Abstract
For a better understanding of epigenetic regulation of cell differentiation, it is important to analyze DNA methylation at a specific site. In this study, we examined changes in the methylation level of CCGG and GATCG sites during mouse folliculogenesis in paraffin-embedded sections of mouse ovaries. For the purpose, we used a new method, histo endonuclease-linked detection of methylation sites of DNA (HELMET), designed to detect methylation sites of DNA with a specific sequence in a tissue section. Unlike the global level of DNA methylation, which was no change in immunohistochemical staining of 5-methylcytosine throughout folliculogenesis, we found that there were hypermethylation of CCGG and GATCG sites in most of the granulosa cells of tertiary follicles compared to that of primary and secondary follicles. Interestingly, TUNEL-positive granulosa cells, which were frequent in mammalian folliculogenesis, became markedly Hpa II-reactive and Sau3A I-reactive, indicating that the CCGG and GATCG sites may be preferentially demethylated during apoptosis.
Keywords: DNA methylation, folliculogenesis, HELMET, apoptosis, epigenetics
I. Introduction
During development of multicellular organisms, different cells acquire different patterns of gene expression. For the process, it is believed that epigenetic factors such as DNA methylation and histone tail modifications are involved [10, 12]. Recently, DNA methylation has been a major focus of studies on epigenetic regulation of gene expression, and it largely depends on the DNA methyltransferase (DNMTs)-catalyzed reaction in which a methyl group is selectively added to cytosine residue of a CpG dinucleotide to form 5-methylcytosine. Through changes in chromatin structure, DNA methylation is supposed to be generally associated with gene silencing, whereas DNA demethylation is related to gene activation [2, 6].
In female germ cells of rats, the methylation of DNA is initiated during the postnatal day (PND) 1–5 and continues throughout oocyte maturation until the preantral follicle stage [10, 12]. In fact, the gametogenesis stage as well as the embryogenesis stage between the early ICM and the post-implantation embryo have heightened DNA methylation activity where DNA methylation patterns are actively erased and reestablished (epigenetic reprogramming) [13, 17]. To catalyze methylation of DNA, two types of DNMTs are known; one (DNMT1) is used for the maintenance of methylation pattern of DNA and the other (DNMT3a and 3b) is for de novo DNA methylation [8, 16]. Moreover, DNMT1o is present as an oocyte specific one, which is accumulated in oocytes and is engaged in the maintenance of imprinted genes during the early development of follicles [8].
In the follicular development, primordial follicle is developed to tertiary follicle through primary and secondary follicles, accompanying a remarkable differentiation of granulosa cells (or follicular epithelial cells). During the process, most of the immature follicles underwent granulosa cell apoptosis and then follicle atresia. We found that activation of the Fas-Fas ligand system is involved in initiating apoptosis in the ovary [7], and more recently several studies also have reported epigenetic-related enzymes or genes involved in the apoptosis of female germ cells including Lsh, a chromatin-remodelling protein of the SNF2-helicase family [5] and Ehmt2, encoding H3K9 mono- and dimethyltransferase [19].
Considering that DNA methylation is closely related to chromatin condensation, we hypothesized that any abnormality of DNA methylation would result in the induction of apoptosis, possibly due to ill-organized heterochromatinization. To correlate the states of DNA methylation with apoptotic oocytes as well as granulosa cells, histochemical approach, rather than biochemical one, would be appropriate. Therefore, we have developed a new histochemical method, histo endonuclease-linked detection of methylated DNA sites (HELMET) [9], to assess DNA methylation level at a specific site such as a CCGG sequence, using a set of restriction enzymes of methylation sensitive (Hpa II)- and insensitive (Msp I)-isoschizomers [1] on a histological section. Using this method, the level of methylation of DNA in individual nuclei was successfully evaluated previously [3, 9, 18].
In the present study, we localized methylated and non-methylated CCGG and GATCG sites in mouse ovarian follicles simultaneously by HELMET. Finally, we clarified the changes in the level of DNA methylation during folliculogenesis and their possible correlation with granulosa cell apoptosis.
II. Materials and Methods
Chemicals and biochemicals
Paraformaldehyde (PFA) was purchased from Merck (Darmstadt, Germany), and 3,3'-diaminobenzidine 4HCl (DAB) was purchased from Dojin Chemical Co. (Kumamoto, Japan). Bovine serum albumin (BSA) (essentially fatty acid and globulin-free), Trizma base and Brij-35 were from Sigma Chemical Co. (St. Louis, MO, USA). Biotin-16-dUTP, digoxigenin-11-dUTP and terminal deoxynucleotidyl transferase (TdT) were from Roche Diagnostics (Mannheim, Germany). Dideoxy ATP (ddATP) and dideoxy TTP (ddTTP) were from Jena Bioscience (Jena, Germany). Hpa II, Msp I, Sau3A I and Mbo I were purchased from Takara Bio Inc. (Shiga, Japan). 4',6-Diamidino-2-phenylindole (DAPI) was from DAKO (Glostrup, Denmark). Permount was purchased from Fisher Scientific Inc. (NJ, USA). All other reagents used in this study were from Wako Pure Chemicals (Osaka, Japan) and were of analytical grade.
Antibodies
Mouse monoclonal anti-5-methylcytosine (2.5 μg/ml) was purchased from Calbiochem (San Diego, CA, USA). Normal goat IgG and sheep IgG were purchased from Dako (Glostrup, Denmark). Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:100), HRP-conjugated goat anti-biotin (1:100) and HRP-conjugated sheep anti-digoxigenin (1:100) were from Chemicon International Inc. (Temecula, CA, USA). FITC-labeled goat anti-biotin (1:100) was from Vector Laboratories (Burlingame, CA, USA) and rhodamine-labeled sheep anti-digoxigenin (1:100) was from Roche Diagnostics (Mannheim, Germany).
Animals and tissue preparation
Female adult ICR mice (8 to 10-week-old) weighing 27–38 g were used in the present study. The experimental protocol was approved by the Animal Ethics Review Committee of Nagasaki University (#0112100012 and #0202200048).
The ovaries dissected out were cut into small pieces, then fixed overnight with 4% PFA in phosphate buffered saline (PBS), pH 7.4, at 4°C and embedded in paraffin in a standard manner. Several serial sections were stained with hematoxylin and eosin (H&E), and used for identification of the differentiation stages of follicle.
Immunohistochemistry for 5-methylcytosine
Immunohistochemical detection of 5-methylcytosine was performed on the paraffin sections of mouse ovaries, as described in detail previously [11, 18, 20]. After they were dewaxed, the sections were immersed in 10 mM citrate buffer, pH 6.0, autoclaved at 120°C for 15 min, and then pre-incubated with 500 μg/ml normal goat IgG dissolved in 1% BSA in PBS, pH 7.2, for 1 hr. Unless otherwise specified, all reactions were conducted at room temperature (RT). Then the sections were reacted overnight with anti-5-methylcytosine (2.5 μg/ml) in 1% BSA in PBS. After washing with 0.075% Brij in PBS three times for 15 min each, HRP-conjugated goat anti-mouse IgG dissolved in 1% BSA in PBS was reacted for 1 hr. After washing with 0.075% Brij in PBS three times for 15 min each, the sites of HRP were visualized with DAB and H2O2 in the presence of nickel and cobalt ions [7]. As a negative control, some sections were reacted with normal mouse IgG instead of the specific monoclonal antibody.
Terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) staining
To identify apoptotic cells, TUNEL was performed, as described previously [7, 9]. Paraffin sections (5 to 6-μm thick) on silane-coated glass slides were dewaxed and digested with 10 μg/ml of proteinase K in PBS at 37°C for 15 min. Then the sections were incubated with TdT buffer (25 mM Tris/HCl buffer, pH 6.6, containing 0.2 M potassium cacodylate and 0.25 mg/ml BSA) alone at RT for 30 min. After the incubation, the slides were reacted with 800 units/ml of TdT dissolved in TdT buffer supplemented with 0.5 μM biotin-16-dUTP, 20 μM dATP, 1.5 mM CoCl2 and 0.1 mM dithiothreitol at 37°C for 90 min. As a negative control, TdT reaction was conducted with 0.5 μM TTP instead of biotin-16-dUTP, or without TdT. After washing with milli-Q water (Millipore, Molsheim, France) (DDW), the signals were detected immunohistochemically with HRP-conjugated goat anti-biotin antibody or FITC-labeled goat anti-biotin antibody. The sites of HRP were visualized by a mixture of DAB, hydrogen peroxide, nickel and cobalt ions, as described above.
HELMET
First, TdT reaction was conducted as described above on paraffin sections of mouse ovaries at 37°C for 2 hr, except for the presence of 20 μM ddATP and 20 μM ddTTP instead of dATP and biotin-16-dUTP, to block the free 3'-OH ends of DNA. After washing with DDW and PBS, successively, the sections were fixed with 4% PFA in PBS for 5 min and then washed twice with PBS and once with DDW for 5 min each. Then the sections were reacted with 100 units/ml of Hpa II or Sau3A I dissolved in 10 mM Tris/HCl buffer, pH 7.5, containing 10 mM MgCl2 and 1 mM dithiothreitol at 37°C for 2 hr. Then the slides were washed three times with DDW for 5 min each. To detect the sites of non-methylated CCGG or GATCG sequence of DNA, TdT reaction with 800 units/ml was conducted with 1.0 μM biotin-16-dUTP or 0.5 μM digoxigenin-11-dUTP at 37°C for 1.5 hr in a moist chamber and then washed three times with PBS for 5 min each.
To visualize the signal, in the case of enzyme immunohistochemistry, the slides were incubated with a mixture of 500 μg/ml of normal goat IgG or normal sheep IgG and 5% BSA in PBS for 1 hr and then reacted with HRP-goat anti-biotin or HRP-sheep anti-digoxigenin dissolved in 5% BSA in PBS for 1 hr, respectively. After washing with 0.075% Brij in PBS three times and then PBS alone for 10 min each, the signals were visualized with a chromogen solution consisting of DAB, H2O2, nickel and cobalt ions, as described above. The slides were dehydrated through serial ethanol solutions and cleared by xylene, and then mounted with Permount. In the case of fluorescence immunohistochemistry, FITC-goat anti-biotin or rhodamine-sheep anti-digoxigenin dissolved in 5% BSA in PBS was reacted for 1 hr and washed similarly. The slides were mounted with 90% (v/v) glycerol in PBS.
To detect the sites of methylated CCGG or GATCG sequence of DNA, the sections were digested with Hpa II or Sau3A I, and then blocked with a mixture of ddATP and ddTTP by TdT, as described above. Then, they were fixed with 4% PFA in PBS for 5 min, and washed twice with PBS for 15 min each. After washing with DDW, the sections were digested with 100 units/ml Msp I or Mbo I dissolved in Tris/HCl buffer, pH 7.9, containing 10 mM MgCl2, 0.5 mM dithiothreitol, 66 mM potassium acetate and 0.1% BSA at 37°C for 2 hr. After washing three times with DDW for 5 min each, TdT reaction was conducted with 0.5 μM biotin-16-dUTP or 0.5 μM digoxigenin-11-dUTP for 37°C for 2 hr in a moist chamber and then washed three times with PBS for 5 min each. Then, the same procedures to that used for the detection of non-methylated CCGG or GATCG sites were taken to visualize the methylated CCGG or GATCG sites.
For double staining of non-methylated and methylated sites, the Hpa II or Sau3A I cutting sites were labeled with biotin-16-dUTP by TdT reaction and blocked with dideoxynucleotides. Then, the procedure to detect methylated sites was taken and the Msp I or Mbo I cutting sites were labeled with digoxigenin-11-dUTP by TdT reaction. Finally, both haptens were visualized by FITC-anti-biotin and rhodamine-anti-digoxigenin, respectively.
The analysis of fluorescence signals was performed with LSM 5 Pascal (V3.2) (Carl Zeiss Co., Jena, Germany). In this study, FITC and rhodamine signals were obtained with an emission filter of 505–530 nm (excitation; 488 nm) and ~560 nm filter (excitation; 543 nm), respectively. The observation was done with Plan-Neofluar 20 × /0.5 and 40 × /0.75 objective lenses, where optical section thickness was 0.5 and 0.7 μm, respectively. ImagJ software was used for measuring the different signals of cell number to do the ratio.
Double staining for TUNEL and HELMET
To correlate TUNEL-positive cells with the state of methylation of CCGG sequence directly, TUNEL was first conducted on paraffin sections of mouse ovaries. After TdT reaction with biotin-16-dUTP, the sections were blocked with dideoxynucleotides as described above and fixed with 4% PFA in PBS for 5 min. Then the sections were washed twice with PBS (15 min each) and once with DDW (5 min), and digested with Hpa II or Msp I as described in the above procedure. For co-localization with non-methylated or methylated sites of CCGG, the cutting sites were labeled with digoxigenin-11-dUTP by TdT and finally, both biotin and digoxigenin moieties were visualized with FITC-anti-biotin and rhodamine-anti-digoxigenin, respectively.
Follicle analysis
A follicle was analyzed if the oocyte had a germinal vesicle present in the section analyzed. All counted follicles were assessed for stage and DNA methylation. Follicles were also classified by their histological details, using standard criteria as primary (including primordial), secondary and tertiary follicles.
Statistical analysis
Experimental data were presented as mean ± standard deviation from at least three independent experiments. The software IBM SPSS 20.0 was used to analyze the data. Differences among the groups were compared by a one-way ANOVA and Student’s t-test or Duncan’s multiple range test after one-way analysis of variance. All analyses were performed using a statistical software package Differences with P < 0.05 or P < 0.01 were considered to be statistically significant.
III. Results
Differential staining of non-methylated and methylated CCGG sites of DNA in adjacent sections of mouse ovary
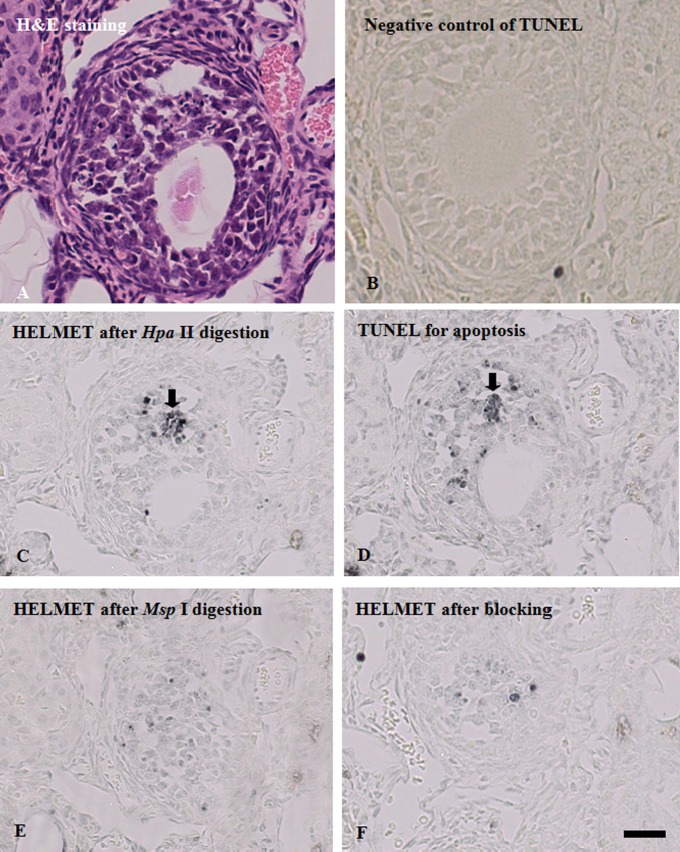
To identify the stage of follicles in mouse ovary, we used H&E stained sections (Fig. 1A). Then with sections, we compared signal distributions of TUNEL, non-methylated CCGG sites and methylated CCGG sites. As shown in Fig. 1D, we found some TUNEL-positive cells as apoptotic cells. When sections were labeled with the dideoxynucleotides after proteinase K digestion, further TdT reaction in TUNEL did not yield any staining (Fig. 1B). Next, the sections that were blocked with the dideoxynucleotides were digested with Hpa II and the non-methylated CCGG sites were labeled with digoxigenin-11-dUTP by TdT and visualized with HRP-anti-digoxigenin (Fig. 1C). Although the majority of granulosa cells were only faintly stained, several cells with a strong staining were found and identified as TUNEL-positive cells (arrow). To detect methylated CCGG sites, an section was digested with Hpa II and blocked with dideoxynucleotides, then digested with Msp I, labeled with digoxigenin-11-dUTP by TdT and visualized with HRP-anti-digoxigenin (Fig. 1F). Then we found that the intensity of the staining varied among the granulosa cells; the cells in tertiary follicle were intensely stained. Interestingly, unlike the staining for non-methylated CCGG sites, the staining for methylated CCGG sites in TUNEL positive cells was very weak. The control sections, in which no Msp I digestion was conducted, showed no staining (Fig. 1E), indicating that blockade with dideoxynucleotides was perfect after Hpa II digestion. In addition, when TUNEL was performed with TTP instead of biotin-16-dUTP or without TdT, no staining was detected.
Fig. 1.
Methylation level of CCGG sites in TUNEL-positive follicle cells in adjacent sections of mouse ovary. Adjacent sections of paraffin-embedded mouse ovary were used for H&E staining, TUNEL, and the detection of non-methylated CCGG sites and methylated CCGG sites, as described in detail in “II. Materials and Methods”. A: H&E staining. B: TUNEL staining. C: Blockade of 3'-OH ends with dideoxynucleotides by TdT. After blockade, the section was labeled with biotin-16-dUTP by TdT and the incorporated biotin was detected with HRP-anti-biotin. No signals were observed. D: Staining for non-methylated CCGG sites. After the blockade procedure described in (C), the section was digested with Hpa II, labeled with biotin-16-dUTP and visualized by enzyme-immunohistochemistry with HRP-anti-biotin. E: Blockade of Hpa II cutting sites with dideoxynucleotides by TdT. The section was digested with Hpa II and the cutting sites were blocked with a dideoxynucleotide mixture. Then the section was processed in a manner similar to that described in (C). F: Staining for methylated CCGG sites. After blockade of Hpa II cutting sites with dideoxynucleotides, the section was digested with Msp I and the cutting sites were labeled with biotin-16-dUTP and visualized with HRP-anti-biotin. Most of TUNEL-positive follicle cells were found in methylation level of CCGG sites. Bar = 100 μm.
Immunohistochemical detection of 5-methylcytosine in mouse ovary
For comparison of the localization pattern of methylated CCGG sites detected by HELMET with that of global 5-methylcytosine, we conducted immunohistochemistry using anti-5-methylcytosine monoclonal antibody. As shown in Fig. 2, the nuclei of most granulosa cells were strongly stained and there was no marked difference in the intensity of the staining among granulosa cells at any differentiation stages of follicles.
Fig. 2.
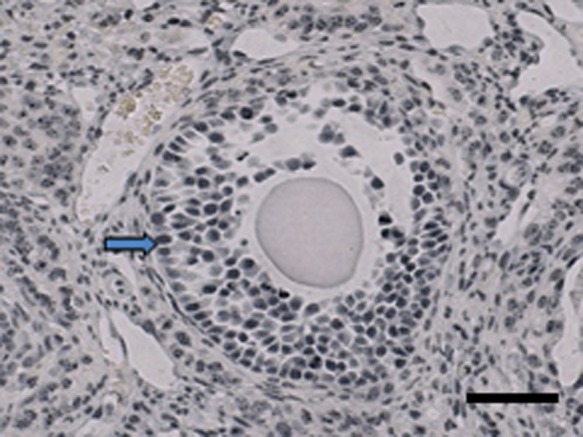
Immunohistochemical detection of 5-methylcytosine in mouse ovary. Paraffin-embedded sections of mouse ovaries were using anti-5-methylcytosine monoclonal antibody, as described in detail in “II. Materials and Methods”. Most of the granulosa nuclei were strongly stained (arrow) and there was no remarkable difference in the intensity of the staining among granulosa cells at various differentiation stages follicles. Bar = 100 μm.
Differential staining of non-methylated and methylated CCGG and GATCG sites of DNA in adjacent sections of mouse ovary
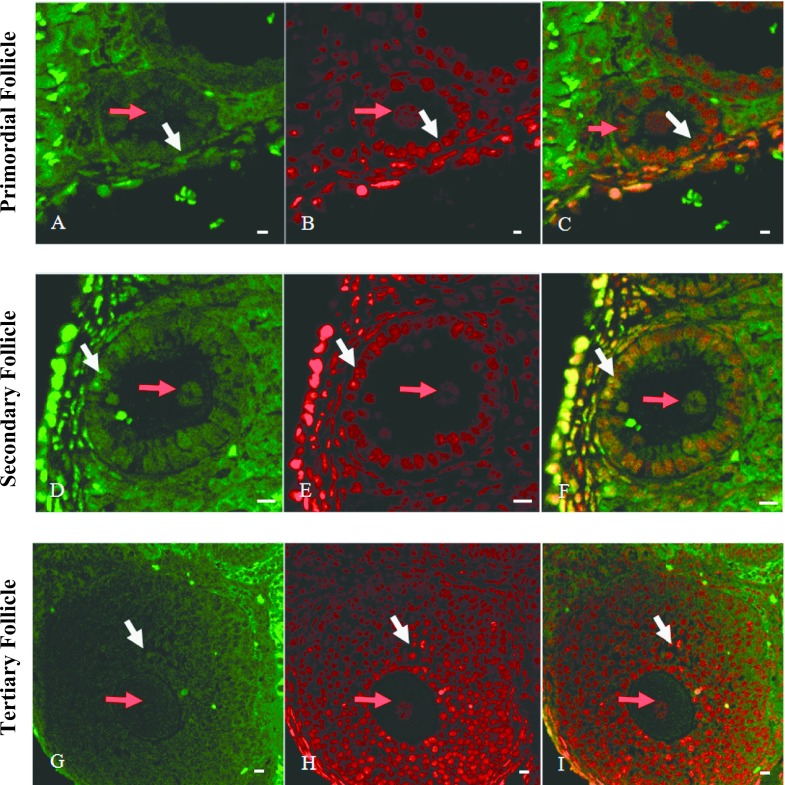
To assess the differences in the ratio of non-methylated to methylated CCGG sequence of DNA among different differentiation stages of granulosa cells, we performed simultaneous staining of both CCGG sequences by HELMET. As shown in Fig. 3, Hpa II-positive non-methylated CCGG was localized in the nuclei of granulosa cells (white arrows) and its staining became to be more intense from primary to secondary follicles (Fig. 3A and D), but attenuates in tertiary follicles (Fig. 3G). On the other hand, the overall staining for methylated CCGG was the most intense in the granulosa cells of tertiary follicle (Fig. 3B, E and H). In merged images (Fig. 3C, F and I), it was clear that granulosa cells were yellow or green, indicating equal or predominant presence of non-methylated CCGG. We also found that Hpa II-positive non-methylated CCGG was localized in the nuclei of oocytes (red arrows), especially in secondary follicle stage.
Fig. 3.
Simultaneous localization of non-methylated and methylated CCGG sequences by HELMET in a paraffin-embedded section of mouse ovary. The paraffin-embedded section was blocked with a dideoxynucleotide mixture by TdT and then the Hpa II cutting sites were labeled with biotin-16-dUTP. After dideoxynucleotide blockade, Msp I cutting sites were labeled with digoxigenin-11-dUTP and then both haptens were visualized with FITC-anti-biotin (A, D and G) and rhodamine-anti-digoxigenin (B, E and H), respectively. Merged images were shown in C, F and I. A, B and C: Primary follicle. D, E and F: Secondary follicle. G, H and I: Tertiary follicle. Red arrow pointed the nuclei of oocytes and white arrow pointed granulosa cells. And non-methylated CCGG was localized in the nuclei of granulosa cells (white arrows) or oocytes (red arrows) and its staining became to be more intense from primary to secondary follicles. Bar = 20 μm.
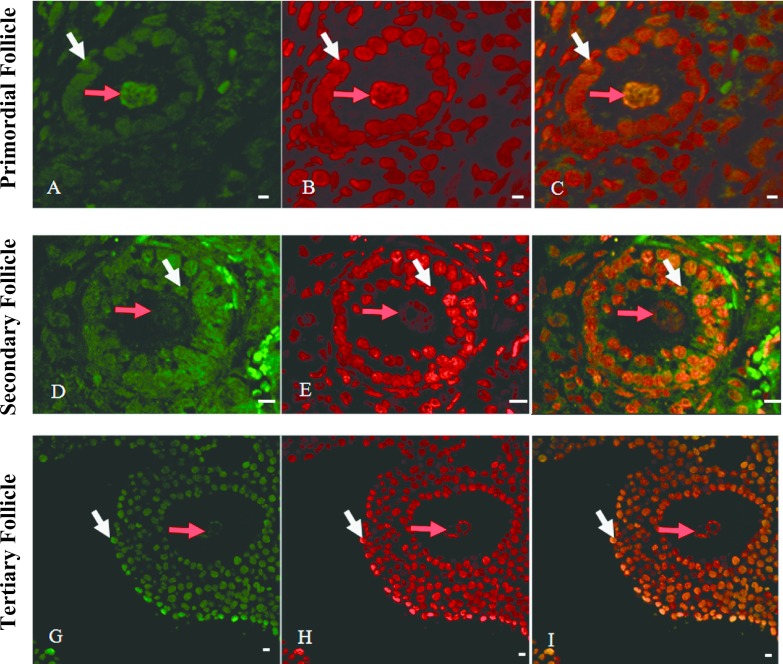
In the meantime, we performed simultaneous staining of non-methylated and methylated GATCG sequences by HELMET and calculated the ratio of non-methylated to methylated GATCG sequence in each nucleus among different differentiation stages of follicles. As shown in Fig. 4, Sau3A I-positive non-methylated GATCG was localized in the nuclei of granulosa cells (white arrows) and its staining pattern was similar to that of non-methylated CCGG (Fig. 4A, D and G). And the same tendency to the methylated CCGG sites was found in the Mbo I-positive methylated GATCG (Fig. 4B, E and H) and the merged images (Fig. 4C, F and I). In addition, we observed the similar change in GATCG in the nuclei of oocytes (red arrows), but the Mbo I-positive methylated GATCG was the most intense in the oocyte of primary follicle.
Fig. 4.
Simultaneous localization of non-methylated and methylated GATCG sequences by HELMET in a paraffin-embedded section of mouse ovary. The paraffin-embedded section was blocked with a dideoxynucleotide mixture by TdT and then the Sau3A I cutting sites were labeled with biotin-16-dUTP. After dideoxynucleotide blockade, Mbo I cutting sites were labeled with digoxigenin-11-dUTP and then both haptens were visualized with FITC-anti-biotin (A, D and G) and rhodamine-anti-digoxigenin (B, E and H), respectively. Merged images are given in C, F and I. Red arrow pointed the nuclei of oocytes and white arrow pointed granulosa cells. And non-methylated GATCG was localized in the nuclei of granulosa cells (white arrows) and its staining became to be more intense from primary to secondary follicles. Bar = 20 μm.
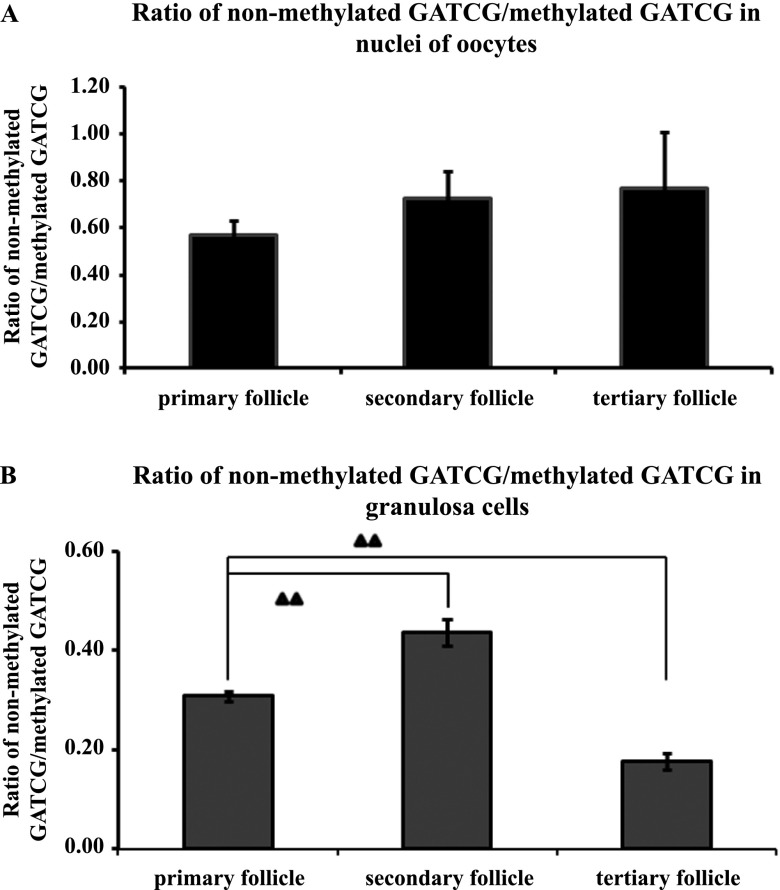
Finally, we calculated the ratio of staining intensity for non-methylated GATCG to methylated GATCG in both nuclei of oocytes and granulosa cells (Fig. 5A and B). Even though an increasing tendency of the ratio from primary follicle to tertiary follicle in nuclei of oocytes was found, it was not significant. On the other hand, the ratios in granulosa cells increased significantly from primary follicle to secondary follicle (0.4) (P < 0.01), but then dropped in tertiary follicle (0.17) (P < 0.01).
Fig. 5.
Ratio of staining intensity for non-methylated to methylated GATCG sequences by HELMET in a paraffin-embedded section of mouse ovary. Panel A represented the ratio in the nuclei of oocytes, where the data were expressed as mean ± standard deviation (n = 6, 18, 6 respectively in primary (including primordial), secondary and tertiary follicles respectively). Panel B indicated the change in the ratio in granulosa cells and the difference was significant. The data were expressed as mean ± standard deviation (n = 10 in primary (including primordial), secondary and tertiary follicles respectively). ▲▲P < 0.01.
IV. Discussion
Epigenetics is defined as the mechanism underlying mitotically or meiotically heritable changes in gene expression that occur without accompanying changes in DNA sequence. The major mechanisms of epigenetics include DNA methylation, histone modifications and expression of various non-coding microRNAs. In the present study, we focused on the change in the level of DNA methylation during mouse ovarian folliculogenesis, using HELMET which permits us to analyze the methylation level of specific sequence sites at an individual cell level. Consequently, we found that the level of methylation at GATCG sites in oocytes tended to decrease from primary to secondary follicles. The level of CCGG and GATCG methylation in granulosa cells significantly decreased from primary to secondary follicles and then increased in tertiary follicle. Moreover, a marked demethylation of CCGG sites was found in TUNEL positive granulosa cells. These results indicate that a failure of follicle stage-dependent increase in CCGG methylation may induce granulosa cell apoptosis, leading to the induction of follicle atresia. In this context, it should be noted that there was no significant difference in immunohistochemical detection of 5-methylcytosine among different stages of follicles. As shown in the above, although the methylation level of DNA assessed by HELMET was clearly changed, total methylation level detected by anti-5-methylcytosine was not fluctuated. This discrepancy may not be surprising when we consider that CCGG sequences, which can be analyzed by HELMET, comprise only 5% of CpG methylation sites. We believe that the methylation level of CCGG sites reflect the effect of DNA methylation on transcriptional regulation of genes more correctly than the total methylation level, because those sites are very frequent in the CpG islands, which are generally associated with gene promoters [14].
It has been already demonstrated that DNA methylation plays an important role in spermatogenesis. The cytosine analogue, 5-aza-2'-deoxycytidine which can decrease DNA methylation levels induced histological abnormalities in the testis [8], accompanying the induction of spermatogenic cell apoptosis [18]. Using HELMET, we reported that the methylation levels at CCGG sites varied with differentiation stages of spermatogenic cells and in addition, apoptotic spermatogenic cells exhibited a marked demethylation at CCGG sites [9], similarly to the case of ovarian granulosa cells.
In primordial germ cells, the level of DNA methylation was known to be very low and the same tendency was found in oogonia [4]. In primordial follicles, the methylation level of oocytes remained unchanged, but granulosa cells were in hypomethylation states [15, 21]. In the present study, our statistical analysis further revealed that the methylation level of GATCG sites in oocytes was not significantly changed among different stages of follicles, while the lowest methylation levels were observed in granulosa cells in secondary follicles. Importantly, it is known that in the stage of secondary follicles, drastic changes in the follicular structure occur; the formation of zona pellucida and the change of simple epithelium to stratified one. The lowest level of methylation of these specific sequences might be related to the activation of a set of gene which are required for such changes.
It was really amazing to find a significant fluctuation in the methylation level of DNA in granulosa cells during ovarian follicle development. The biological significance is now largely unclear, but the future study on chromatin structure would shed a light at the point.
V. Conflicts of Interest
The authors declare that there are no conflicts of interest.
VI. Acknowledgments
This research was supported in part by grants from the National Natural Science Foundation of China (81202175), Natural Science Foundation of Fujian Province (2012J05149), outstanding youth project in colleges and universities of Fujian Province (JA13132) and a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (No. 18390060 and 16K15173 to Koji, T).
VII. References
- 1.Anway M. D., Cupp A. S., Uzumcu M. and Skinner M. K. (2005) Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308; 1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein B. E., Meissner A. and Lander E. S. (2007) The mammalian epigenome. Cell 128; 669–681. [DOI] [PubMed] [Google Scholar]
- 3.Chen X. H., Song N., Matsumoto K., Nanashima A., Nagayasu T., Hayashi T., Ying M., Endo D., Wu Z. R. and Koji T. (2013) High expression of trimethylated histone H3 at lysine 27 predicts better prognosis in non-small cell lung cancer. Int. J. Oncol. 43; 1467–1480. [DOI] [PubMed] [Google Scholar]
- 4.Chung Y. G., Ratnam S. and Chillet J. R. (2003) Abnormal regulation of DNA methyltransferase expression in cloned mouse embryos. Biol. Reprod. 69; 146–153. [DOI] [PubMed] [Google Scholar]
- 5.De La Fuente R., Baumann C., Fan T., Schmidtmann A., Dobrinski I. and Muegge K. (2006) Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat. Cell Biol. 8; 1448–1454. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg A. D., Allis C. D. and Bernstein E. (2007) Epigenetics: a landscape takes shape. Cell 128; 635–638. [DOI] [PubMed] [Google Scholar]
- 7.Hakuno N., Koji T., Yano T., Kobayashi N., Tsutsumi O., Taketani Y. and Nakane P. K. (1996) Fas/APO-l/CD95 system as a mediator of granulosa cell apoptosis in ovarian follicle atresia. Endocrinology 137; 1938–1948. [DOI] [PubMed] [Google Scholar]
- 8.Kelly T. L. and Trasler J. M. (2004) Reproductive epigenetics. Clin. Genet. 65; 247–260. [DOI] [PubMed] [Google Scholar]
- 9.Koji T., Kondo S., Hishikawa Y., An S. and Sato Y. (2008) In situ detection of methylated DNA by histo endonuclease-linked detection of methylated DNA sites: a new principle of analysis of DNA methylation. Histochem. Cell Biol. 130; 917–925. [DOI] [PubMed] [Google Scholar]
- 10.McGee E. A., Perlas E., LaPolt P. S., Tsafriri A. and Hsueh A. J. (1997) Follicle-stimulating hormone enhances the development of preantral follicles in juvenile rats. Biol. Reprod. 57; 990–998. [DOI] [PubMed] [Google Scholar]
- 11.Nakajima K., Shibata Y., Hishikawa Y., Suematsu T., Mori M., Fukuhara S., Koji T., Sawase T. and Ikeda T. (2014) Coexpression of ang1 and tie2 in odontoblasts of mouse developing and mature teeth—a new insight into dentinogenesis. Acta Histochem. Cytochem. 47; 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obata Y. and Kono T. (2002) Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J. Biol. Chem. 277; 5285–5289. [DOI] [PubMed] [Google Scholar]
- 13.Reik W., Dean W. and Walter J. (2001) Epigenetic reprogramming in mammalian development. Science 293; 1089–1093. [DOI] [PubMed] [Google Scholar]
- 14.Saxonov S., Berg P. and Brutlag D. L. (2006) A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U S A 103; 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W. and Reik W. (2012) The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48; 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma S., Kelly T. K. and Jones P. A. (2010) Epigenetics in cancer. Carcinogenesis 31; 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith Z. D., Chan M. M., Mikkelsen T. S., Gu H. C., Gnirke A., Regev A. and Meissner A. (2012) A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484; 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song N., Endo D., Song B., Shibata Y. and Koji T. (2016) 5-aza-2'-deoxycytidine impairs mouse spermatogenesis at multiple stages through different usage of DNA methyltransferases. Toxicology 361–362; 62–72. [DOI] [PubMed] [Google Scholar]
- 19.Tachibana M., Nozaki M., Takeda N. and Shinkai Y. (2007) Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 26; 3346–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tobinaga S., Matsumoto K., Nagayasu T., Furukawa K., Abo T., Yamasaki N., Tsuchiya T., Miyazaki T. and Koji T. (2015) Keratinocyte growth factor gene electroporation into skeletal muscle as a novel gene therapeutic approach for elastase-induced pulmonary emphysema in mice. Acta Histochem. Cytochem. 48; 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomizawa S., Nowacka-Woszuk J. and Kelsey G. (2012) DNA methylation establishment during oocyte growth: mechanisms and significance. Int. J. Dev. Biol. 56; 867–875. [DOI] [PubMed] [Google Scholar]