Abstract
Quick and accurate molecular testing is necessary for the better management of many inherited diseases. Recent technological advances in various next generation sequencing (NGS) platforms, such as target panel-based sequencing, has enabled comprehensive, quick, and precise interrogation of many genetic variations. As a result, these technologies have become a valuable tool for gene discovery and for clinical diagnostics. The AmpliSeq Inherited Disease Panel (IDP) consists of 328 genes underlying more than 700 inherited diseases. Here, we aimed to assess the performance of the IDP as a sensitive and rapid comprehensive gene panel testing. A total of 88 patients with inherited diseases and causal mutations that were previously identified by Sanger sequencing were randomly selected for assessing the performance of the IDP. The IDP successfully detected 93.1% of the mutations in our validation cohort, achieving high overall gene coverage (98%). The sensitivity for detecting single nucleotide variants (SNVs) and short Indels was 97.3% and 69.2%, respectively. IDP, when coupled with Ion Torrent Personal Genome Machine (PGM), delivers comprehensive and rapid sequencing for genes that are responsible for various inherited diseases. Our validation results suggest the suitability of this panel for use as a first-line screening test after applying the necessary clinical validation.
Keywords: inherited diseases, mutations, gene panel, targeted NGS, AmpliSeq Inherited Disease Panel, Saudi Human Genome Program database
1. Introduction
The morbidity, mortality, and disability that are associated with inherited diseases can be greatly reduced or prevented through improving the accuracy and speed of molecular testing. Genetic advances over the past decades led to the development of different mutation screening techniques, which have been applied in research or integrated into diagnostic laboratory protocols [1,2,3,4,5]. Sanger sequencing, for instance, is widely used for the genetic evaluation of inherited diseases, owing to its ability to detect point mutations and small insertion/deletion sequence changes with high accuracy. However, this method is typically utilized to evaluate a small fragment of a single gene, and in some (less common) cases, to test a limited number of genes simultaneously. Even with the wide availability of this method, it can be very expensive, labor intensive, and time consuming when being used to interrogate diseases with genetic diversity [6,7]. This is often the case with inherited diseases, in which various genes can be involved in the development of a monogenic disorder. The fast development and the increasing availability of several next generation sequencing (NGS) technologies, namely, whole genome, whole exome, and target panel-based sequencing allowed for comprehensive, high resolution, and accurate interrogation of the genome or portions of it in a short time. These features made NGS a valuable tool for discovery research and for clinical genetic diagnostics [8,9].
Targeted NGS is becoming the preferred method for identifying causal mutations in genetically heterogeneous disorders, such as cardiomyopathies, retinopathies, hearing loss, epilepsies, immunodeficiency, and ataxias [10,11,12,13,14,15,16]. A wide range of panels comprising either preselected or customized assortment of genes associated with a single or a group of conditions are commercially available (either as a service or ready-to-use) and are widely used to detect germline or somatic mutations [17,18,19,20]. Ion Torrent sequencing platform has been shown to perform well in terms of run time and total cost [21]. These are two important considerations when developing assays for use in routine clinical diagnostics. In 2012, Ion Torrent released its first version of the comprehensive panel for hereditary diseases, known as AmpliSeq Inherited Disease Panel (IDP, http://www.edgebio.com/sites/default/files/IonAmpliSeq_InheritedDiseasePanel_Flyer_CO25400_May%2023%202012.pdf). The recent version consists of 328 genes underlying over 700 inherited diseases. Therefore, we aimed here to assess the performance of the IDP as a sensitive and rapid comprehensive genetic assay. We randomly selected patients with inherited diseases in whom causal mutations were previously identified via Sanger sequencing for validating the detection yield of IDP.
2. Materials and Methods
2.1. Samples
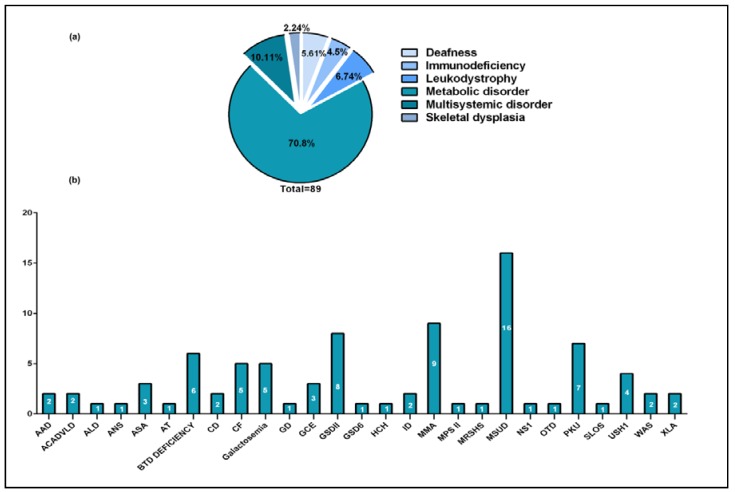
We randomly selected samples from 88 Saudi patients with previously confirmed genetic diagnosis after obtaining full informed consent from all of the participants. Written informed consents were obtained from all of the study subjects, which is in adherence with the declaration of Helsinki, and according to King Faisal Specialist Hospital & Research Centre (KFSHRC) Institutional Review Board (IRB) and Research Advisory Committee (RAC) rules and regulations under the following approved projects: (RAC#2020011, approved-present), (RAC#2050022, approved-present) (RAC#2100001, approved-present). The conditions and corresponding genes that were tested in this study are listed in Table 1. A breakdown of our validation cohort by disease category and the number of samples tested for each condition is summarized in Figure 1. DNA was extracted from whole blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). The quantity of extracted DNA was estimated using the broad range Qubit® 2.0 kit (Invitrogen, Carlsbad, CA, USA), according to manufacturer’s instructions.
Table 1.
List of inherited conditions tested in this study.
| Clinical Diagnosis | Abbreviation | Category | Genes Included in the Panel | OMIM# |
|---|---|---|---|---|
| Agammaglobulinemia, X-linked, Type I | XLA | Immunodeficiency | BTK | 300755 |
| Argininosuccinate Lyase Deficiency | ASA | Metabolic disorder | ASL | 207900 |
| Arylsulfatase A Deficiency | AAD | Leukodystrophy | ARSA | 250100 |
| Ataxia Neuropathy Spectrum/Alpers Syndrome | ANS | Multisystemic disorder | POLG | 203700 |
| Ataxia-telangiectasia | AT | Multisystemic disorder | ATM | 208900 |
| Biotinidase Deficiency | BTD deficiency | Metabolic disorder | BTD | 253260 |
| Canavan Disease | CD | Leukodystrophy | ASPA | 271900 |
| Cystic Fibrosis | CF | Multisystemic disorder | CFTR | 219700 |
| Galactosemia | Galactosemia | Metabolic disorder | GALT | 230400 |
| Gaucher Disease | GD | Multisystemic disorder | GBA | 230800 |
| Glycine Encephalopathy | GCE | Metabolic disorder | GCSH, GLDC, AMT | 605899 |
| Glycogen Storage Disease Type VI | GSD6 | Metabolic disorder | GBE1 | 232700 |
| Hunter Syndrome/Mucopolysaccharidosis, Type II (MPS II) | MPS II | Metabolic disorder | IDS | 309900 |
| Hypochondroplasia | HCH | Skeletal dysplasia | FGFR3 | 146000 |
| Inherited Deafness | ID | Deafness | GJB2, GJB3, GJB6, COL11A2, KCNQ4 | 220290 |
| Maple syrup Urine Disease | MSUD | Metabolic disorder | BCKDHA, BCKDHB, DBT, DLD | 248600 |
| Marshall syndrome | MRSHS | Skeletal dysplasia | COL11A1 | 154780 |
| Methylmalonic Acidemia | MMA | Metabolic disorder | MMAA, MMAB, MMACHC, MUT | 251100 |
| Noonan Syndrome (Types 1, 3, 4, 5 ,6) | NS | Multisystemic disorder | KRAS, NRAS, PTPN11, RAF1, SOS1 | 163950/609942/610733/611533/613224 |
| Ornithine Transcarbamylase Deficiency | OTD | Metabolic disorder | OTC | 311250 |
| Phenylketonuria | PKU | Metabolic disorder | PAH | 261600 |
| Pompe Disease/Glycogen Storage Disease II (GSD II) | GSDII | Metabolic disorder | GAA | 232300 |
| Smith-Lemli-Optiz Syndrome | SLOS | Metabolic disorder | DHCR7 | 270400 |
| Usher Syndrome Type 1 | USH1 | Deafness | PCDH15, USH1C, CDH23, MYO7A | 276900/601067 |
| Very Long Chain Acyl-Coenzyme A Dehydrogenase Deficiency | ACADVLD | Metabolic disorder | ACADVL | 201475 |
| Wiskott-Aldrich Syndrome | WAS | Immunodeficiency | WAS | 301000 |
| X-Linked Adrenoleukodystrophy | ALD | Leukodystrophy | ABCD1 | 300100 |
Figure 1.
Disease categories and number of samples tested for each condition. (a) Samples breakdown by disease category. (b) Number of samples studied for each disease.
2.2. Library Building and Sequencing
Ten nanograms of DNA were used in three primer pools, in conjunction with Ion AmpliSeq Library Kit 2.0 (Thermo Fisher, Carlsbad, CA, USA) for 14 cycles. Pooled PCR amplicons were first digested using FuPa reagent (Thermo Fisher), and then ligated with universal adaptors. A maximum of two samples were pooled for emulsion PCR (ePCR) using the Ion OneTouch™ 200 Template Kit (Thermo Fisher). The ePCR template Ion Sphere particles were enriched using the Ion OneTouch ES (Thermo Fisher), following the manufacturer’s instructions. To assess the enrichment status, we used Ion Sphere Quality Control Kit (Thermo Fisher), and the products were measured using Qubit® 2.0 Fluorometer (Thermo Fisher) following manufacturer’s instructions. Template positive Ion Sphere particles were sequenced using the Ion PGM™ 200 Sequencing Kit (Thermo Fisher) on the Ion Personal Genome Machine (PGM) and 318TM semiconductor chip.
2.3. Data Analysis
The bed files containing targeted regions corresponding to the IDP panel were generated based on human genome 19 (hg19) build and used for sequence reads alignment and analysis. Resulting aligned reads (BAM files) were visualized and interrogated with Integrative Genomics Viewer software (http://www.broadinstitute.org/igv). To generate good quality reads, adaptor sequences were trimmed and low quality reads were excluded before using Torrent Suite v4.0 with Variant Calling plugin for the initial analysis (Thermo Fisher, https://github.com/iontorrent/TS). The obtained variants were then annotated using public and in-house databases, as previously described [22]. The Variant calling plugin was set to include variants with a minimum coverage of 20×, following the software developers’ recommendations. On average, each variant calling file (VCF) contained around 800–1200 variants/sample, which was reduced after initial filtration to an average of 20 variants/sample.
2.4. Tertiary Analysis and Variant Validation
Tertiary analysis was carried out, as previously described [22,23], after applying more stringent quality filtering criteria of a minimum coverage of 50× and a quality score of at least 350 for single nucleotide variants (SNVs) and 700 for Indels. Briefly, after passing the initial quality check, the VCFs underwent a step-wise filtering process by two independent trained researchers that were both blind to the phenotype and the original mutation of each sample. The first step of the filtering process involved excluding intronic variants, synonymous variants, or variants present in international and/or local databases (Saudi Human Genome Program database-SHGP) (with MAF > 1%). In the second step, variants’ deleteriousness was assessed using three different prediction software (SIFT [24], PolyPhen-2 [25] and Mutation Taster [26]). Those predicted as “tolerated”, “neutral”, or “benign” were removed. In most cases only homozygous changes were selected, yielding a maximum of three variants/sample. Finally, the causative pathogenic variant was selected from the shortlist of variants, according to the clinical diagnosis. The final results were compared to the initial Sanger sequencing data to estimate the concordance.
3. Results
3.1. Sequencing Quality, Coverage and Overall Panel Performance
Sequencing and read mapping quality of the runs are summarized in Table S1. The overall average number of reads at Q17 was about 3.06 million per run, with an average base yield of about 392 Mbp. The panel generates a total of 10,309 amplicons with an average read length of 135 bp at Q0 and 118 bp at Q20. On average, 95% of these reads were aligned to the target regions (the target set of genes). The overall average depth was 191×. Target base coverage at 1×, 20×, and 50× are 98.36%, 93.29%, and 89.34%, respectively (Table S1). Also, 95% of the amplicons were free of strand bias. The average coverage per gene was ~98 (Table S2). Of the total amplicons, 253 (2.5%) had coverage less than 90%, and 105 (1%), from 78 genes, had suboptimal performance (Table S3). Overall, this shows good sequencing parameters for the runs that were included in the analysis.
3.2. Variant Calling
For the 88 samples that were analyzed, an average of 1058 variants per sample were called and annotated. Recalling that the total length of the target region is 1,509,563, this translates to seven variants per 10,000 bases per sample. For the combined set of samples, the total number of unique variants was 11,118, with a rate of 7.36 variants per 1000 bases.
3.3. Variant Detection Yield
Validation yield was determined by evaluating the concordance between the original mutation and the NGS results for the corresponding case. However, two of the samples were excluded from the analysis due to either amplicon failure or design. Using our analysis pipeline, the IDP successfully detected 93.1% (97.3% for SNVs and 69.2% for Indels) of the mutations in our validation cohort (Table 2). Of note, the detection rate when including the two failed samples is 91%, (96% for SNVs and 64.3% for Indels). In addition, mutations that were masked in Table 2 were revealed to the researchers for the purpose of the analysis. Moreover, missing variants were attributed to low coverage, or the homopolymer effect (Table S4).
Table 2.
List of mutations detected by AmpliSeq Inherited Disease Panel (IDP).
| Original Mutation | IDP Panel | |||||
|---|---|---|---|---|---|---|
| Case ID | Gene | Transcript ID | cDNA | Protein | Original Mutation Found (Y/N) | Genotype Matching (Y/N) |
| 13-0045 | ABCD1 | NM_000033 | c.1581C>A | p.Y527X | Y | Y |
| 13-0051 | ACADVL | NM_000018 | c.65C>A | p.S22X | Y | Y |
| 14-3258 | ACADVL | NM_000018 | c.65C>A | p.S22X | Y | Y |
| 13-0097 | AMT | NM_001164710 | c.533G>A | p.R178H | Y | Y |
| 13-0098 | AMT | NM_001164710 | c.280C>T | p.R94W | Y | Y |
| 13-0059 | ARSA | NM_000487 | c.1055A>G | p.N352S | Y | Y |
| 13-0095 | ARSA | NM_000487 | #Missense | Y | Y | |
| 13-0040 | ASL | NM_001024943 | c.1000C>T | p.Q334X | Y | Y |
| 13-0131 | ASL | NM_001024943 | c.556C>T | p.R186W | Y | Y |
| 13-0132 | ASL | NM_001024943 | c.544C>T | p.R182X | Y | Y |
| 13-0133 | ASPA | NM_000049 | #Frameshift insertion | N | NA | |
| 14-3064 | ASPA | NM_000049 | #Frameshift deletion | N | NA | |
| 13-0020 | ATM | NM_000051 | c.381_381delA | p.V128* | Y | Y |
| 13-0009 | BCKDHA | NM_000709 | c.905A>C | p.D302A | Y | Y |
| 13-0013 | BCKDHA | NM_000709 | c.905A>C | p.D302A | Y | Y |
| 13-0093 | BCKDHA | NM_000709 | c.1270C>T | p.Q424X | Y | Y |
| 13-0094 | BCKDHA | NM_000709 | c.647-1G>C | NA | Y | Y |
| 13-0096 | BCKDHA | NM_000709 | c.347A>G | p.D116G | Y | Y |
| 13-0120 | BCKDHA | NM_000709 | c.808G>A | p.A270T | Y | Y |
| 13-0135 | BCKDHA | NM_000709 | c.659_662delCGTA | p.Y221Qfs*108 | Y | Y |
| 13-0091 | BCKDHB | NM_000056 | c.286_288delGAA | p.E96del | N | NA |
| 13-0092 | BCKDHB | NM_000056 | c.1A>T | p.M1L | N | NA |
| 13-0129 | BCKDHB | NM_000056 | c.1006G>A | p.G336S | Y | Y |
| 13-0053 | BTD | NM_000060 | #Missense | Y | Y | |
| 13-0061 | BTD | NM_000060 | #Frameshift deletion | Y | Y | |
| 13-0062 | BTD | NM_000060 | #Missense | Y | Y | |
| 13-0087 | BTD | NM_000060 | #Missense | Y | Y | |
| 13-0088 | BTD | NM_000060 | #Frameshift deletion | Y | Y | |
| 13-0019 | BTK | NM_000061 | c.763C>T | p.R255X | Y | Y |
| 13-0028 | BTK | NM_000061 | c.982C>T | p.Q328X | Y | Y |
| 13-0043 | CFTR | NM_000492 | c.1418_1418delG | p.G473Efs*54 | N | NA |
| 13-0054 | CFTR | NM_000492 | c.1520_1522delTCT | p.F508del | Y | Y |
| 14-3079 | CFTR | NM_000492 | c.3700A>G | p.I1234V | Y | Y |
| 14-3072 | CFTR | NM_000492 | c.416A>T | p.H139L | Y | Y |
| 14-3071 | CFTR | NM_000492 | c.3700A>G | p.I1234V | Y | Y |
| 13-0090 | CDH23 | NM_001171930 | #Missense | Y | Y | |
| 13-0102 | COL11A1 | NM_080630 | c.2354G>A | p.G785E | Y | Y |
| 13-0011 | DBT | NM_001918 | c.61_61delC | p.R21Afs*12 | Y | Y |
| 13-0046 | DBT | NM_001918 | c.773-2A>G | NA | Y | Y |
| 13-0099 | DBT | NM_001918 | c.1195T>C | p.S399P | Y | Y |
| 13-0104 | DBT | NM_001918 | c.1281+3A>G | NA | Y | Y |
| 13-0107 | DBT | NM_001918 | c.137A>G | p.K46R | Y | Y |
| 13-0127 | DBT | NM_001918 | c.773-2A>G | NA | Y | Y |
| 13-0108 | DHCR7 | NM_001163817 | c.861C>G | p.N287K | Y | Y |
| 13-0100 | FGFR3 | NM_000142 | c.1138G>A | p.G380R | Y | Y |
| 13-0010 | GAA | NM_000152 | #Nonsense | Y | Y | |
| 13-0012 | GAA | NM_000152 | #Nonsense | Y | Y | |
| 13-0055 | GAA | NM_000152 | #Nonsense | Y | Y | |
| 13-0103 | GAA | NM_000152 | #Nonsense | Y | Y | |
| 13-0109 | GAA | NM_000152 | #Missense | Y | Y | |
| 13-0110 | GAA | NM_000152 | #Missense | Y | Y | |
| 13-0124 | GAA | NM_000152 | c.655G>A | p.G219R | Y | Y |
| 13-0128 | GAA | NM_000152 | #Nonsense | Y | Y | |
| 13-0064 | GALT | NM_000155 | #Frameshift deletion | Y | Y | |
| 13-0125 | GALT | NM_001258332 | #Missense | Y | Y | |
| 13-0134 | GALT | NM_001258332 | #Missense | Y | Y | |
| 14-3078 | GALT | NM_001258332 | #Missense | Y | Y | |
| 14-3067 | GALT | NM_001258332 | #Missense | Y | Y | |
| 13-0101 | GBA | NM_000157 | c.152G>T | p.S51I | Y | Y |
| 13-0130 | GBE1 | NM_000158 | #Missense | Y | Y | |
| 13-0116 | GJB2 | NM_004004 | c.299T>C | p.W77R | Y | Y |
| 13-0116 | GJB2 | NM_004004 | c.506G>A | p.C169Y | Y | Y |
| 13-0111 | GLDC | NM_000170 | c.2113G>A | p.V705M | Y | Y |
| 13-0060 | IDS | NM_000202 | # Nonsense | Y | Y | |
| 13-0112 | MMAA | NM_172250 | #Nonsense | Y | Y | |
| 13-0126 | MMAA | NM_172250 | #Missense | Y | Y | |
| 13-0063 | MUT | NM_000255 | #Frameshift deletion | Y | Y | |
| 13-0105 | MUT | NM_000255 | c.329A>G | p.Y110C | Y | Y |
| 13-0121 | MUT | NM_000255 | c.278G>A | p.R93H | Y | Y |
| 14-3081 | MUT | NM_000255 | c.1160C>T | p.T387I | Y | Y |
| 14-3080 | MUT | NM_000255 | c.2200C>T | p.Q734X | Y | Y |
| 14-3070 | MUT | NM_000255 | c.2200C>T | p.Q734X | Y | Y |
| 14-3065 | MUT | NM_000255 | c.1677-1G>C | NA | Y | Y |
| 13-0044 | MYO7A | NM_000260 | c.5880-5882delCTT | p.F1961del | Y | Y |
| 13-0117 | MYO7A | NM_000260 | c.2005C>T | p.R669X | Y | Y |
| 13-0066 | OTC | NM_000531 | # Missense | Y | Y | |
| 13-0057 | PAH | NM_000277 | #Missense | Y | Y | |
| 13-0113 | PAH | NM_000277 | #Nonsense | Y | Y | |
| 13-0114 | PAH | NM_000277 | #Missense | N | NA | |
| 13-0122 | PAH | NM_000277 | #Missense | Y | Y | |
| 13-0123 | PAH | NM_000277 | #Nonsense | Y | Y | |
| 13-0136 | PAH | NM_000277 | #Missense | Y | Y | |
| 14-3073 | PAH | NM_000277 | #Missense | Y | Y | |
| 13-0058 | POLG | NM_001126131 | c.2419C>T | p.R807C | Y | Y |
| 13-0115 | PTPN11 | NM_002834 | c.188A>G | p.Y63C | Y | Y |
| 13-0026 | WAS | NM_000377 | c.91G>A | p.E31K | Y | Y |
| 13-0027 | WAS | NM_000377 | c.100C>T | p.R34X | Y | Y |
Mutations were either homozygous (regular), heterozygous (bolded) or hemizygous (underlined). #Exact mutation cannot be disclosed here being part of another ongoing unpublished study.
4. Discussion
In this study, we report the successful validation of IDP as a comprehensive and sensitive assay for detecting causal mutations in a variety of inherited diseases. Using IDP with PGM, we achieved (~98%) sequence coverage of the targeted regions, with an average depth of 191X. A total of (1058) variants were detected in each sample before filtration. We were able to detect (93.1%) of the originally reported causal mutations. The remaining (6.9%) of causal variants were not detected due to the inadequate coverage of challenging DNA regions with homology or high GC-content (Table S4).
A wide collection of disease-focused or comprehensive gene panels for inherited diseases is commercially available and is being used in clinical laboratories with various NGS platforms [17,27]. Examples of comprehensive inherited disease panels, other than the one assessed here, include Otogenetics and TruSight Inherited Disease Panel (Table S5). The panel from Otogenetics comprises the largest number of genes (~4500), however, the subsequent data analysis and the interpretation could be very challenging and time consuming. Besides, this panel is available only as a service. On the other hand, both the IDP and TruSight are available as predesigned ready-to-use panels. The TruSight covers 552 genes focused only on severe recessive child-onset diseases, whereas the IDP surveys 328 genes that are implicated in > 700 child or adult-onset inherited diseases. Both IDP and TruSight offer fast time-to-results. Delivering accurate results in a short turn-around time is imperative for any diagnostic test, as results may impact the treatment decision or prevent unnecessary interventions.
When it comes to choosing the most appropriate genetic testing strategy, clinicians often face the challenge of deciding which NGS-based approach (targeted vs, Whole Exome Sequencing, WES) to pursue as first-tier genetic screening. Exome sequencing utilization in clinical settings (clinical exome sequencing) allows for the unbiased evaluation of roughly all 21,000 genes. This is crucial in situations of diagnostic uncertainty, in diseases with significant genetic and phenotypic heterogeneity or even to minimize the effect of diagnostic error as testing is not restricted to genes implicated in a certain disorder. One important advantage of exome sequencing is the capacity to identify alterations in both well characterized and novel genes, allowing for data re-analysis in the light of new gene-disease associations. Another advantage is that it can improve the management of patients by alerting physicians to unanticipated comorbidities that may alter the course of treatment or impact prognosis. On the other hand, technical limitations of this approach include incomplete gene coverage (especially in problematic regions), variant validation, and interpretation [28,29]. Another limitation to this approach is that it generates a long list of variants most of which are variants of uncertain significance (VUS) that are usually overlooked or are filtered out. Some of these variants could be clinically relevant (may represent actual mutations), however, unfeasible to validate. An additional ethical issue is, the disclosure of incidental/secondary findings, which are not uncommon [30,31].
Targeted gene panels have the potential to overcome some of the current exome sequencing limitations. They offer a superior coverage of up to 100% when coupled with Sanger sequencing [32]. Unlike exome sequencing, gene panel analysis generates substantially less variants, thus making the validation and interpretation much more efficient. This can minimize the chances of missing VUS with potential clinical relevance. More importantly, because the analysis is restricted to genes that are related to the primary clinical condition, the issue of incidental findings is reduced to a minimal concern in targeted-panels [27,33]. However, the major limitation of targeted-gene panels is that a panel could become obsolete if its content is not constantly updated to catch up with the fast pace of new genes discovery.
With regard to the choice of NGS-based genetic testing, there seems to be a general consensus on using targeted-panels as first-tier genetic testing, particularly for diseases with distinct phenotypes and a good knowledge of the underlying genes. On the other hand, exome or genome sequencing are recommended to be reserved for those cases in which molecular diagnosis could not be established via targeted-panel testing [34,35]. Recently, a targeted sequencing approach using 13 different gene panels covering the majority of OMIM reported genes, demonstrated a high degree of clinical sensitivity and specify providing evidence for the advantages of utilizing targeted-panels over exome sequencing as first-tier genetic testing approach [22].
Due to the random selection of samples, the performance of the IDP could not be evaluated for all disease categories and certainly not all phenotypes. Our samples represented various disease categories with the majority being classified as metabolic disorders (Figure 1a). However, it is important to note that half of the Mendelian conditions that are prevalent in Saudi Arabia (Thalassemia, lysosomal storage disorders, hearing loss, organic acidemias, and retinal dystrophies) are covered by the IDP [36].
The assay in its current format is not intended to identify copy number variants, however, it is possible to incorporate algorithms for assessing this type of alteration into the bioinformatics pipeline after preforming the necessary validation [37]. Sensitivity validation results for this panel met the high degree that is required for research use. However, additional important quality measures, such as (run-to-run or laboratory-to-laboratory) reproducibility, should be evaluated before clinical implementation of the assay [38].
5. Conclusions
This study demonstrated the suitability of the IDP as a rapid and comprehensive approach for screening a large number of genes that are responsible for over 700 different inherited diseases. It is worth mentioning that reported detection yield of gene panels for inherited diseases varies widely (24–95%), placing the rate achieved in our study at the upper range [22,23,39,40,41,42]
Inherited diseases are expected to be frequently encountered in consanguineous populations. For instance, in Saudi Arabia, inherited conditions, such as Thalassemia, lysosomal storage disorders, hearing loss, organic acidemias, and retinal dystrophies are common [36,43]. In response to that, the Ministry of Health established two national molecular screening programs; newborn and premarital [44,45]. The incorporation of a comprehensive gene panel (such as the IDP and other available panels [22]) as a second-tier testing approach into any ongoing public screening programs would enhance their performance by improving diagnostic accuracy and expanding the range of conditions for which screening is available.
Acknowledgments
The IDP gene panel experiments and analyses were all carried out as part of the Saudi Human Genome Program (SHGP), both at King Abdulaziz City for Science and Technology (KACST) and King Faisal Specialist Hospital & Research Center (KFSHRC).
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/5/267/s1, Table S1: IDP panel sequencing quality metrics; Table S2: Coverage per gene; Table S3: Amplicons with low overall performance; Table S4: List and details of undetected variants; Table S5: Examples of commercial NGS-panels.
Author Contributions
M.A.-S., M.Al., H.A.Z. and Z.A.-H. contributed with patients recruitment, diagnosis and clinical assessment. F.I. and K.R. contributed with the identification of the original mutations (positive controls). N.A.A.T. supervised the IDP gene-panel sequencing. A.E.M., R.K., and A.A.I. carried out the IDP gene panel sequencing. A.E.M., A.I.T., and M.Ab. analyzed the gene-panel sequencing data. M.A. and T.F. performed the bioinformatics data analysis. N.A.A.T., M.Ab. and B.B. contributed to manuscript writing. B.A.-M. drafted the manuscript. N.A.A.Tassan critically reviewed the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
None of the authors have any competing interests in the manuscript.
References
- 1.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith L.M., Sanders J.Z., Kaiser R.J., Hughes P., Dodd C., Connell C.R., Heiner C., Kent S.B., Hood L.E. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 3.Beck J.S., Kwitek A.E., Cogen P.H., Metzger A.K., Duyk G.M., Sheffield V.C. A denaturing gradient gel electrophoresis assay for sensitive detection of p53 mutations. Hum. Genet. 1993;91:25–30. doi: 10.1007/BF00230217. [DOI] [PubMed] [Google Scholar]
- 4.Ogino S., Kawasaki T., Brahmandam M., Yan L., Cantor M., Namgyal C., Mino-Kenudson M., Lauwers G.Y., Loda M., Fuchs C.S. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J. Mol. Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumazawa R., Tomiyama H., Li Y., Imamichi Y., Funayama M., Yoshino H., Yokochi F., Fukusako T., Takehisa Y., Kashihara K., et al. Mutation analysis of the PINK1 gene in 391 patients with parkinson disease. Arch. Neurol. 2008;65:802–808. doi: 10.1001/archneur.65.6.802. [DOI] [PubMed] [Google Scholar]
- 6.Huang X.C., Quesada M.A., Mathies R.A. DNA sequencing using capillary array electrophoresis. Anal. Chem. 1992;64:2149–2154. doi: 10.1021/ac00042a021. [DOI] [PubMed] [Google Scholar]
- 7.Kambara H., Takahashi S. Multiple-sheathflow capillary array DNA analyser. Nature. 1993;361:565–566. doi: 10.1038/361565a0. [DOI] [PubMed] [Google Scholar]
- 8.Tucker T., Marra M., Friedman J.M. Massively parallel sequencing: The next big thing in genetic medicine. Am. J. Hum. Genet. 2009;85:142–154. doi: 10.1016/j.ajhg.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su Z., Ning B., Fang H., Hong H., Perkins R., Tong W., Shi L. Next-generation sequencing and its applications in molecular diagnostics. Expert. Rev. Mol. Diagn. 2011;11:333–343. doi: 10.1586/erm.11.3. [DOI] [PubMed] [Google Scholar]
- 10.Nemeth A.H., Kwasniewska A.C., Lise S., Parolin Schnekenberg R., Becker E.B., Bera K.D., Shanks M.E., Gregory L., Buck D., Zameel Cader M., et al. Next generation sequencing for molecular diagnosis of neurological disorders using ataxias as a model. Brain. 2013;136:3106–3118. doi: 10.1093/brain/awt236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sikkema-Raddatz B., Johansson L.F., de Boer E.N., Almomani R., Boven L.G., van den Berg M.P., van Spaendonck-Zwarts K.Y., van Tintelen J.P., Sijmons R.H., Jongbloed J.D., et al. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum. Mutat. 2013;34:1035–1042. doi: 10.1002/humu.22332. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y., Wang Z., Wang Z., Chen D., Chai Y., Pang X., Sun L., Wang X., Yang T., Wu H. Targeted next-generation sequencing in Uyghur families with non-syndromic sensorineural hearing loss. PLoS ONE. 2015;10:e0127879. doi: 10.1371/journal.pone.0127879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nijman I.J., van Montfrans J.M., Hoogstraat M., Boes M.L., van de Corput L., Renner E.D., van Zon P., van Lieshout S., Elferink M.G., van der Burg M., et al. Targeted next-generation sequencing: A novel diagnostic tool for primary immunodeficiencies. J. Allergy Clin. Immunol. 2014;133:529–534. doi: 10.1016/j.jaci.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 14.Besnard T., Garcia-Garcia G., Baux D., Vache C., Faugere V., Larrieu L., Leonard S., Millan J.M., Malcolm S., Claustres M., et al. Experience of targeted usher exome sequencing as a clinical test. Mol. Genet. Genom. Med. 2014;2:30–43. doi: 10.1002/mgg3.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemke J.R., Riesch E., Scheurenbrand T., Schubach M., Wilhelm C., Steiner I., Hansen J., Courage C., Gallati S., Burki S., et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. 2012;53:1387–1398. doi: 10.1111/j.1528-1167.2012.03516.x. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-San Jose P., Corton M., Blanco-Kelly F., Avila-Fernandez A., Lopez-Martinez M.A., Sanchez-Navarro I., Sanchez-Alcudia R., Perez-Carro R., Zurita O., Sanchez-Bolivar N., et al. Targeted next-generation sequencing improves the diagnosis of autosomal dominant retinitis pigmentosa in Spanish patients. Invest. Ophthalmol. Vis. Sci. 2015;56:2173–2182. doi: 10.1167/iovs.14-16178. [DOI] [PubMed] [Google Scholar]
- 17.Yohe S., Hauge A., Bunjer K., Kemmer T., Bower M., Schomaker M., Onsongo G., Wilson J., Erdmann J., Zhou Y., et al. Clinical validation of targeted next-generation sequencing for inherited disorders. Arch. Pathol. Lab. Med. 2015;139:204–210. doi: 10.5858/arpa.2013-0625-OA. [DOI] [PubMed] [Google Scholar]
- 18.Vasli N., Bohm J., Le Gras S., Muller J., Pizot C., Jost B., Echaniz-Laguna A., Laugel V., Tranchant C., Bernard R., et al. Next generation sequencing for molecular diagnosis of neuromuscular diseases. Acta Neuropathol. 2012;124:273–283. doi: 10.1007/s00401-012-0982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Sullivan J., Mullaney B.G., Bhaskar S.S., Dickerson J.E., Hall G., O'Grady A., Webster A., Ramsden S.C., Black G.C. A paradigm shift in the delivery of services for diagnosis of inherited retinal disease. J. Med. Genet. 2012;49:322–326. doi: 10.1136/jmedgenet-2012-100847. [DOI] [PubMed] [Google Scholar]
- 20.Liu S., Wang H., Zhang L., Tang C., Jones L., Ye H., Ban L., Wang A., Liu Z., Lou F., et al. Rapid detection of genetic mutations in individual breast cancer patients by next-generation DNA sequencing. Hum. Genom. 2015;9:2. doi: 10.1186/s40246-015-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glenn T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 22.Saudi Mendeliome G. Comprehensive gene panels provide advantages over clinical exome sequencing for mendelian diseases. Genome Biol. 2015;16:134. doi: 10.1186/s13059-015-0693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Mousa H., Abouelhoda M., Monies D.M., Al-Tassan N., Al-Ghonaium A., Al-Saud B., Al-Dhekri H., Arnaout R., Al-Muhsen S., Ades N., et al. Unbiased targeted next-generation sequencing molecular approach for primary immunodeficiency diseases. J. Allergy Clin. Immunol. 2016;137:1780–1787. doi: 10.1016/j.jaci.2015.12.1310. [DOI] [PubMed] [Google Scholar]
- 24.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. Sift web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using polyphen-2. Curr. Protoc. Hum. Genet. 2013 doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz J.M., Rodelsperger C., Schuelke M., Seelow D. Mutationtaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 27.Rehm H.L. Disease-targeted sequencing: A cornerstone in the clinic. Nat. Rev. Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ku C.S., Cooper D.N., Polychronakos C., Naidoo N., Wu M., Soong R. Exome sequencing: Dual role as a discovery and diagnostic tool. Ann. Neurol. 2012;71:5–14. doi: 10.1002/ana.22647. [DOI] [PubMed] [Google Scholar]
- 29.Xue Y., Ankala A., Wilcox W.R., Hegde M.R. Solving the molecular diagnostic testing conundrum for mendelian disorders in the era of next-generation sequencing: Single-gene, gene panel, or exome/genome sequencing. Genet. Med. 2015;17:444–451. doi: 10.1038/gim.2014.122. [DOI] [PubMed] [Google Scholar]
- 30.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M., et al. Clinical exome sequencing for genetic identification of rare mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z., et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y., Ruivenkamp C.A., Hoffer M.J., Vrijenhoek T., Kriek M., van Asperen C.J., den Dunnen J.T., Santen G.W. Next-generation diagnostics: Gene panel, exome, or whole genome? Hum. Mutat. 2015;36:648–655. doi: 10.1002/humu.22783. [DOI] [PubMed] [Google Scholar]
- 33.Blackburn H.L., Schroeder B., Turner C., Shriver C.D., Ellsworth D.L., Ellsworth R.E. Management of incidental findings in the era of next-generation sequencing. Curr. Genom. 2015;16:159–174. doi: 10.2174/1389202916666150317232930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ACMG Board of Directors Points to consider in the clinical application of genomic sequencing. Genet. Med. 2012;14:759–761. doi: 10.1038/gim.2012.74. [DOI] [PubMed] [Google Scholar]
- 35.LaDuca H., Farwell K.D., Vuong H., Lu H.M., Mu W., Shahmirzadi L., Tang S., Chen J., Bhide S., Chao E.C. Exome sequencing covers > 98% of mutations identified on targeted next generation sequencing panels. PLoS ONE. 2017;12:e0170843. doi: 10.1371/journal.pone.0170843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alkuraya F.S. Genetics and genomic medicine in saudi arabia. Mol. Genet. Genom. Med. 2014;2:369–378. doi: 10.1002/mgg3.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grasso C., Butler T., Rhodes K., Quist M., Neff T.L., Moore S., Tomlins S.A., Reinig E., Beadling C., Andersen M., et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J. Mol. Diagn. 2015;17:53–63. doi: 10.1016/j.jmoldx.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santani A., Murrell J., Funke B., Yu Z., Hegde M., Mao R., Ferreira-Gonzalez A., Voelkerding K.V., Weck K.E. Development and validation of targeted next-generation sequencing panels for detection of germline variants in inherited diseases. Arch. Pathol. Lab. Med. 2017;141:787–797. doi: 10.5858/arpa.2016-0517-RA. [DOI] [PubMed] [Google Scholar]
- 39.Broendberg A.K., Christiansen M.K., Nielsen J.C., Pedersen L.N., Jensen H.K. Targeted next generation sequencing in a young population with suspected inherited malignant cardiac arrhythmias. Eur. J. Hum. Genet. 2018;26:303–313. doi: 10.1038/s41431-017-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrigan M., Duignan E., Malone C.P., Stephenson K., Saad T., McDermott C., Green A., Keegan D., Humphries P., Kenna P.F., et al. Panel-based population next-generation sequencing for inherited retinal degenerations. Sci. Rep. 2016;6:33248. doi: 10.1038/srep33248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mori T., Hosomichi K., Chiga M., Mandai S., Nakaoka H., Sohara E., Okado T., Rai T., Sasaki S., Inoue I., et al. Comprehensive genetic testing approach for major inherited kidney diseases, using next-generation sequencing with a custom panel. Clin. Exp. Nephrol. 2017;21:63–75. doi: 10.1007/s10157-016-1252-1. [DOI] [PubMed] [Google Scholar]
- 42.Pua C.J., Bhalshankar J., Miao K., Walsh R., John S., Lim S.Q., Chow K., Buchan R., Soh B.Y., Lio P.M., et al. Development of a comprehensive sequencing assay for inherited cardiac condition genes. J. Cardiovasc. Transl. Res. 2016;9:3–11. doi: 10.1007/s12265-016-9673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alfadhel M., Benmeakel M., Hossain M.A., Al Mutairi F., Al Othaim A., Alfares A.A., Al Balwi M., Alzaben A., Eyaid W. Thirteen year retrospective review of the spectrum of inborn errors of metabolism presenting in a tertiary center in Saudi Arabia. Orphanet J. Rare Dis. 2016;11:126. doi: 10.1186/s13023-016-0510-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afifi A.M., Abdul-Jabbar M.A. Saudi newborn screening. A national public health program: Needs, costs, and challenges. Saudi. Med. J. 2007;28:1167–1170. [PubMed] [Google Scholar]
- 45.Memish Z.A., Saeedi M.Y. Six-year outcome of the national premarital screening and genetic counseling program for sickle cell disease and β-thalassemia in saudi arabia. Ann. Saudi. Med. 2011;31:229–235. doi: 10.4103/0256-4947.81527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.