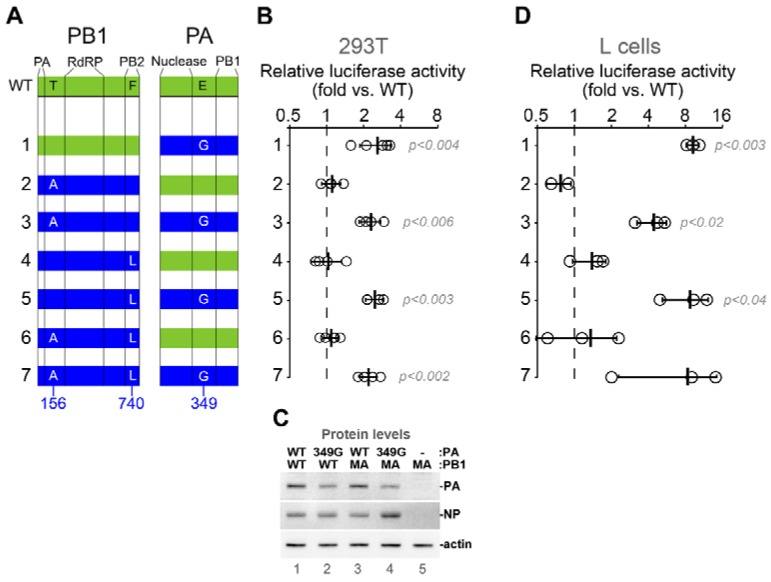
Figure 4.
PA E349G substitution enhances viral RNA polymerase activity. (A) Schematic representation of PB1 and PA proteins showing approximate boundaries of major domains in the primary sequence (vertical lines) and positions of amino acids mutated in this study. Green and blue shading indicates wild-type and mutant proteins, respectively. (B) Relative luciferase activity was measured in the replicon assay using wild-type CA/07-derived PB2 and NP constructs in combination with PB1 and PA constructs that correspond to the numbered combinations depicted in (A). Open circles indicate values for each independent replicate normalized to the wild-type replicon values obtained in parallel (dashed line). p-values are calculated using the paired Student’s t test (n ≥ 4). (C) Western blot analysis of the whole cell lysates of 293T cells transfected for the select replicon assays shown in (B). PB1 and PA variants transfected as indicated. (D) The assay described in (B) was performed in mouse L cells using a firefly luciferase reporter driven by mouse POL1 promoter. p-values are calculated using the paired Student’s t test (n = 3). RdRP: RNA-dependent RNA polymerase. WT: wild type.