and siderophores and also positively responds to C6-HSL molecules by enhanced biofilm formation and EPS production in concentration dependent manner. Response of Gram positive, S. sciuri isolated from milk collecting utensil to AHL, generally produced by Gram negative bacteria may be because of sharing same econiche by both Gram positive and Gram-negative bacteria on milk utensils used in milk industry. Since both Gram positive and Gram-negative bacteria share same econiche on milk utensils, Gram positive S. sciuri may have frequently encountered AHL produced by Gram negative bacteria and therefore over the years S. sciuri may have develop mechanism for response to AHL QS molecules synthesized by Gram negative bacteria. Researchers develop variety of Quorum sensing inhibitor (QSI) molecules which interfere with either agr QS system or AHL QS system to tackle biofilm formation, antibiotic resistance and expression of virulence factor by Gram positive or Gram-negative bacteria respectively (LaSarre and Federle 2013; Naik et al. 2017). S. sciuri strain NN14 showed resistance to antibacterial molecule (R)-2-(2-hydroxynaphthalen-1-yl)-thiazolidine-4-carboxylic acid in presence of C6-HSL molecules, therefore to control biofilm formation on dairy utensils by methicillin resistant Gram positive emerging pathogen S. sciuri, researchers should not only target agr quorum sensing system but should target both AHL QS and agr QS together for better results. Since infection caused by methicillin resistant emerging pathogen S. sciuri through consumption of dairy products, will be difficult to treat, therefore controlling biofilm on dairy utensils by targeting Quorum sensing system is very good strategy for controlling dairy food borne infections. Present study confirmed the cross-talk between Gram positive and Gram-negative bacteria and helped us to understand persistence of emerging pathogen S. sciuri in dairy industry and how both Gram positive bacteria and Gram-negative bacteria respond to each other when present in same econiche.
Conclusion
The present study reports on multi-antibiotic resistant emerging pathogen S. sciuri, which responds positively to the N-hexanoyl-homoserine lactone (C6-HSL) molecule with increase in their exopolysaccharide producing and biofilm forming potential in a concentration dependent manner. We confirmed the cross-talk between Gram positive and Gram-negative bacteria through AHL QS. S. sciuri strain NN14 resist anti Staphylococcal compound in presence of AHL molecule, therefore to control biofilm formation and persistence of multidrug resistant emerging pathogen S. sciuri in Dairy/food industry, researchers should target both AHL QS and agr QS together for better results.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
SERB-DST Young Scientist Project (File Number: YSS/2014/000258). Thanks to Ms. Purva Bhangui and Dr. Shyamalina Haldar, Goa University.
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3123-0) contains supplementary material, which is available to authorized users.
References
- Ateba CN, Mbewe M, Moneoang MS, Bezuidenhout CC. Antibiotic resistant Staphylococcus aureus isolated from milk in the Mafikeng area, North West province, South Africa. S Afr J Sci. 2010;106:1–6. doi: 10.4102/sajs.v106i11/12.243. [DOI] [Google Scholar]
- Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- Becker K, Heilmann C, Peters G. Coagulase-negative Staphylococci. Clin Microbiol Rev. 2012;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswa P, Doble M. Production of acylated homoserine lactone by Gram-positive bacteria isolated from marine water. FEMS Microbiol Lett. 2013;343:34–41. doi: 10.1111/1574-6968.12123. [DOI] [PubMed] [Google Scholar]
- de Almeida FA, Pimentel-Filho NJ, Pinto UM, Montavani HC, de Oliveira LL, Vanrtti MCD. Acyl homoserine lactone-based quorum sensing stimulates biofilm formation by Salmonella enteritidis in aerobic conditions. Arch Microbiol. 2017;199:475–486. doi: 10.1007/s00203-016-1313-6. [DOI] [PubMed] [Google Scholar]
- Dharmik PG, Gomashe AV. Isolation, characterization and antibiotic susceptibility profile of Staphylococcus aureus from raw milk samples in Nagpur district, India. J Appl Nat Sci. 2011;3:319–322. doi: 10.31018/jans.v3i2.208. [DOI] [Google Scholar]
- Gonzalez JE, Keshavan ND. Messing with bacterial quorum sensing. Microbiol Molec Microbiol Rev. 2006;70:859–875. doi: 10.1128/MMBR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray B, Hall P, Gresham H. Targeting agr- and agr-like quorum sensing systems for development of common therapeutics to treat multiple Gram-positive bacterial infections. Sensors. 2013;13:5130–5166. doi: 10.3390/s130405130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedin G, Widerstrom M. Endocarditis due to Staphylococcus sciuri. Eur J Clin Microbiol Infect Dis. 1998;17:673–675. doi: 10.1007/BF01708356. [DOI] [PubMed] [Google Scholar]
- Holden MTG, Chhrabra SR, de Nys R, Stead P, Bainton NJ, Hill PJ, Manefield M, Kumar N, Labette M, England D, Rice S, Givskov M, Salmond GPC, Stewart GSAB, Bycroft BW, Kjelleberg S, Williams P. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol Microbiol. 1999;33:1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x. [DOI] [PubMed] [Google Scholar]
- Horii T, Suzuki Y, Kimura T, Kanno T, Maekawa M. Intravenous catheter-related septic shock caused by Staphylococcus sciuri and Escherichia vulneris. Scand J Infect Dis. 2001;33:930–932. doi: 10.1080/00365540110076750. [DOI] [PubMed] [Google Scholar]
- Irie Y, Parsek MR. Quorum sensing and microbial biofilms. In: Romeo T, editor. Bacterial biofilms. Current topics in microbiology and immunology. Berlin: Springer; 2008. pp. 67–84. [DOI] [PubMed] [Google Scholar]
- Jain AB, Vaidya A, Ravichandran V, Kashaw SK, Agrawal RK. Recent developments and biological activities of thiazolidinone derivatives: a review. Bioorg Med Chem. 2012;20:3378–3395. doi: 10.1016/j.bmc.2012.03.069. [DOI] [PubMed] [Google Scholar]
- LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Molec Biol Rev. 2013;77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt JH, Kadouri DE, O’Toole GA. Growing and analysing static biofilms. Curr Protoc Microbiol. 2011 doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohantana A, Mazumder PB, Bhattacharjee A, Deb B. Identification of Enterotoxigenic coagulase negative Staphylococcus spp. isolated from bovine milk and milk products of southern Assam, India. J Pure Appl Microbiol. 2015;9:49–56. [Google Scholar]
- Naik MM, Pandey A, Dubey SK. Biological characterization of lead-enhanced exopolysaccharide produced by a lead resistant Enterobacter cloacae strain P2B. Biodegradation. 2012;23:775–783. doi: 10.1007/s10532-012-9552-y. [DOI] [PubMed] [Google Scholar]
- Naik MM, Bhangui P, Bhat C. The First report on Listeria monocytogenes producing siderophores and responds positively to N-acyl homoserine lactone (AHL) molecules by enhanced biofilm formation. Arch Microbiol. 2017 doi: 10.1007/s00203-017-1416-8. [DOI] [PubMed] [Google Scholar]
- Novick RP, Muir TW. Virulence gene regulation by peptides in Staphylococci and other Gram positive bacteria. Curr Opin Microbiol. 1999;2:40–45. doi: 10.1016/S1369-5274(99)80007-1. [DOI] [PubMed] [Google Scholar]
- Piessens V, De VS, Verbist B, Braem G, Van NA, De VL, Heyndrickx M, Van CE. Characterization of coagulase-negative staphylococcus spp. from cow milk and environment based on bap, icaA, and mecA genes and phenotypic susceptibility to antimicrobials and teat dips. J Dairy Sci. 2012;95:7028–7038. doi: 10.3168/jds.2012-5400. [DOI] [PubMed] [Google Scholar]
- Qazi S, Middleton B, Muharram SH, Cockayne A, Hill P, O’Shea P, Chhabra SR, Cámara M, Williams P. N-acyl homoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect Immun. 2006;74:910–919. doi: 10.1128/IAI.74.2.910-919.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shrout JD, Tolker-Nielsen T, Givskov M, Parsek MR. The contribution of cell–cell signaling and motility to bacterial biofilm formation. MRS Bull. 2011;36:367–373. doi: 10.1557/mrs.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath PHA, Mair NS, Sharpe ME, Holt JG. Bergeys manual of systematic bacteriology. Baltimore: Williams and Wilkins; 1986. [Google Scholar]
- Soumya KR, Philip S, Sugathan S, Mathew J, Radhakrishnan EK. Virulence factors associated with coagulase negative Staphylococci isolated from human infections. 3 Biotech. 2017;7:140. doi: 10.1007/s13205-017-0753-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanovic S, Dakic I, Djukic S, Lozuk B, Svabic-Vlahovic M. Surgical wound infection associated with Staphylococcus sciuri. Scand J Infect Dis. 2002;34:685–686. doi: 10.1080/00365540110076949a. [DOI] [PubMed] [Google Scholar]
- Stepanovic S, Jezek P, Vukovic D, Dakic I, Petras P. Isolation of members of the Staphylococcus sciuri group from urine and their relationship to urinary tract infections. J Clin Microbiol. 2003;41:5262–5264. doi: 10.1128/JCM.41.11.5262-5264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanovic S, Dakik I, Morrison D, Hauschild T, Jezek P, Petras P, Martel A. Identification and characterization of clinical isolates of members of the Staphylococcus sciuri group. J Clin Microbiol. 2005;43:956–958. doi: 10.1128/JCM.43.2.956-958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker HC, Brahmbhatt MN, Nayak JB. Isolation and identification of Staphylococcus aureus from milk and milk products and their drug resistance patterns in Anand, Gujarat. Vet World. 2013;6:10–13. doi: 10.5455/vetworld.2013.10-13. [DOI] [Google Scholar]
- Wallet F, Stuit L, Boulanger E, Roussel-Delvallez M, Dequiedt P, Courcol RJ. Peritonitis due to Staphylococcus sciuri in a patient on continuous ambulatory peritoneal dialysis. Scand J Infect Dis. 2000;32:697–698. doi: 10.1080/003655400459667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.