Abstract
Animal brucellosis is a reemerging disease in China, particular in northwest China. The Brucella species (even genus) are highly conserved; therefore the use of Multilocus sequencing typing (MLST: based on conserved housekeeping loci) is more suitable for discrimination at species or biovar level on Brucella. In this study, MLST was used to analyze the characterization of Brucella from sheep and yaks during 2015 and 2016. All 66 isolates were collected from northwest China, including Inner Mongolia, Xinjiang, Qinghai, and Gansu provinces. Isolates were cultured on Brucella agar medium and identified by MLST. MLST identified five ST types: ST8 (n = 55), ST7 (n = 2), ST3 (n = 5), ST1 (n = 2), and ST14 (n = 2). This analysis revealed that B. melitensis isolates exhibited high single genotypes (ST8) in the most northwest China. MLST of isolates provides helpful information on understanding genetic characterization of Brucella in northwest China.
1. Introduction
Brucellosis, caused by Brucella spp., is a common zoonotic disease worldwide [1]. It has been considered as a reemerging infectious disease in China because of the increasing incidences in the past several years [2]. Brucella melitensis is responsible for the major causative agent of brucellosis in sheep and human. Sheep and goats are major herbivores in northwest China and are primarily kept by poor rural farmers in pastoral areas [3, 4]. Therefore, brucellosis has an important zoonosis in northwest China.
Multilocus sequencing typing (MLST), as a genotyping tool for assessing genetic diversity and relationships, was widely used to identify and analyze diversity of bacteria and epidemiology characterization [5–8]. Although the data of MLST of Brucella stranis in China can not adequately reflect its epidemiological characteristics and relationship between disease emerging and development, the prevalence of genotypes of Brucella strains from Inner Mongolia has changed over time in the three stages [9]. The aims of the present study were to identify genotypes of Brucella in northwest China and we tried to analyze relationship between disease prevalence and genotypic diversity. Therefore, MLST was used to analyze 66 Brucella isolates from sheep and yaks during 2015 and 2016.
2. Materials and Methods
2.1. Brucella Strains and DNA Extraction
In the previous study [10], sixty-six isolates were collected from northwest China; 13, 26, 17, and 10 were from Gansu, Inner Mongolia, Xinjiang, and Qinghai provinces, respectively (Table 1). Brucella were reproduced in BBL™ Brucella Broth (BD, USA) with 5% horse serum at 37°C. The 50 ml mid-log phase culture was harvested by centrifugation at 10,000 ×g for 5 min and resuspended in 10 ml PBS (0.01 M, pH 7.2). Total genomic DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Germany) according to the manufacturer's instructions. DNA extracted from all isolates was stored at −20°C.
Table 1.
Geographical distribution and genotype of Brucella isolates in northwest China.
| Province | Host | Locality ID | Region | Species | Genotype | Isolate size |
|---|---|---|---|---|---|---|
| Gansu | Sheep | 1 | Qingyang | B. melitensis | ST7 | 4 |
| ST8 | 6 | |||||
| 2 | Lanzhou | B. melitensis | ST8 | 1 | ||
| 3 | Wuwei | B. melitensis | ST8 | 2 | ||
| Inner Mongolia | Sheep | 4 | Otog | B. melitensis | ST8 | 6 |
| 5 | Ulanqab | B. suis | ST14 | 1 | ||
| B. melitensis | ST8 | 11 | ||||
| 6 | Linhe | B. abortus | ST1 | 2 | ||
| B. suis | ST14 | 1 | ||||
| B. melitensis | ST8 | 5 | ||||
| Xinjiang | Sheep | 7 | Urumchi | B. melitensis | ST8 | 7 |
| 8 | Shihezi | B. melitensis | ST8 | 3 | ||
| 9 | Hetian | B. melitensis | ST8 | 5 | ||
| 10 | Bayingolin | B. melitensis | ST8 | 2 | ||
| Qinghai | Yaks | 11 | Tianjun | B. abortus | ST3 | 3 |
| Sheep | B. melitensis | ST3 | 1 | |||
| 12 | Haiyan | B. melitensis | ST3 | 1 | ||
| ST8 | 3 | |||||
| 13 | Yushu | B. melitensis | ST8 | 1 | ||
| 14 | Zhiduo | B. melitensis | ST8 | 1 |
Province: region of sample collection; host: host from which the bacteria were isolated; locality ID corresponds to the counties of isolation; region: region from which the samples and isolates were collected.
2.2. MLST Genotyping
MLST genotyping was performed by analyzing nine distinct genomic loci, including seven housekeeping genes (gap, aroA, glk, dnaK, gyrB, trpE, and cobQ), one outer membrane protein (omp25), and one intergenic fragment (int-hyp) [11]. PCR amplification was performed as described previously [9]. Sequences obtained from purified PCR products were aligned using MEGA 5 according to published MLST sequences in GenBank (accession numbers AM694191–AM695630) [11]. A local comparison database was established after downloading of relevant data, and distinct alleles identified at the nine selected loci were each given a numerical designation according to the sequences of the defined alleles. Each sequence type over all loci (ST) was predicted by comparisons and analyses based on a local comparison database established using MEGA 5 and a web-based MLST service (Brucella Base, https://pubmlst.org/brucella/). DNA preparations from the B. melitensis 16 M, B. abortus 544, and B. suis 1330 reference strains were used as controls.
2.3. Analysis of MLST DATA
To clarify the molecular characteristics and evolutionary relationships of brucellosis in northwest China, all 66 isolates were analyzed using BioNumerics version 7.6. Using the same software, clustering analysis was performed using minimal spanning tree [12, 13]. The resulting genotypes were compared using the web-based MLST database (https://pubmlst.org/brucella/). Genotypic diversification of Brucella in northwest China was analyzed using this study and published data [9, 12–14].
3. Results
3.1. MLST Results
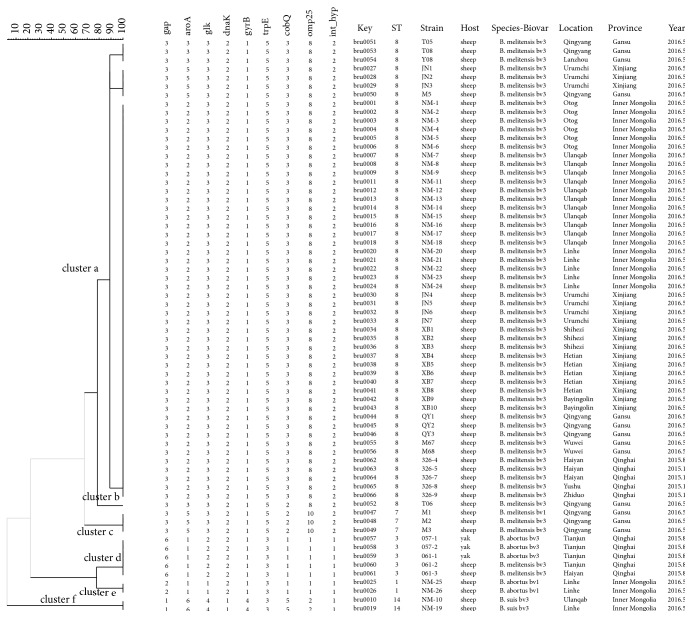
MLST analysis showed that five known MLST genotypes were identified: ST3 (6-1-2-2-1-3-1-1-1; n = 5), ST1 (2-1-1-2-1-3-1-1-1; n = 2), ST8 (3-3-3-2-1-5-3-8-2; n = 44), ST7 (3-5-3-2-1-5-3-8-2; n = 3), and ST14 (1-6-4-1-4-3-5-2-1; n = 2). Clustering analysis by using BioMumerics software showed that the 66 isolates formed six main clusters (a–f). B. melitensis was distributed in cluster a–c, and genotype ST8 plays a dominant role in isolates (53/66). Genotypes ST1 belonged to B. abortus; genotype ST14 belonged to B. suis; genotypes ST7 and ST8 belonged to B. melitensis, while 5 isolates identify as ST3, including two B. melitensis strains and three B. abortus strains, which were isolated from Tianjun county, Qinghai province (Figure 1). Therefore, MLST is more suitable for discrimination at species or biovar level on Brucella species. Analysis of MLST was conducted in 69 Brucella strains involving nucleotide sequences of 4396 positions. The result showed that 28 segregating sites were presented among those loci.
Figure 1.
Dendrogram based on the MLST genotyping assay showing relationships of 66 Brucella isolates. Key: serial number for the 66 isolates; ST: MLST genotype fo isolates; strain: the number conferred to isolates; host: the hosts from which the bacteria were isolated; location: sample specific location (Table 1 in detail); province: the regions of sample collection; year: the time of isolation of bacteria.
3.2. Molecular Epidemiology of Brucella in Northwest China
Among 66 isolates, B. melitensis isolates were identified as genotypes ST3, ST7, and ST8; B. abortus isolates were ST1 and ST3; and B. suis isolates were ST14. ST8 was a dominant genotype in those B. melitensis isolates, and it is a widespread Brucella genotype in northwest China. The results also showed that B. abortus infected sheep, but worryingly, B. suis biovar 3 with genotype ST14 emerged in sheep in Inner Mongolia. On the other hand, two isolates from sheep in Qinghai province were B. melitensis biovar 3, while MLST result presented genotype ST3, which was identified from isolates in yaks.
4. Discussion
In the present study, we used MLST methods to genotype Brucella isolates from northwest China. A total of 66 Brucella isolates were examined by MLST typing. In the previous study, all isolates were B. melitensis, B. abortus, and B. suis. B. melitensis was dominant epidemic strain in animal and B. abortus and B. suis also infected sheep; B. suis biovar 3 especially emerged in Inner Mongolia. B. melitensis genotype ST8 not only was the predominant genotype in sheep but also responded for human brucellosis. These results reveal that human brucellosis in northwest China is closely related to infectious sheep.
In the south of Gansu province, B. melitensis isolates have two genotypes (ST7 and ST8). It was indicated that the epidemiology of Brucella in there was diversified. MLVA analysis of China isolates suggested that there were three predominant but different lines of Brucella transmission in China, while a common thing is that the transmission lines are from north to south [15], but MLST analysis has a comprehensive understanding of Brucella epidemic in China. So far, one of the regrets is that there is little data related to genotype study involved Brucella in the central and western regions in China. Therefore, change and transmission line of brucellosis are unclear from the south of Gansu to the central and western regions in China, and the causes and consequence of Brucella diversity should be investigated by molecular epidemiology analysis of emerging Brucella in the central and western regions in China.
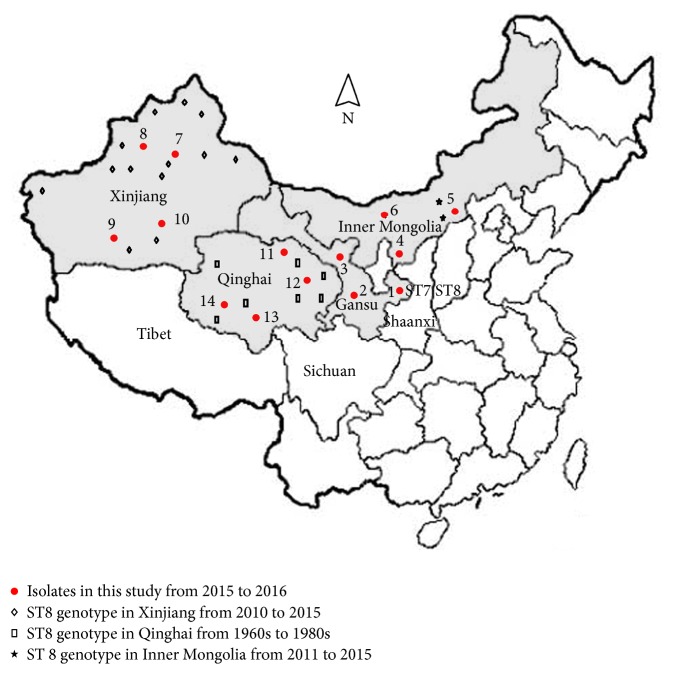
Collection of MLST data of isolates from northwest China: ST8 was a dominant genotype of B. melitensis which were responsible for sheep and human brucellosis (Figure 2) [9, 13, 14]. MLST analysis of Brucella strains isolated from the 1980s in Qinghai province revealed that genotype ST8 of B. melitensis was widely spread in sheep, blue sheep, and human [13]. B. melitensis ST8 genotype was also identified from isolates in sheep and human in Gansu, Xinjiang, and Inner Mongolia [12, 14].
Figure 2.
Geographic distribution of Brucella genotype ST8 in northwest China. Dotted (red) countries: sampling area (Table 1 in detail); other shapes: the other data collected in different provinces in northwest China from [12–14].
In the previous study, brucellosis in China was divided into three periods, high incidence (1950–1960s), decline (1970–1980s), and reemergence (1990–2000s) [9]. On the other hand, brucellosis has been rising every year since the beginning of the 21st century, especially outbreaks in parts of the northwest China in recent years [2, 16]. In Inner Mongolia, 61.11% (11/18), 14.29% (3/21), 47.62% (10/21), and 100% (116/116) genotype ST8 Brucella were isolated in 1950–1960s, 1970–1980s, 1990–2000s, and 2010–2015 stages, respectively [9, 12]. Those results revealed that genotype ST8 B. melitensis is an important epidemiological marker for trend of brucellosis of epidemics in northwest China. It is further suggested that the ST8 is an extensive endemic in northwest region of China. Conversely, lower genetic diversity and crowding effects may favor transmission and select faster replicating organisms with major zoonotic potential [16–18], which will increase threatening of brucellosis for animals and human. Recently, a research involving MLST analysis of Brucella isolates in China between 1953 and 2013 showed that a total of 206 isolates have been identified as 32 MLST genotypes (STs), which included 13 new STs (ST71-83), although ST8 was a dominant genotype [19]. It is implied that the study of Brucella genotype in larger scale epidemic strains should be carried out in order to understand the characteristics of Brucella epidemic and genetic variation.
5. Conclusion
MLST was used to analyze 66 Brucella isolates from the northwest China. 83% (55/66) were genotype ST8. Analysis of genotype of Brucella strains from the northwest China further confirmed that ST8 was a dominant Brucella genotype responding for sheep and human disease.
Acknowledgments
This study was supported by The Chinese Center for Disease Control and Prevention and the Center for Animal Disease Prevention and Control in Qinghai Province. This study was financially supported partially by the National Key Research and Development Plan (2016YFD0500907 and 2017YFD0500903), the National Modern Agricultural Industry Technology Fund for Scientists in Sheep Industry System, China (CARS-39-04B), and the Fundamental Research Funds for CAAS (1610312017014).
Contributor Information
Xiaoan Cao, Email: caoxiaoan@caas.cn.
Zhizhong Jing, Email: jingzhizhong@caas.cn.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- 1.Pappas G., Papadimitriou P., Akritidis N., Christou L., Tsianos E. V. The new global map of human brucellosis. 2006;6(2):91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- 2.Lai S., Zhou H., Xiong W., et al. Changing epidemiology of human brucellosis, China, 1955–2014. 2017;23(2):184–194. doi: 10.3201/eid2302.151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou Q., Sun X., Zhang J., Liu Y., Wang Y., Jin Z. Modeling the transmission dynamics of sheep brucellosis in Inner Mongolia Autonomous Region, China. 2013;242(1):51–58. doi: 10.1016/j.mbs.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F., Li Z., La X., et al. Multiple-locus variable-number tandem-repeat analysis of Brucella isolates from patients in Xinjiang China. 2015;8:15716–15723. [PMC free article] [PubMed] [Google Scholar]
- 5.Castro-Nallar E., Hasan N. A., Cebula T. A., et al. Concordance and discordance of sequence survey methods for molecular epidemiology. 2015;2015(2, article e761) doi: 10.7717/peerj.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri R. R., Henderson I. R. The evolution of the Escherichia coli phylogeny. 2012;12(2):214–226. doi: 10.1016/j.meegid.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira A. C., Dias R., de Sá M. I. C., Tenreiro R. Whole-genome mapping reveals a large chromosomal inversion on Iberian Brucella suis biovar 2 strains. 2016;192:220–225. doi: 10.1016/j.vetmic.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Shome R., Krithiga N., Shankaranarayana P. B., et al. Genotyping of Indian antigenic, vaccine, and field Brucella spp. Using multilocus sequence typing. 2016;10(3):237–244. doi: 10.3855/jidc.6617. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y., Ke Y., Wang Y., et al. Changes of predominant species/biovars and sequence types of Brucella isolates, Inner Mongolia, China. 2013;13(1, article 514) doi: 10.1186/1471-2334-13-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao X., Li Z., Liu Z., et al. Molecular epidemiological characterization of Brucella isolates from sheep and yaks in northwest China. 2018;65(2):e425–e433. doi: 10.1111/tbed.12777. [DOI] [PubMed] [Google Scholar]
- 11.Whatmore A. M., Perrett L. L., MacMillan A. P. Characterisation of the genetic diversity of Brucella by multilocus sequencing. 2007;7, article 34 doi: 10.1186/1471-2180-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z. G., Wang M., Liu R. H., et al. Multilocus sequence typing of Brucella clinical isolated strains in Inner Mongolia. 2016;32, article 6 [Google Scholar]
- 13.Ma J.-Y., Wang H., Zhang X.-F., et al. MLVA and MLST typing of Brucella from Qinghai, China. 2016;5(1, article 26) doi: 10.1186/s40249-016-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun M.-J., Di D.-D., Li Y., et al. Genotyping of Brucella melitensis and Brucella abortus strains currently circulating in Xinjiang, China. 2016;44:522–529. doi: 10.1016/j.meegid.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 15.Tian G.-Z., Cui B.-Y., Piao D.-R., et al. Multi-locus variable-number tandem repeat analysis of Chinese Brucella strains isolated from 1953 to 2013. 2017;6(1, article 89) doi: 10.1186/s40249-017-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno E. Retrospective and prospective perspectives on zoonotic brucellosis. 2014;5, article 213 doi: 10.3389/fmicb.2014.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDaniel C. J., Cardwell D. M., Moeller R. B., Gray G. C. Humans and cattle: a review of bovine zoonoses. 2014;14(1):1–19. doi: 10.1089/vbz.2012.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Román K., Castillo R., Gilman R. H., et al. A foodborne outbreak of brucellosis at a police station cafeteria, Lima, Peru. 2013;88(3):552–558. doi: 10.4269/ajtmh.12-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piao D., Liu X., Di D., et al. Genetic polymorphisms identify in species/biovars of Brucella isolated in China between 1953 and 2013 by MLST. 2018;18(1, article 7) doi: 10.1186/s12866-018-1149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]