Abstract
Introduction
Studies are needed to assess the quality of transcriptome analysis in paired human tissue samples preserved by different methods and different gene amplification platforms to enable data comparisons across experimenters.
Methods
RNA was extracted from kidney biopsies, either submerged in RNA-stabilizing solution (RSS) or stored in formalin-fixed, paraffin-embedded (FFPE) blocks. RNA quality and integrity were compared. Gene expression of the common rejection module and other immune cell genes were quantified for both tissue preservation methods in the same sample using conventional quantitative polymerase chain reaction (QPCR) by 2 different commercial platforms, (fluidigm [FD]) or barcoded-oligos (nanostring [NS]).
Results
RNA quality was inferior in FFPE tissues. Despite this, gene expression for 19 measured genes on the same sample, stored in FFPE or RSS, were strongly correlated on the FD (r = 0.81) or NS platforms (r = 0.82). For the same samples, interplatform gene expression correlations were excellent (r = 0.80) for RSS and moderate (r = 0.66) for FFPE. Significant differences in gene expression were confirmed on both platforms (FD: P = 1.1E-03; NS: P = 2.5E-04) for biopsy-confirmed acute rejection.
Conclusion
Our study provided supportive evidence that despite a low RNA quality of archival FFPE kidney transplantation tissue, small quantities of this tissue can be obtained from existing paraffin blocks to provide a viable and rich biospecimen source for focused gene expression assays. In addition, reliable and reproducible gene expression evaluation can be performed on these FFPE tissues using either a QPCR-based or a barcoded-oligo approach, which provides opportunities for collaborative analytics.
Keywords: FFPE, gene expression, kidney transplant, nanostring, QPCR, rejection
Massive data from high-throughput transcriptional profiling of almost 1.9 million samples is publically available in the Gene Expression Omnibus. Gene expression microarrays and RNA sequencing methods are usually used for this high-throughput discovery phase.1 Although often criticized for the presence of false positives, the transcriptome data provides a snapshot or time-course spectrum of biological perturbations in human diseases.2 Validation of genes discovered through these aforementioned methods for biomarker discovery and/or validation or mechanistic studies requires repeat measurements on the same tissue sample, as well as independent samples with the same phenotype. The validation studies are also important to control for demographic and clinical confounders that may have had a significant impact on gene-set perturbations. Due to a paucity of human tissue samples and the cost of experiments, these validation studies are often performed with low-throughput, but robust, assays such as quantitative polymerase chain reaction (QPCR).3 Additional important considerations also include the quality of the tissue RNA, its adequacy for different platforms, and the depth and complexity of RNA interrogation technology, which highlights the critical importance of tissue mRNA preservation.4, 5, 6 Addressing these questions are of paramount importance for the conduct of precision medicine in human diseases. Several approaches to preserve tissue samples have been tested.4, 7 Snap freezing in liquid nitrogen is not always convenient and is costly to maintain. Several RNA stabilizing solutions (RSSs) retain RNA integrity once the biosample is submerged.8 Formalin-fixed, paraffin-embedded (FFPE) tissues capture biology and have been extensively used for immunohistochemistry and in situ hybridization.9 They are a rich source of biological information, although the degradative nature of formalin fixation on nucleic acids has been a major barrier to widespread adoption for transcriptomic analysis.10 However, modern molecular techniques with improved fixation, extraction, amplification, and quantification of genetic materials have made DNA and RNA analysis possible on biospecimens previously believed to be unsatisfactory or unavailable.11, 12, 13 QPCR is the conventional approach and workhorse for low-throughput gene expression validation because it is robust and has ease of experimental setup and data analysis.14, 15 Recently, a new platform for low-throughput gene expression based on molecular barcoding to quantify mRNA that does not require amplification has become available.16, 17, 18 Previous studies have favorably compared the barcoded-oligo assay to QPCR in other clinical settings19, 20 Recently, a study by Adam et al. quantified the expression of a literature-derived, antibody-mediated rejection 34-gene panel in fresh-preserved and FFPE kidney tissues with QPCR and the barcoded-oligo assay, respectively.19 Their findings demonstrated reasonable correlation (r = 0.487; P < 0.001) between the 2 assays.19 However, there has not been a true 2 × 2 (4-way) study that compares the preservation method (FFPE and fresh-preserved) and mRNA quantification platform (QPCR and barcoded-oligo assay).
In this carefully planned and executed National Institutes of Health−funded study (U01 AI113362-01), we evaluated the integrity of RNA in kidney transplant (tx) biopsies (bx) preserved in RSS or FFPE tissue blocks. The amplification performance of selected target genes by the QPCR platform (fluidigm [FD]) and the platform that uses barcoded-oligos (nanostring [NS]) on both types of tissues was assessed. Finally, we examined the usefulness of the 2 types of tissues and the 2 platforms on a gene biomarker panel for inflammation and acute rejection (AR) of kidney transplantation. Although this study focused on kidney transplant biopsy analysis, the data presented and the strategies are applicable to any organ or tissue source of interest that is handled similarly.
Methods
Patient Enrollment and Study Design
Twenty renal tx recipients were enrolled into the study (Table 1), divided into 2 phases. First, 10 consecutive for-cause kidney transplant biopsy were selected, in which matched tissues in RNA preservative and routinely processed FFPE blocks were available from the same patient at the same time point, regardless of the histological diagnosis. In addition to processing tissue for FFPE, approximately one-quarter of each bx from these patients was submerged in RNAlater. These 10 bx were evaluated for the cross-biospecimen (RSS vs. FFPE) quality and RNA amplification, with the latter being examined between the Fluidigm Biomark system (South San Francisco, CA) and nCounter system (NanoString Technologies, Seattle, WA) platforms for selected gene expression for 19 target genes. Second, 10 additional bx with a diagnosis of AR (n = 5; determined by either cause or 6-month protocol bx) and with normal morphology (n = 5; 6-month protocol bx) were selected to test the performance of the individual and combined common rejection module (CRM) score expression of selected genes. FFPE tissue was used only because matching RNAlater preserved tissue did not exist for these samples, based on their previously noted ability to discriminate organ transplant biopsy with AR21 (Table 1).
Table 1.
Patient information
| Case No. | Unique Pt ID | Age (yr)/sex | Primary disease | Transplant type | Time post transplant | Indication of biopsy | Pathology diagnosis |
|---|---|---|---|---|---|---|---|
| Study no. 1 | |||||||
| 1 | 1 | 50/M | Hypertension | LURT | 6 mos | Protocol | Mild nonspecific changes |
| 2 | 5 | 33/F | Unknown | DDRT | 6 mos | Cause, rising serum Cr | Borderline changes |
| 3 | 10 | 41/M | FSGS | DDRT | 2 yr | Cause, rising serum Cr | ACR, 1B; moderate MVI |
| 4 | 11 | 50/M | Hypertension, HIV | DDRT | 6 mos | Cause, AKI | ACR, 2A |
| 5 | 59 | 39/M | DM, HIV | SPK | 6 mos | Protocol | Mild IFTA |
| 6 | 60 | 30/M | FSGS | DDRT | 6 mos | Protocol | moderate IFTA |
| 7 | 61 | 60/F | Hypertension | DDRT | 9 yr | Cause, rising serum Cr, proteinuria | Tx glomerulopathy with severe MVI |
| 8 | 62 | 49/M | Unknown | DDRT | 8 yr | Cause, rising serum Cr | ACR, 1A |
| 9 | 63 | 40/F | DM | SPK | 6 mos | Protocol | Borderline changes |
| 10 | 64 | 43/F | Glomerulonephritis | DDRT | 6 mos | Protocol | Moderate MVI |
| Study no. 2 | |||||||
| 11 | 3 | 74/M | Hypertension, DM | LRRT | 6 mos | Protocol | Normal |
| 12 | 54 | 66/F | Hypertension | DDRT | 6 mos | Protocol | Normal |
| 13 | 55 | 36/F | Unknown | LRRT | 6 mos | Protocol | Normal |
| 14 | 56 | 37/F | DM | SPK | 6 mos | Protocol | Normal |
| 15 | 57 | 58/F | Hereditary nephritis | LURT | 6 mos | Protocol | Mild nonspecific inflammation |
| 16 | 9 | 26/F | Lupus nephritis | LRRT | 20 d | Cause, rising serum Cr | ACR, 1A |
| 17 | 31 | 67/F | PKD | DDRT | 1 yr | Protocol | ACR, 1A |
| 18 | 33 | 40/F | Hypertension | DDRT | 6 mos | Protocol | ACR, 2A |
| 19 | 39 | 50/M | Hypertension | LURT | 10 d | Cause, rising serum Cre | ACR, 1A |
| 20 | 43 | 41/M | DM | SPK | 6 mos | Protocol | ACR, 1A |
ACR, acute cellular rejection; AKI, acute kidney injury; Cr, creatinine; DDRT, deceased donor renal transplant; DM, diabetes mellitus; FSGS, focal segmental glomerulosclerosis; IFTA, interstitial fibrosis and tubular atrophy; LRRT, living related renal transplant; LURT, living unrelated renal transplant; MVI, microvascular inflammation; PKD, polycystic kidney disease; SPK, simultaneous pancreas kidney transplant; Tx, transplant.
Total RNA Extraction From FFPE Embedded and RNAlater Submerged Tissue
We used 4- × 10-μm-thick sections from 1 core of a 16-gauge needle biopsy to extract total RNA from FFPE samples. We initially evaluated the minimal input RNA needed by assessing the RNA quantity from 3 different approaches of 4, 7, and 9 sections, and determined that using 4 FFPE sections was sufficient for obtaining sufficient RNA for QPCR (data not shown), using the PureLink FFPE Total RNA Isolation Kit (Thermo Fisher, Catalog no. K1560-02, Thermo Fisher Scientific, Foster City, CA). RNAlater submerged tissue was obtained from one-quarter to one-half of a 16-gauge needle bx (Qiagen, Valencia, CA) and stored at −80°C; total RNA was extracted using a master mix of 790-μl TRIzol and 10-μl glycogen. Tissue samples were homogenized, incubated at 15°C to 25°C for 5 minutes, and 160-μl chloroform was added for phase separation. The mixture was incubated again at 25°C for 2 minutes, followed by centrifugation at 4°C and used for RNA extraction using the RNeasy Micro Kit (Qiagen Catalog no. 4004). RNA quantity and integrity were determined with the Thermo Scientific NanoDrop ND-2000 UV-Vis Spectrophotometer (Thermo Fisher Scientific) and Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA), respectively.
cDNA Synthesis and QPCR for the FD Platform
A total of 50-ng RNA was reversed transcribed into complementary DNA using Superscript II (Invitrogen, Thermo Fisher Scientific) and then amplified in a target-specific amplification step for 19 genes, namely, BASP, CD20, CD31 (PECAM1), CD4, CD6, CD68, CD8A, COL4A1, CXCL10, CXCL9, FoxP3, INPP5D, ISG20, LCK, NKG7, PRPRC (CD45), PSMB9, RUNX3, and TAP1 using TaqMan PreAmp Master Mix (cat. no. 4488593; Thermo Fisher Scientific) and TaqMan Primers and Probes (Supplementary Table S1), for a total of 18 amplification cycles. QPCR reactions were performed in the BioMark FD system using 18S gene as a housekeeping gene and Human XpressRef Universal Total RNA (Qiagen, Germantown, MD) as a reference RNA for a total of 40 cycles. Resulting chip data were initially analyzed for quality control using the BioMark Analysis Software Version 2.0 (FD), and cycle threshold (Ct) values were exported into Excel (Microsoft, Redmond, WA). Normalization of the data was done in 2 steps. (i) Ct values of individual genes were normalized against the Ct value of 18S for each gene to get delta Ct values. (ii) Delta Ct values of each sample were normalized against delta Ct values of the reference sample to get delta delta Ct values that were subsequently used to calculate fold change (RQ) values for each gene in each sample.
Barcoded-Oligos Design and Assay by the NS Platform
Barcoded codesets were designed for each of the 19 genes and 5 reference genes (GAPDH, GUSB, HPRT1, LDHA, and TBP) by NanoString Technologies (Supplementary Table S2). Fifty nanograms of each RNA sample were added to the codeset in a hybridization buffer and incubated at 65°C for 16 hours. The codeset consisted of reporter and capture probes that hybridized the target sequences of interest, forming a tripartite complex. After the assay, the raw counts for each assay were collected using the NS data analysis software, nSolver (NanoString Technologies). Normalization of the data was performed using nSolver for the following 2 methods. (i) Positive control normalization: gene expression data were normalized to the mean of the positive control probes for each assay. (ii) RNA content normalization: gene expression data were normalized to the geometric mean of housekeeping genes in the codeset.
Statistical Data Analysis
Because of its steady expression, 18S ribosomal RNA has been a popular reference RNA in gene expression analyses. For the QPCR platform, 18S ribosomal RNA was used as the reference gene. However, for the nCounter system of NS, the system could not handle a high abundance of 18S ribosomal RNA. We chose 5 common reference RNAs (GUSB, HPRT1, LDHA, TBP, GAPDH) as reference RNAs and used the mean signal as reference value. We tested gene expression correlation by comparing Ct values of 18S ribosomal RNA with mean Ct values of the 5 reference RNAs (GUSB, HPRT1, LDHA, TBP, and GAPDH) using 10 FFPE samples. There was a strong correlation with the Pearson’s correlation (r) of 0.97 (R2 = 0.87) (P < 0.0001) in between the Ct value of 18S ribosomal RNA and the mean Ct value of the previously described 5 reference genes. This demonstrated that gene expression analyses performed with either 18S ribosomal RNA or the 5 common reference genes were comparable. Following reference gene normalization, the QPCR platform data were log2 transformed. Unsupervised and supervised hierarchical clustering were performed using GENE-E (https://software.broadinstitute.org/GENE-E) using 1– Pearson’s correlation and the average as the metric and linkage methods, respectively. Correlation values were calculated using Pearson’s and Spearman’s rank-based correlation methods (GraphPad Prism, La Jolla, CA), in which the correlation coefficient, r, ranged from −1 to +1. A significant difference in 2 sets of data was determined by performing an unpaired t test with a 2-tailed P value option using GraphPad Prism. A P value of <0.05 was considered significant.
Results
Comparison of RNA Quantity and Quality From RNAlater Submerged Versus FFPE-Embedded Kidney Transplant Biopsy Tissue
RNA quantity measured from one-quarter of the 16-gauge needle bx cores (n = 10) and submerged in RNAlater solution yielded 2.8 ± 1.9 μg total RNA; 4- × 10-μM-thick slices of the FFPE tissue (n = 10) from the matching kidney transplant biopsy yielded 1.7 ± 1.0 μg RNA. RNA quality based on the 260:280 ratio of the extracted RNA was similar to 1.98 ± 0.12 for RNA extracted from RNAlater tissue compared with 1.95 ± 0.06 for RNA extracted from FFPE sections. RNA integrity on RNA samples isolated from the RSS was excellent with RNA integrity number (RIN) values of 9.24 ± 0.51, although this was considerably lower for RNA isolated from FFPE tissues, with RIN values of 2.53 ± 0.94 (P = 2.10E-10) (Figure 1).
Figure 1.
The RNA integrity number (RIN) of total RNA extracted from kidney tissue preserved in RNA-stabilizing solution (RNAlater) was compared with that of formalin-fixed, paraffin-embedded (FFPE) tissue (a–c). Representative chromatogram for 3 FFPE samples with respective RIN numbers. (d–f) Representative chromatogram for 3 RNAlater samples.
Comparisons of the FD and NS Technologies Based on Cost of Instrument, Assay Time, and Assay Cost
The wide availability of cataloged primer and probesets enabled FD to generate data within a few days, whereas for of NS, the time to generate data ranged somewhere from within a week to 4 to 5 weeks if barcoded codesets need to be designed and synthesized. The robustness of both platforms is comparable, and both platforms are capable of multiplexing. As of early 2017, using these platforms for a project to assess gene expression level of 20 genes on 48 samples would cost approximately $21 USD by the FD platform, whereas NS is more expensive, with an estimated cost of approximately $67 USD per sample. The costs and performance capabilities at the time of the preparation of this paper are summarized in Table 2.
Table 2.
Comparison of Fluidigm BioMark Versus Nanostring nCounter system
| Parameters | Nanostring | Fluidigm |
|---|---|---|
| Cost of instrument (estimated/USD)a | 225,000 (nCounter Max) | 235,000 (BioMark) |
| Cost per sample (estimated/USD)a | 67 | 21 |
| Multiplexing option | 96 well plate and others | 96 well plate and others |
| Input RNA | 50 ng | 50 ng |
| Probes used nanostring/amplicons | 50 bases | 50−100 bp |
| Sensitivity | ∼1 copy per cell | ∼1 copy per cell |
| Detection limit | Single copy mRNA | Single copy mRNA |
| Throughput per day | Up to 39,600 data points | Up to 50,688 data points |
| Time to result | ∼22 h for one 12-sample cartridge. This includes hybridization, sample preparation, and time with the Digital Analyzer | ∼8 h for one 96 × 96 chip (∼up to 48 that include cDNA synthesis, target specific amplification, priming of the chip and QPCR run. |
| Dynamic range | ∼6 logs | ∼ 6−7 logs |
| Analysis software | Uses nSolver Analysis Software. Freely available | Uses Singular Toolset Software. Freely available |
| No. of genes that can be assessed | 800 genes | Flexible, ideal for 20−200 genes |
| Types of samples | Fresh or frozen cells and tissue, FFPE, biological fluids | Fresh or frozen cells and tissue, FFPE, biological fluids |
bp, base pair; FFPE, formalin-fix, paraffin-embedded; QPCR, quantitative polymerase chain reaction.
Listed costs are as of January 2017 and are subject to change by vendors.
Correlation of Gene Expression Across Different Tissue Preservation and Profiling Methods
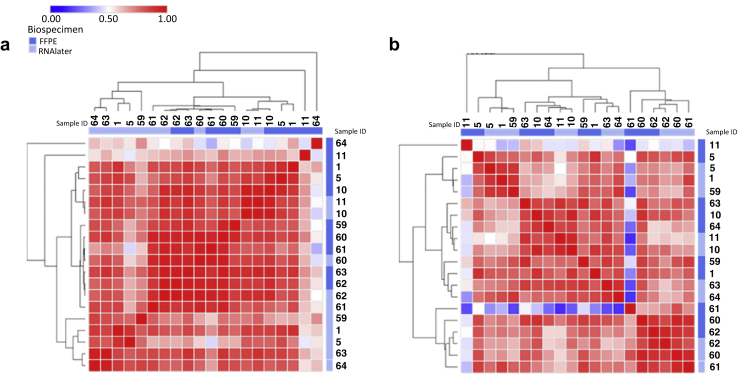
The correlation between the mean expression of all genes profiled on the FFPE and RNAlater fractions was 0.81 (95% confidence interval [CI]: 0.76−0.86; P < 0.0001) and 0.82 (95% CI: 0.76−0.86; P < 0.0001) on the NS and FD platforms, respectively (Supplementary Figures S1a−c and Supplementary Figure S2a). Heat map of data that resulted from unsupervised hierarchical clustering of the paired samples correlations as quantified by NS and FD are shown in Figure 2. These data suggested that despite high correlation, there were subtle differences reflected in gene expression data generated by RSS versus FFPE and also in FD versus ND. Results by Spearman’s rank-based analysis revealed similar findings (Supplementary Table S3), which suggested that the high correlation was independent of platform specific normalization methods.
Figure 2.
Heat map resulted with the data from unsupervised hierarchical analysis for the Pearson correlation of absolute mRNA transcript abundance as determined by (a) a Fluidigm quantitative polymerase chain reaction and (b) a Nanostring nCounter for all pair-wise combinations of samples.
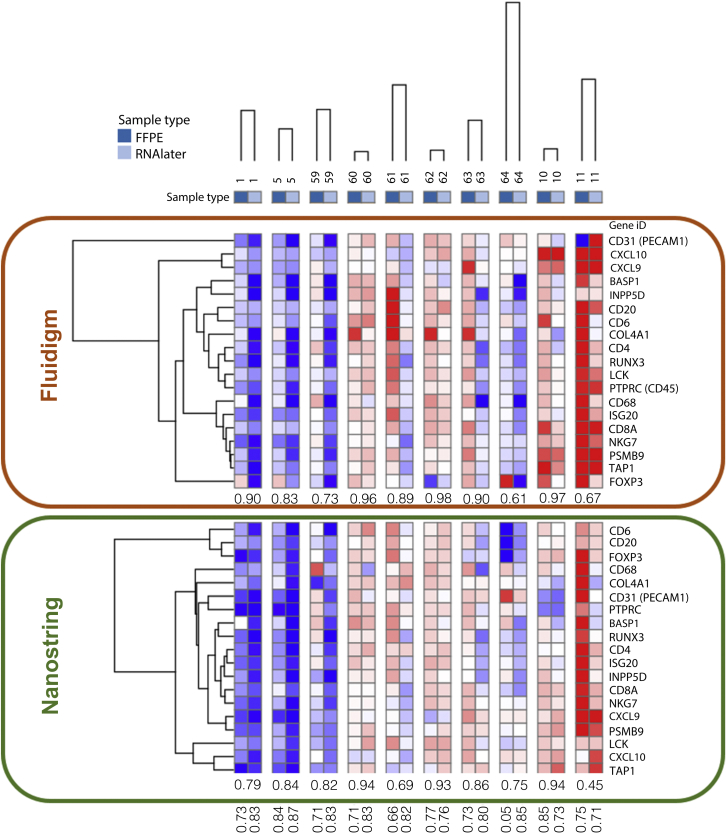
The correlation of amplifying the same gene set in FFPE tissues alone across both platforms was further assessed. Correlation of all available pairs of data on FFPE tissues when interrogated by FD and NS was 0.79 (95% CI: 0.73−0.85; P = 0.0001) (Supplementary Figure S1d, and Supplementary Figure S2b). Again, Spearman’s correlations resulted in similar findings (Supplementary Table S3). The heat map of the data that resulted from supervised hierarchical clustering is shown in Figure 3. In the heat map, gene expression values for each gene across all samples is presented in color gradient, ranging from dark red (maximum expression) to dark blue (minimum expression).
Figure 3.
Heat map with supervised hierarchical clustering of mRNA expression quantified by Nanostring and Fluidigm on matched formalin-fix, paraffin-embedded, and RNAlater biosamples. Gene expression correlation between biospecimen and platform are labeled horizontally and vertically, respectively.
Comparison of Results to Provide Differences in Gene Expression Levels in Between Different Phenotypes
We next evaluated the performance of 11 individual CRM genes (BASP, CD6, CXCL10, CXCL9, INPP5D, ISG20, LCK, NKG7, PSMB9, RUNX3, and TAP1) and 8 other cell-specific markers (CD20, CD6, CD68, CD8A, COL4A FOXP3, and CD45). The CRM score was computed across an additional 10 independent FFPE samples (5 AR and 5 non-AR) as a measure to distinguish the AR phenotype, measured on both the FD and NS platforms. Gene expression was consistently greater in all 19 transcripts in AR compared with non-AR samples across both platforms, with statistically significant (P < 0.05) differential expression for the following CRM genes on the FD platform (Figure 4a): CXCL10, CXCL9, ISG20, LCK, NKG7, PSMB9, RUNX3, TAP1, and the following cell-surface markers: CD20, CD4, CD6, CD8A, FOXP3, and CD45 (PTPRC), whereas all genes except for CD31 (PECAM1) were significantly differentially expressed when quantified on the NS platform (Figure 4b). Both platforms were able to reliably differentiate AR samples based on significantly elevated CRM scores in AR (9.96 ± 2.31 in AR vs. 1.64 ± 0.80 in non-AR on FD, P = 1.1E-3; and 7.28 ± 1.19 in AR vs. 1.04 ± 0.33 in non-AR on NS; P = 2.5E-04) (Figure 4c).
Figure 4.
Gene expression of the selected gene-set quantified by (a) Fluidigm and (b) Nanostring from formalin-fix, paraffin-embedded samples obtained from stable grafts and allografts undergoing acute rejection. (c) Scatterplot of common rejection module (CRM) score comparison between acute rejection and stable grafts as measured on Fluidigm and Nanostring. Mean and SEM are shown.
Discussion
Small quantities of available tissue samples make it necessary to address the accessibility and applicability of archival tissue for investigational studies. Proper sample management for collection, handling, fixation, and processing are quintessential for successful studies.
Targeted transcriptional analysis from FFPE tissues was successfully shown on the NS platform,16, 22 with moderate correlation with QPCR in the setting of antibody-mediated rejection in kidney tx tissues.19 Compared with standard QPCR, the NS platform had a higher correlation with histologic findings that was attributed to the tissue variability of fresh-preserved tissue compared with FFPE sections. However, that study did not quantitate mRNA expression in both FFPE and RSS tissues simultaneously on both types of platforms,19 and no studies to date have addressed if gene amplification by multiplex QPCR on the FD platform will have similar results to the NS platform. In addition, there have been no direct comparisons of the workflow and cost of running kidney bx samples on both FD and NS platforms, which is valuable information for investigators beginning to take on these kinds of studies.
It was hypothesized that because the FFPE sample was dissected from the same region where the histology reads were provided, correlation of gene expression values would be higher than those seen from gene expression analysis performed on a separate piece of tissue submerged in RSS (e.g., RNAlater). This question was also addressed in this study because the same tissue sample was compared head to head, across the same set of genes, with either the FFPE or the RSS specimen. In addition, the amplification efficiency and the correlation of the gene expression results in the context of AR were also compared directly on both the FD and the NS platforms. These data were not previously evaluated in this manner, and hence, provided valuable information to the academic tx community.
FFPE tissue offers many benefits, including stability at room temperature that allows for ease of long-term storage in well-maintained large biorepositories.23, 24 Archived FFPE samples are generally well annotated and accompanied by an abundance of longitudinal clinical data, making them valuable study materials. Despite the clear benefits, FFPE tissue interrogation has not been clearly adopted to date, due to known degradation of genomic material and obstacles with efficient RNA extraction and sufficient quality for experimentation.25, 26 The inferior quality of RNA isolated from FFPE compared with RSS was confirmed in our study, yet we showed that this did not impact the amplification of target genes by either the FD or the NS platforms overall. Because the ideal QPCR amplicon length is >55 bases, the degree of RNA degradation from FFPE appears to retain this fragment length in most of the samples processed. Nevertheless, it is important to note that the oldest FFPE biopsy in this study was 1.5 years old. Evaluating the same approach on bx tissue stored for a much longer time warrants further investigation to assess the additive effects of degradation of FFPE embedded tissues over long periods of time.
The applicability of either the FD or the NS platforms for evaluation of gene expression in the context of FFPE tissue alone in kidney transplant biopsy is an important conclusion of this study. This is a significant observation because targeted low-throughput gene expression methodologies such as the FD and NS platforms provide the next step, cost-effective, rapid validation of smaller, informative gene sets.14, 15 However, we observed that in samples in which the degradation of RNA was more significant (sample #64; RIN = 2.4), there were poor correlations between the 2 platforms. The FD platform reported missing data for very low amplification results, whereas the NS platform imputed a baseline background value for missing data. We observed that there was significantly better correlation in between the 2 platforms for the RSS tissue in this sample (Supplementary Figure S2b). In addition, although the RIN number was low for sample #64), it was not the only parameter that assessed amplification efficiency in the FFPE sample, because some of the other FFPE samples with low RIN numbers and similar sample storage length had better amplification results. Due to data imputation for low and/or missing sample amplification by the NS platform, we found much better correlations for FFPE and RSS for these samples with NS, although some of these data might require additional scrutiny for “false positive” data quality control on the NS platform (Supplementary Figure S2a). Removal of this outlier sample from the analysis resulted in comparable performance between the FD and NS platforms on both tissue types. The FD platform yielded data with relatively higher correlations (especially for samples 11 and 61) (Supplementary Figure S2A). These variations in gene expression might be related to intrinsic factors that are connected to platform efficiency and the process of direct detection without the need of amplification in the NS platform, but which are needed for preamplification on the FD platform.17, 27, 28
We also demonstrated that gene expression signatures could readily differentiate histologically confirmed AR and non-AR tissue samples in archived FFPE blocks on either platforms. We used the combined scoring across the recently published CRM genes for quantifying kidney tx graft inflammation in the context of AR.21, 29 Because of the available resources for this project, we only performed biopsies from AR and normal grafts. This leaves us with a question of whether these assays are useful in separating kidney tx injuries with subtle changes (e.g., AR from borderline changes), T-cell−mediated rejection from antibody-mediated rejection, and so on. Future studies will have to be performed with a larger cohort of carefully chosen samples to further establish the usefulness of these tissues and assays.
In conclusion, in this study, we compared not only the usefulness of FFPE with tissues preserved in RSS, but also the efficacy of different methods of gene expression analysis of tx kidney bx using 2 commercially available platforms, using either conventional QPCR or barcoded-oligo based technology, with detailed comparisons of performance, handling, and the cost of both methodologies, to allow the reader to critically assess the choice of either platform for their own research.
Disclosure
All the authors declared no competing interest.
Acknowledgements
The authors would like to acknowledge the funding support from the National Institutes of Health: UO1/CTOT21 (FV), R21 TR001761 (MMS), U24AI118675 (ZL), and T32DK007219 (MN). The authors would also like to thank the support from Dr. Sindhu Chandran, UCSF Medical Center, for helping with sample collection.
Footnotes
Table S1. Primer information for primers used for Fluidigm assay.
Table S2. Nanostring codeset information.
Table S3. Spearman’s correlation of gene expression between formalin-fixed, paraffin-embedded (FFPE) and RNAlater tissue as measured on Fluidigm and Nanostring.
Figure S1. (A−C) Assessment of the correlation of amplifying the same gene set in formalin-fixed, paraffin-embedded (FFPE) and tissues preserved in RNA stabilizing solution (RNAlater) was done. (D) Assessment of the correlation of amplifying the same gene set in FFPE by Nanostring and quantitative polymerase chain reaction (QPCR) was performed.
Figure S2. Correlation of total gene expression of 19 gene-set between (A) formalin-fixed, paraffin-embedded (FFPE) and RNAlater submerged tissue on Nanostring and Fluidigm platform and between (B) Nanostring and Fluidigm on FFPE and RNAlater submerged tissues.
Supplementary material is linked to the online version of the paper at www.kireports.org
Contributor Information
Minnie M. Sarwal, Email: minnie.sarwal@ucsf.edu.
Zoltan Laszik, Email: zoltan.laszik@ucsf.edu.
Supplementary Material
Primer information for primers used for Fluidigm assay.
Nanostring codeset information.
Spearman’s correlation of gene expression between formalin-fixed, paraffin-embedded (FFPE) and RNAlater tissue as measured on Fluidigm and Nanostring.
(A−C) Assessment of the correlation of amplifying the same gene set in formalin-fixed, paraffin-embedded (FFPE) and tissues preserved in RNA stabilizing solution (RNAlater) was done. (D) Assessment of the correlation of amplifying the same gene set in FFPE by Nanostring and quantitative polymerase chain reaction (QPCR) was performed.
Correlation of total gene expression of 19 gene-set between (A) formalin-fixed, paraffin-embedded (FFPE) and RNAlater submerged tissue on Nanostring and Fluidigm platform and between (B) Nanostring and Fluidigm on FFPE and RNAlater submerged tissues.
References
- 1.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tarca A.L., Romero R., Draghici S. Analysis of microarray experiments of gene expression profiling. Am J Obstet Gynecol. 2006;195:373–388. doi: 10.1016/j.ajog.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawitan Y., Michiels S., Koscielny S. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics. 2005;21:3017–3024. doi: 10.1093/bioinformatics/bti448. [DOI] [PubMed] [Google Scholar]
- 4.Bradley W.H., Eng K., Le M. Comparing gene expression data from formalin-fixed, paraffin embedded tissues and qPCR with that from snap-frozen tissue and microarrays for modeling outcomes of patients with ovarian carcinoma. BMC Clin Pathol. 2015;15:17. doi: 10.1186/s12907-015-0017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams M.A. Stabilizing the code-methods to preserve RNA prove their worth. Biomark Insights. 2010;5:139–143. doi: 10.4137/BMI.S6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Barbacioru C., Hyland F. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedorowicz G., Guerrero S., Wu T.D. Microarray analysis of RNA extracted from formalin-fixed, paraffin-embedded and matched fresh-frozen ovarian adenocarcinomas. BMC Med Genomics. 2009;2:23. doi: 10.1186/1755-8794-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basiye F.L., Schoone G.J., Beld M. Comparison of short-term and long-term protocols for stabilization and preservation of RNA and DNA of Leishmania, Trypanosoma, and Plasmodium. Diagn Microbiol Infect Dis. 2011;69:66–73. doi: 10.1016/j.diagmicrobio.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 9.Scicchitano M.S., Dalmas D.A., Boyce R.W. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J Histochem Cytochem. 2009;57:849–860. doi: 10.1369/jhc.2009.953497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan M., Sedmak D., Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staff S., Kujala P., Karhu R. Preservation of nucleic acids and tissue morphology in paraffin-embedded clinical samples: comparison of five molecular fixatives. J Clin Pathol. 2013;66:807–810. doi: 10.1136/jclinpath-2012-201283. [DOI] [PubMed] [Google Scholar]
- 12.Kashofer K., Viertler C., Pichler M. Quality control of RNA preservation and extraction from paraffin-embedded tissue: implications for RT-PCR and microarray analysis. PloS One. 2013;8:e70714. doi: 10.1371/journal.pone.0070714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turashvili G., Yang W., McKinney S. Nucleic acid quantity and quality from paraffin blocks: defining optimal fixation, processing and DNA/RNA extraction techniques. Exper Mol Pathol. 2012;92:33–43. doi: 10.1016/j.yexmp.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 15.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Chen X., Deane N.G., Lewis K.B. Comparison of Nanostring nCounter(R) data on FFPE colon cancer samples and Affymetrix microarray data on matched frozen tissues. PloS One. 2016;11:e0153784. doi: 10.1371/journal.pone.0153784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norton N., Advani P.P., Serie D.J. Assessment of tumor heterogeneity, as evidenced by gene expression profiles, pathway activation, and gene copy number, in patients with multifocal invasive lobular breast tumors. PloS One. 2016;11:e0153411. doi: 10.1371/journal.pone.0153411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pesenacker A.M., Wang A.Y., Singh A. A regulatory T-cell gene signature is a specific and sensitive biomarker to identify children with new-onset type 1 diabetes. Diabetes. 2016;65:1031–1039. doi: 10.2337/db15-0572. [DOI] [PubMed] [Google Scholar]
- 19.Adam B., Afzali B., Dominy K.M. Multiplexed color-coded probe-based gene expression assessment for clinical molecular diagnostics in formalin-fixed paraffin-embedded human renal allograft tissue. Clin. Transplant. 2016;30:295–305. doi: 10.1111/ctr.12689. [DOI] [PubMed] [Google Scholar]
- 20.Richard A.C., Lyons P.A., Peters J.E. Comparison of gene expression microarray data with count-based RNA measurements informs microarray interpretation. BMC Genomics. 2014;15:649. doi: 10.1186/1471-2164-15-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khatri P., Roedder S., Kimura N. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen T., Wallden B., Schaper C. Analytical validation of the PAM50-based Prosigna breast cancer prognostic gene signature assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 2014;14:177. doi: 10.1186/1471-2407-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kokkat T.J., Patel M.S., McGarvey D. Archived formalin-fixed paraffin-embedded (FFPE) blocks: a valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank. 2013;11:101–106. doi: 10.1089/bio.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bass B.P., Engel K.B., Greytak S.R. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen? Arch Pathol Lab Med. 2014;138:1520–1530. doi: 10.5858/arpa.2013-0691-RA. [DOI] [PubMed] [Google Scholar]
- 25.von Ahlfen S., Missel A., Bendrat K. Determinants of RNA quality from FFPE samples. PloS One. 2007;2:e1261. doi: 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie R., Chung J.Y., Ylaya K. Factors influencing the degradation of archival formalin-fixed paraffin-embedded tissue sections. J Histochem Cytochem. 2011;59:356–365. doi: 10.1369/0022155411398488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polytarchou C., Oikonomopoulos A., Mahurkar S. Assessment of circulating microRNAs for the diagnosis and disease activity evaluation in patients with ulcerative colitis by using the nanostring technology. Inflamm Bowel Dis. 2015;21:2533–2539. doi: 10.1097/MIB.0000000000000547. [DOI] [PubMed] [Google Scholar]
- 28.Northcott P.A., Shih D.J., Remke M. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123:615–626. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigdel T.K., Bestard O., Tran T.Q. A computational gene expression score for predicting immune injury in renal allografts. PloS One. 2015;10:e0138133. doi: 10.1371/journal.pone.0138133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer information for primers used for Fluidigm assay.
Nanostring codeset information.
Spearman’s correlation of gene expression between formalin-fixed, paraffin-embedded (FFPE) and RNAlater tissue as measured on Fluidigm and Nanostring.
(A−C) Assessment of the correlation of amplifying the same gene set in formalin-fixed, paraffin-embedded (FFPE) and tissues preserved in RNA stabilizing solution (RNAlater) was done. (D) Assessment of the correlation of amplifying the same gene set in FFPE by Nanostring and quantitative polymerase chain reaction (QPCR) was performed.
Correlation of total gene expression of 19 gene-set between (A) formalin-fixed, paraffin-embedded (FFPE) and RNAlater submerged tissue on Nanostring and Fluidigm platform and between (B) Nanostring and Fluidigm on FFPE and RNAlater submerged tissues.