Abstract
miR-146a inhibits inflammatory responses in human keratinocytes and in different mouse models of skin inflammation. Little is known about the role of miR-146b in the skin. In the present study, we confirmed the increased expression of miR-146a and miR-146b (miR-146a/b) in lesional skin of psoriasis patients. The expression of miR-146a was about 2-fold higher than that of miR-146b in healthy human skin and it was more strongly induced by stimulation of pro-inflammatory cytokines in keratinocytes and fibroblasts. miR-146a/b target genes regulating inflammatory responses or proliferation were altered in the skin of psoriasis patients, among which FERMT1 was verified as direct target of miR-146a. In silico analysis of genome-wide data from >4,000 psoriasis cases and >8,000 controls confirmed a moderate association between psoriasis and genetic variants in miR-146a gene. Transfection of miR-146a/b suppressed and inhibition enhanced keratinocyte proliferation and the expression of psoriasis-related target genes. Enhanced expression of miR-146a/b-influenced genes was detected in cultured keratinocytes from miR-146a−/− and skin fibroblasts from miR-146a−/− and miR-146b−/− mice stimulated with psoriasis-associated cytokines as compared to wild type mice. Our results indicate that besides miR-146a, miR-146b is expressed and might be capable of modulation of inflammatory responses and keratinocyte proliferation in psoriatic skin.
INTRODUCTION
Psoriasis is a common inflammatory skin disease characterized by red and scaling skin plaques. Patients with psoriasis are at higher risk of multiple comorbidities, including diabetes, cardiovascular diseases, psoriatic arthritis, Crohn’s disease, lymphomas and depression, and as a result, have a shorter lifespan (Armstrong et al., 2015; Guttman-Yassky et al., 2011a). Th1, Th17 and Th22 cells and cytokines they produce have been suggested to be responsible for the development of the inflammatory environment and characteristic hyperproliferation of keratinocytes in psoriasis (Guttman-Yassky et al., 2011b; Krueger, 2012). Recent clinical studies, in which psoriasis patients were successfully treated with different IL-17 antagonists, point out the pivotal role of IL-17 in the pathogenesis of psoriasis (Griffiths et al., 2015; Langley et al., 2014; Papp et al., 2012a; Papp et al., 2012b). Genetic studies demonstrate that psoriasis is associated with markers near genes involved in Th17 differentiation and responsiveness, for example, IL23R, RUNX3 and STAT3 (Kim et al., 2015; Tsoi et al., 2012) as well as genes from the NF-κB signaling pathway regulating innate immune responses, such as CARD14, CARM1 and NFKBIZ (Baurecht et al., 2015; Tsoi et al., 2015; Tsoi et al., 2012).
microRNAs (miRNAs) are gene expression regulators that trigger target mRNA degradation and inhibit translation and thereby regulate most of the biological processes in higher eukaryotes (Boldin et al., 2012; Izaurralde, 2015). The miR-146 family consists of two members, miR-146a and miR-146b (miR-146a/b), which are encoded by independent genes located on human chromosomes 5 and 10, respectively. Mature miR-146a/b differ from each other in only two nucleotides, which suggests that they target very similar sets of transcripts (Taganov et al., 2006). Both miR-146a/b have been shown to be upregulated in the skin of psoriasis patients (Lovendorf et al., 2015; Sonkoly et al., 2007). Numerous studies demonstrate that miR-146a has an anti-inflammatory function. The expression of miR-146a is upregulated by NF-κB and it directly inhibits several factors from the NF-κB pathway, including IL-1 receptor-associated kinase 1 (IRAK1) (Taganov et al., 2006) and caspase recruitment domain-containing protein 10 (CARD10) (Crone et al., 2012; Rebane et al., 2014). miR-146a−/− mice develop autoimmunity caused by the incapacity of T regulatory (Treg) cells to suppress Th1 type responses (Boldin et al., 2011; Lu et al., 2010). In human primary keratinocytes, miR-146a inhibits the expression of many pro-inflammatory factors, including CCL5 and IL-8 (Meisgen et al., 2014; Rebane et al., 2014). miR-146a has been shown to act as anti-inflammatory miRNA in mouse models of AD (Rebane et al., 2014), irritant contact dermatitis (Urgard et al., 2016) and psoriasis (Srivastava et al., 2016). The single nucleotide polymorphism (SNP) rs2910164 located in miR-146a precursor has been suggested to be associated with psoriasis in Han Chinese population (Zhang et al., 2014) and among Caucasians (Srivastava et al., 2016). The expression of miR-146b has been shown to be activated by the signal transducer and activator of transcription (STAT)3 (Curtale et al., 2013). The functions of miR-146b are less described, however, considering high similarity, miR-146a/b seem to have important and complementary role in proper functioning of immune system (Ahn et al., 2013; Curtale et al., 2013).
In the current study, we analyzed the expression of miR-146a/b and target genes in the skin of patients with plaque psoriasis, investigated the functions and the expression regulation of miR-146a/b in keratinocytes and fibroblasts, and explored the association of single nucleotide polymorphisms (SNPs) in miR-146a/b and selected target genes with psoriasis. Our results indicate that besides miR-146a, miR-146b may play a role in the maintenance of skin homeostasis and the regulation of inflammatory responses and keratinocyte proliferation in psoriatic skin.
RESULTS
The expression of miR-146a/b in keratinocytes and in the skin of psoriasis patients
Previous miRNA profiling studies have suggested increased expression of miR-146a/b in lesional skin of psoriasis patients (Lovendorf et al., 2015; Sonkoly et al., 2007). First, we confirmed by RT-qPCR that miR-146a/b are more highly expressed in lesional as compared to non-lesional skin of psoriasis patients and the skin of control individuals (Figure 1a, Supplementary Table S1), however, no correlation with psoriasis severity was found (see Supplementary Figure S1 online). The expression levels of miR-146a/b were higher in the skin as compared to cultured primary keratinocytes and fibroblasts, but lower than in peripheral blood mononuclear cells (PBMCs) (Figure 1b). The expression of miR-146a was about 2.4-fold higher than miR-146b in control skin samples (Figure 1c). In situ hybridization (ISH) analysis showed increased expression of miR-146a in the skin from psoriasis patients with the highest staining in the stratum spinosum area of the epidermis in the psoriatic lesional skin. The miR-146b signal was weaker than that of miR-146a and was detected in both the epidermis and the dermis of lesional skin of psoriasis patients (Figure 1d). miR-146a was induced by IL-1β, TNF-α and IL-17A in human proliferating keratinocytes, in 3D reconstituted epidermis and in fibroblasts (Figure 1e–g). miR-146b expression was increased in response to IFN-γ in keratinocytes and in reconstituted epidermis (Figure 1e) and in response to IL-22 only in reconstituted epidermis (Figure 1f) where the expression of IL-22 receptor components IL10RB and IL22RA1 (Akdis et al., 2016) were increased (Supplementary Figure S2a). In fibroblasts, the expression of miR-146b was moderately enhanced in response to IL-1β (Figure 1g).
Figure 1. The expression of miR-146a/b in the skin, keratinocytes and fibroblasts.
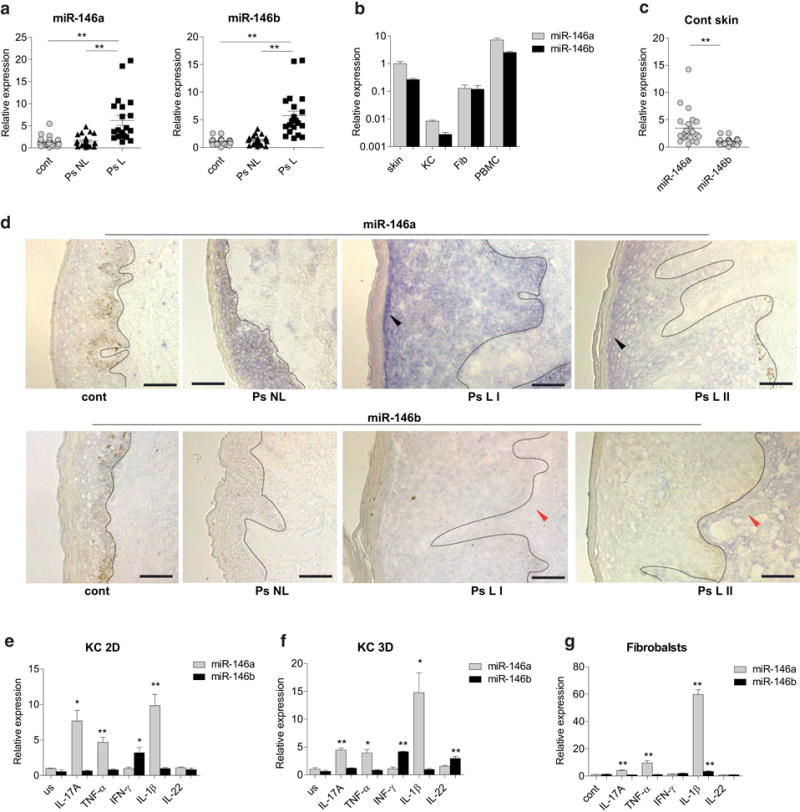
(a–c) Relative expression of miR-146a/b in lesional (L) and non-lesional (NL) skin from psoriasis (Ps) patients (a), human primary keratinocytes (KC), fibroblasts (Fib) or peripheral blood mononuclear cells (PBMCs)(b) and control skin samples (c) was measured by RT-qPCR and is shown compared to the normal skin (cont). (b) (n=5). (d) ISH images of cont, Ps NL and two Ps L skin biopsies are shown. Blue color shows the expression of miR-146a/b, bar=75 μm. The basement line between the epidermis and dermis is indicated with the dotted line. Stronger signals of miR-146a in the stratum spinosum and miR-146b in the dermis are indicated with black and red arrows, respectively. (e-g) Proliferating keratinocytes (KC 2D), reconstituted epidermis (KC 3D) or fibroblasts were stimulated with for 48 h or left unstimulated (us) and subjected to RT-qPCR analysis. Data is shown compared to the mean expression of miR-146a in us cells (=1), (n=4). (a-c, e-g) Student’s t-test, * P < 0.05, ** P < 0.01.
Genes regulated by miR-146a are differentially expressed in the skin of psoriasis patients
We next tested whether the expression of miR-146a-influenced inflammation-related genes is changed in the skin of psoriasis patients. Figure 2a demonstrates that IRAK1, CCL5 and IL-8 were upregulated, and CARD10 was downregulated in lesional skin of psoriasis patients. To identify other psoriasis-related processes influenced by miR-146a, we performed pathway analysis of published array data (Rebane et al., 2014) and found that the genes involved in the regulation of cell proliferation are enriched among the set of 102 genes downregulated by miR-146a in unstimulated keratinocytes. Most of these genes were also suppressed by miR-146a in keratinocytes stimulated with TNF-α and IFN-γ (Figure 2b, Supplementary Tables S2 online). Using Targetscan-based search approach, we identified that five of these genes were putative or previously published direct targets of miR-146a (Figure 2b and Supplementary Table S3 online). Among these, a novel proliferation-associated putative direct target fermitin family member 1 (FERMT1, also known as kindlin-1) was found to be increased in non-lesional and lesional skin of psoriasis patients. Earlier verified miR-146a direct target, NUMB (Hung et al., 2013) was enhanced in lesional skin of psoriasis patients (Figure 2c). The relative levels of miR-146a/b and the target genes revealed no significant correlation in psoriatic lesional skin (Supplementary Figure S3).
Figure 2. The expression of miR-146a-influenced genes is changed in psoriasis patients.
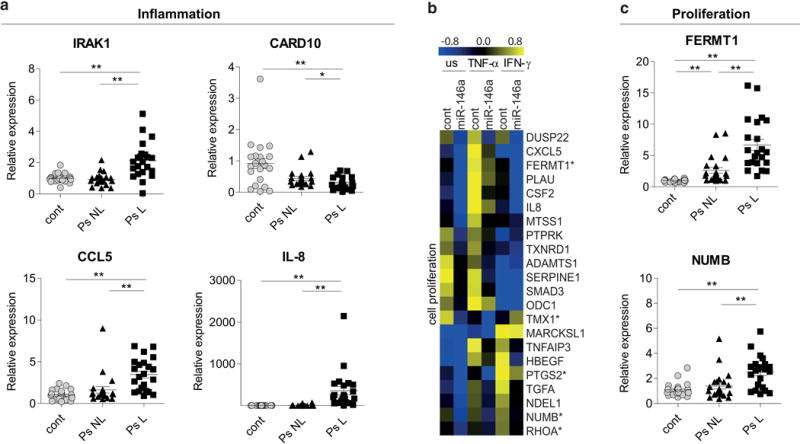
(a and c) The relative mRNA expression in lesional (L) and non-lesional (NL) skin from psoriasis (Ps) patients is shown compared to the mean of controls (cont). Data represent the mean ± SEM. Student’s t-test, * P < 0.05, ** P < 0.01. (b) Keratinocytes were transfected either with control (cont) or pre-miR-146a (miR-146a) for 24 h and then stimulated with IFN-γ or TNF-α for 48 h or left unstimulated (us). Log2 values of expression signals are mean-centered for each gene separately. The proposed direct targets are marked with an asterisk.
miR-146a inhibits the proliferation of human primary keratinocytes
miR-146a has been shown to suppress viability of keratinocytes in a cell counting assay, in which the impact on proliferation and cell death cannot be discriminated (Zhang et al., 2014). To more precisely describe miR-146a effect, we transfected keratinocytes with miR-146a mimics and performed both the proliferation and cell death assays. Figure 3a demonstrates that the overexpression of miR-146a strongly reduced the proliferation of unstimulated keratinocytes and the inhibition of miR-146a increased keratinocyte proliferation in used conditions (Figure 3b). In the subsequent cell death assay, we did not detect difference in unstimulated cells transfected with miR-146a or the control. Pro-inflammatory cytokines TNF-α and IFN-γ caused activation-induced cell death as then about 11% of the control transfected cells were detected Annexin-V positive, which was reduced when miR-146a was transfected (Figure 3c and Supplementary Figure S4). Thus, after 96 h of transfection, about 90% of unstimulated keratinocytes were viable, while in the presence of TNF-α or IFN-γ, only about 80% of the cells were viable. The transfection of miR-146a led to the increase in viability as then 84.6 ± 2.1% and 87.8 ± 0.9% of the cells were viable in TNF-α or IFN-γ-treated cells, respectively (Figure 3d).
Figure 3. miR-146a inhibits proliferation and activation-induced apoptosis of human primary keratinocytes.
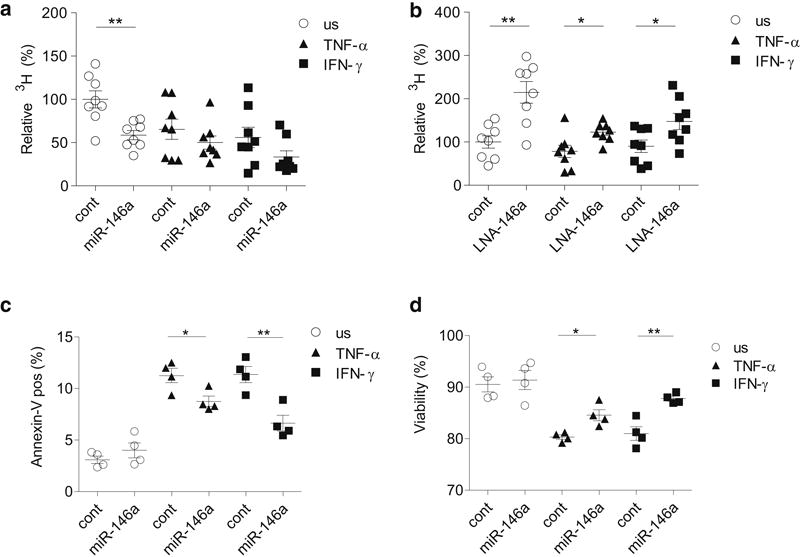
Keratinocytes were transfected either with control (cont) or pre-miR-146a (miR-146a) (a, c) or with control LNA (cont) or miR-146a inhibitor (LNA-146a) (b) for 24 h and then stimulated as indicated for 24 h (a, b) or 72 h (c, d) or left unstimulated (us). 3H thymidine was added for another 14h (a and b). The percentage of Annexin-V positive cells (c) and the viability (d), Annexin-V and 7AAD negative cells) of unstimulated (us) or stimulated primary KCs is presented. Data represent the mean ± SEM, Student’s t-test, * P < 0.05, ** P < 0.01.
FERMT1 is a direct target of miR-146a
Among genes suppressed by miR-146a, FERMT1 is known as a positive regulator of keratinocyte proliferation needed for formation of normal skin structure (Duperret et al., 2014; Herz et al., 2006). Using Targetscan, we found two miR-146a binding sites located within the 3′ untranslated region (3′ UTR) of FERMT1 mRNA (Figure 4a), which we inserted together with flanking areas into the luciferase reporter vector and performed luciferase assays. As presented in Figure 4b, the transfection of miR-146a resulted in suppression of the luciferase activity of the reporters containing FERMT1 3′ UTR regions with miR-146a binding sites but not with the mutant binding sites. In line with the array results (Figure 2b), the transfection of miR-146a suppressed the expression of endogenous FERMT1 at mRNA and in less extent at protein level when stimulated with TNF-α, IFN-γ or IL-17A (Figure 4c and d). siRNA inhibition for FERMT1 but not CARD10 and IRAK1 (Supplementary Figure S5) led to the suppression of viability of keratinocytes (Figure 4e). Immunofluorescence (IF) analysis confirmed increased expression of the FERMT1 protein in lesional skin of psoriasis patients with highest expression in the basal layer of the epidermis (Figure 4f), while the keratinocyte differentiation marker Keratin 10 (KRT10) was more highly expressed in stratum spinosum. These results demonstrate that FERMT1 is a direct target of miR-146a and has increased expression in psoriatic skin.
Figure 4. FERMT1 is a novel miR-146a target.
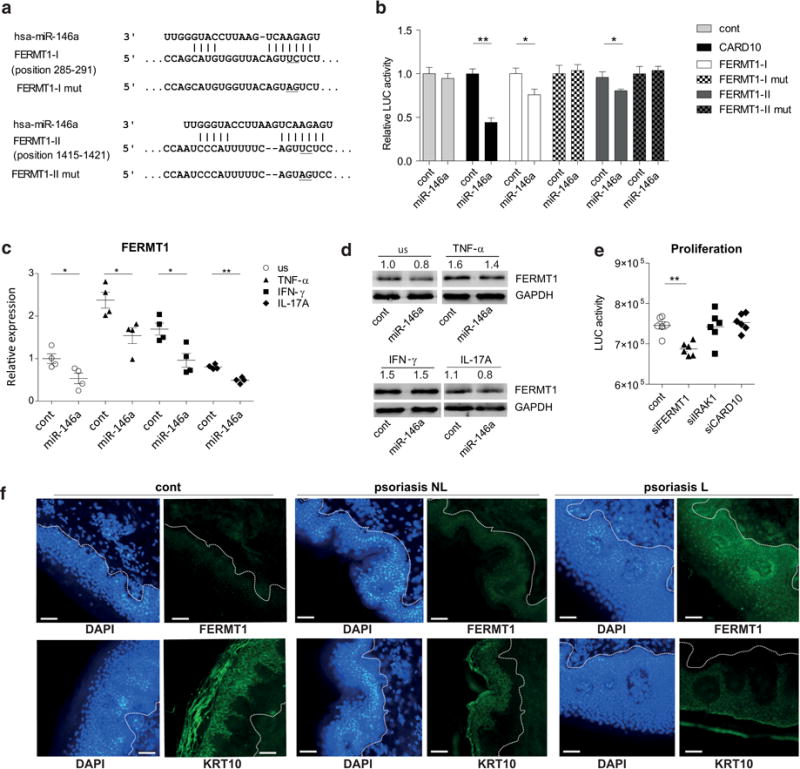
(a) miR-146a and the mutated binding sites (underlined). Positions indicate the distance from the beginning of FERMT1 3′UTR. (b) The relative firefly luciferase (LUC) activity is normalized to the value of control miRNA and empty vector (cont; =1). Data represent the mean ± SEM (n=8). (c and d) Keratinocytes were transfected with cont or pre-miR-146a (miR-146a) for 24 h and then stimulated as indicated for 48 h or left unstimulated (us). (e) The proliferation was measured with keratinocytes transfected with cont or specific siRNAs for 24 h. (b, c and e) Data represent the mean ± SEM. Student’s t-test, * P < 0.05, ** P < 0.01. (f) Immunofluorescence and DAPI staining of lesional (L) and non-lesional (NL) skin of psoriasis (Ps) patients and controls (cont). Bars correspond to 50 μm.
miR-146b inhibits psoriasis-associated miR-146a target genes and proliferation of primary keratinocytes
Mature miR-146b differs from miR-146a in two nucleotides that are located outside the seed area, which suggests that miR-146a/b target very similar set of genes. Thus, we next transfected keratinocytes with miR-146a or miR-146b mimics, stimulated the cells with IFN-γ or TNF-α or left unstimulated and measured relative mRNA expression of psoriasis associated miR-146a target genes. In most of the conditions, the transfection of miR-146b led to the significant suppression of CARD10, IRAK1, CCL5, IL-8, FERMT1 and NUMB in a similar extent as the transfection with miR-146a (Figure 5). A comparable suppression of the target genes expressed by miR-146a/b was also observed in the fibroblasts (Supplementary Figure S6). Similarly to miR-146a, the transfection of miR-146b led to the suppression of proliferation of keratinocytes (Figure 5b).
Figure 5. miR-146b inhibits psoriasis-related miR-146a target genes and proliferation of keratinocytes.
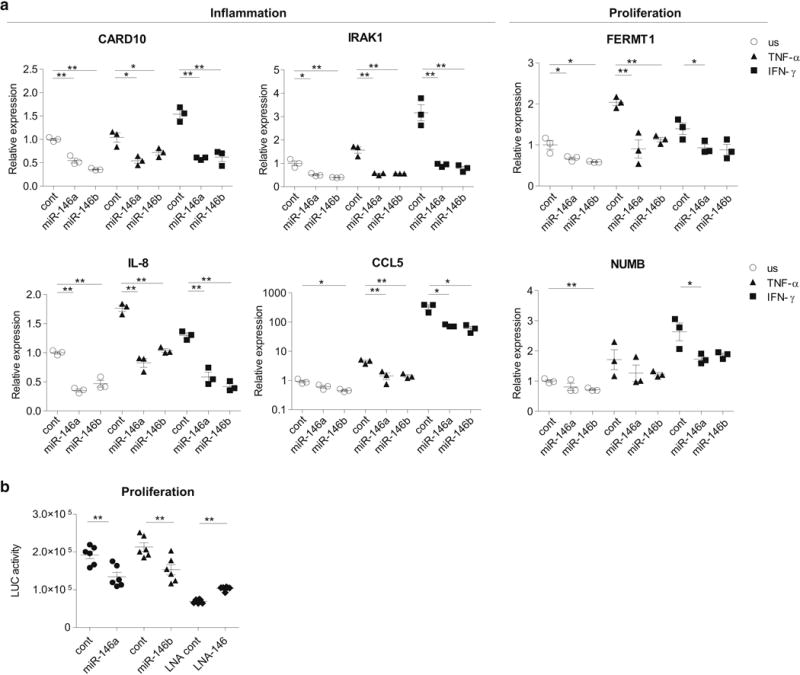
Keratinocytes were transfected either with control (cont) or miR-146a/b mimics or with the control LNA inhibitor (cont) or LNA inhibitor for miR-146a/b (LNA-146). Where indicated, the cells were stimulated with IFN-γ or TNF-α for 48 h or left unstimulated (us). Data represent mean ± SEM. Student’s t-test, * P < 0.05, ** P < 0.01. (a) Relative expression compared to unstimulated cells (=1) is shown. (b) The proliferation assay was performed 24 h after the transfection.
Association analysis of MIR146A and target gene variants with psoriasis
To investigate whether psoriasis is associated with genetic variations within or close to miR-146a/b encoding genes (MIR146A and MIR146B) and previously verified direct target genes IRAK1, CARD10, CCL5 and NUMB (Crone et al., 2012; Hung et al., 2013; Rebane et al., 2014; Taganov et al., 2006) as well as FERMT1, we carried out in silico candidate gene analysis using existing genome-wide association studies (GWAS) data (Baurecht et al., 2015). The in silico analysis showed suggestive evidence for presence of two independent psoriasis susceptibility loci in the MIR146A gene region. One marker mapped 853 bp upstream from miR-146a encoding sequence, with the lead variant rs2961920 (OR=1.12, P=0.0015) being in perfect linkage disequilibrium (LD) (r2=1) with a functional polymorphism rs2910164 (Luo et al., 2011) reported to be associated with various autoimmune diseases (Li et al., 2015) and with psoriasis in Han Chinese population with the same effect direction (Zhang et al., 2014). An independent association was observed for rs184776122 (OR=0.80, P = 0.005) 37 kb downstream of the MIR146A gene (Supplementary Table S4 online). Recently, rs2910164-CC genotype was reported to have a protective association with psoriasis in HLA-C*06 (a main risk allele in psoriasis) negative patients with Caucasian origin (Srivastava et al., 2016). We performed similar association analysis as well as stratification by HLA-Cw*0602 using our dataset. We observed a significant protective effect of the rs2910164-C allele with similar effect size as reported in (Srivastava et al., 2016). In contrast, we could not confirm the protective recessive effect of rs2910164-CC and instead observed a protective heterozygote effect of rs2910164-GC (Supplementary Table S5). Stratified analysis by HLA-Cw*0602 showed the mentioned rs2910164-C and the rs2910164-GC effects above in both strata (Supplementary Table S5). Analysis of the combination of HLA-Cw*0602 and rs2910164 as proposed by Srivastava et al. revealed the well-known strong association of HLA-Cw*0602 with psoriasis and an additional moderate protective effect of rs2910164-GC genotype in both HLA-Cw*0602 positive and negative patients (Supplementary Table S5). Out of the 5 target genes, only rs7293163, mapped to the cell division cycle 42 effector protein (CDC42EP1) gene close to the CARD10 gene, showed a moderate association with psoriasis (OR=1.14, P = 0.00017) (Supplementary Table S4 online).
The regulation of miR-146a/b and target genes
To study the capacity of endogenous miR-146a/b to modulate their target genes, we next performed a series of experiments with human and mouse keratinocytes. The stimulation with psoriasis-related cytokines TNF-α, IL-17A and IFN-γ revealed that miR-146a/b targets are more strongly induced at early (6, 12 and 24 h) and miR-146a/b at late (48 or 72 h) time-points, when the targets were reduced (CCL5 and IL-8) or stimulated to lesser extent (FERMT1) in human keratinocytes (Supplementary Figure S7). As expected, in most of the conditions, the LNA inhibitor targeting both miR-146 family members lead to the increased expression of the target genes in unstimulated keratinocytes and in the cells stimulated with TNF-α and IL-17A (Supplementary Figure S8). To distinguish the effect of miR-146a and miR-146b, we then used keratinocytes and skin fibroblasts from wild type (WT), miR-146a−/− and miR-146b−/− mice (Supplementary Figure S9a), stimulated the cells with TNF-α or IL-17A and measured the levels of target genes. The increased expression of mouse orthologue of IL-8, Cxcl1, Fermt1 and CARD10 was detected in all conditions in keratinocytes from miR-146a−/− mice as compared to WT mice, while no significant increase in Irak1 and Ccl5 was detected in any used conditions (Supplementary Figure 10a). Although miR-146a/b had comparable expression in whole mouse skin, the expression of miR-146a was detected at least 12-fold higher in cultured keratinocytes as compared to miR-146b (Supplementary Figure S9a and b). Accordingly, we could not detect any significant difference in the expression of miR-146a/b target genes in any condition in cultured keratinocytes from miR-146b−/− and WT mice. To analyze whether miR-146b has potential to influence psoriasis-related inflammatory responses in the skin through other cell types, we next used skin fibroblasts from WT, miR-146−/− and miR-146b−/− mice and performed stimulation experiments as we did in keratinocytes. Although miR-146b expression was apparently lower than that of miR-146a also in fibroblasts (Supplementary Figure S9c), significantly increased expression of Cxcl1 and Irak1 in all conditions, Card10 in cells stimulated with TNF-α or IL-17A and Ccl5 in cells stimulated TNF-α was detected in fibroblasts from miR-146b−/− mice as compared to WT mice. Fibroblasts from miR-146a−/− mice expressed increased expression of Card10 and Irak1 in TNF-α- and IL-17A-stimulated fibroblasts and Ccl5 in fibroblasts stimulated with TNF-α (Supplementary Figure 10b). As similarly to the human fibroblasts, the expression of Fermt1 and Numb1 was almost undetectable in mouse skin fibroblasts, these genes were excluded from the analysis.
DISCUSSION
Lesional skin in psoriasis is characterized by persistent inflammation and hyperproliferation of keratinocytes, however, the molecular mechanisms behind these features are still not fully understood. The function of miR-146a in psoriasis and association of the single nucleotide polymorphism (SNP) rs2910164 located in miR-146a precursor has been studied before (Srivastava et al., 2016; Zhang et al., 2014). In the current study, we demonstrate that besides miR-146a, miR-146b is expressed and probably able to regulate inflammatory responses in keratinocytes and skin fibroblasts and thereby may influence the pathogenesis of psoriasis. In addition, we show that a positive regulator of keratinocyte proliferation, FERMT1 is a direct target of miR-146a and that the effect of miR-146a on keratinocyte proliferation is most probably independent from the suppression of inflammatory responses and activation-induced cell death. Large-scale in silico analysis of GWAS data confirmed a moderate association between psoriasis and multiple genetic variants in the MIR146A gene. Together, our results confirm miR-146a anti-inflammatory function in the skin and indicate that in case of psoriasis, the expression of miR-146a/b is increased in response to disease-associated cytokines, which then leads to the inhibition of inflammatory responses and hyper-proliferation of keratinocytes through the effect on multiple targets (see also Supplementary Figure S11).
Previous miRNA profiling studies suggested that the expression of miR-146a/b is increased in the lesional skin of psoriasis patients (Lovendorf et al., 2015; Sonkoly et al., 2007). We confirmed this result and demonstrated that miR-146a level is elevated in the epidermis and in less extent in the dermis of psoriatic skin, while miR-146b is increased in both the epidermis and the dermis. In human keratinocytes and fibroblasts, the expression of miR-146a was about two-fold higher and was induced by pro-inflammatory cytokines TNF-α, IL-β and IL-17A, while the expression of miR-146b was induced in response to IFN-γ and IL-22 in keratinocytes and in less extent by IL-1β in fibroblasts. This is in line with previous studies demonstrating that the expression of miR-146a is induced by NF-κB and miR-146b is STAT3 and/or STAT1 dependent (Ahn et al., 2013; Curtale et al., 2013).
Among miR-146a target genes that were suppressed in keratinocytes (Rebane et al., 2014), we detected increased expression of IRAK1, CCL5, IL-8, NUMB and FERMT1 and downregulation of CARD10 in psoriatic plaques when compared to healthy skin. However, there was no correlation of the expression levels of miR-146a/b and the targets in lesional skin samples. This indicates that the expression of miR-146a/b and most of the target genes is modulated by similar cellular signals in the psoriatic skin and suggests that miR-146a/b act through multiple genes and not through a single dominant target. Accordingly, miR-146a has been suggested to suppress proliferation of keratinocytes through the targeting of EGFR (Zhang et al., 2014) or IL-8 (Srivastava et al., 2016). We show here that miR-146a/b also have capacity to inhibit the expression of a positive regulators of keratinocyte proliferation FERMT1 (also known as Kindlin-1) (Herz et al., 2006; Rognoni et al., 2014) and NUMB. Previously, it has been shown that mutations in the FERMT1 gene cause Kindler syndrome, a progressive cutaneous atrophy characterized by skin blistering, premature skin aging and increased risk of skin cancer (Duperret et al., 2014). NUMB has been shown to be suppressed by miR-146a in multiple cancers, including melanoma and oral carcinoma (Forloni et al., 2014; Hung et al., 2013).
Although we did not find reverse correlation of miR-146a/b and analyzed targets in psoriatic skin, there is strong evidence that endogenous level of miR-146a is sufficient for the suppression of inflammatory responses and that local administration of miR-146a mimics has anti-inflammatory effect in the skin. Accordingly, 146a−/− mice developed more severe disease in mouse models of AD and psoriasis, while pre-injection of miR-146a alleviated the inflammation in mouse models of irritant contact dermatitis and psoriasis (Rebane et al., 2014; Srivastava et al., 2016; Urgard et al., 2016). As miR-146a/b have similar expression level in vivo in human and mouse skin, miR-146a/b together might have remarkably stronger influence than studies performed with miR-146a−/− mice demonstrate. On the other hand, the observation that the expression of miR-146a/b in normal skin and psoriatic lesions is 10-50-fold lower than that of the most highly expressed miR-203 and miR-125b further supports the therapeutic potential of miR-146a/b overexpression (Lovendorf et al., 2015; Sahmatova et al., 2016).
It has been shown before (Taganov et al., 2006) and we demonstrate here that miR-146a/b inhibit multiple same targets. However, our experiments do not allow to estimate whether the effects of miR-146a/b are with the same strength. Although in human keratinocytes, the expression level of miR-146a/b differed about two times, in cultured mouse keratinocytes, the expression of miR-146a was at least 12-fold higher than that of miR-146b. Accordingly, keratinocytes from miR-146a−/− mice expressed increased levels of Card10, Cxcl1, Fermt1 and Numb in all used conditions, but there was no difference in keratinocythe from miR-146b−/− mice as compared to WT mice. Although the miR-146b level was apparently 4-fold lower as compared to miR-146a also in mouse skin fibroblasts, the increased expression of miR-146a/b target genes in fibroblasts from miR-146b−/− mice was detected in most conditions. The expression of studied genes in fibroblasts from miR-146a−/− mice was also found to be increased in multiple settings. These results indicate that miR-146a might have more important role in keratinocytes and miR-146b in fibroblasts. Still, further studies are needed to better characterize the roles and functions of the miR-146 family in inflammatory skin diseases and psoriasis.
In line with the results from the functional experiments, our in silico genetic analysis confirmed that there is a suggestive association of genetic variations in miR-146a region and psoriasis, but no association of miR-146b was found. Among the analyzed target genes, only rs7293163 in the CARD10 region showed a significant association with psoriasis. It should be noted that the moderate genetic association with psoriasis might be also due the miR-146a functions in the immune cells, which we did not study here.
In conclusion, the capacity of miR-146a/b simultaneously to inhibit inflammatory responses, activation-induced cell death and proliferation of keratinocytes and fibroblasts suggests that miR-146a/b are mainly pacifying miRNAs that contribute to the skin homeostasis and controlling of inflammatory responses in both healthy and diseased skin. In addition, our results are in line with recently published in vivo mouse studies (Srivastava et al., 2016; Urgard et al., 2016) suggesting that overexpression or local administration of miR-146a/b mimics may have potential for the treatment of psoriasis.
MATERIALS AND METHODS
Patients
30 patients with plaque psoriasis and 30 control subjects were included in the study, of which 22 patient and 22 control samples were used for RT-qPCR analysis (see Supplemental Table S1). 8 patient and 8 control samples were used for ISH and IF, of which two representative ISHs and one IF are shown. Association of SNPs in miR146a/b and the target genes (± 50 kb) was analyzed in silico using GWAS data of 4,489 psoriasis cases and 8,240 controls. Details of the meta-analysis have been previously reported (Baurecht et al., 2015). All participating studies were approved by relevant institutional review boards, and all participants provided written or oral consent. For more detailed description of the study subjects see Supplementary Materials and Methods online.
Cell culture
Pooled, normal human epidermal keratinocytes (Promocell, Heidelberg, Germany) were cultured in Keratinocyte-SFM with supplements (Life Technologies, Grand Island, NY). Human primary fibroblasts and PBMCs were isolated and cultured as described earlier (Reemann et al., 2014; Tserel et al., 2011). Isolation and short-term culture of keratinocytes and fibroblasts was performed according to (Lichti et al., 2008). For stimulation and transfection conditions see Supplementary Materials and Methods online.
Mouse strains
miR-146a−/− on C57BL/6J background corresponding WT mice were purchased from the Jackson Laboratory (Bar Harbor, Me). The miR-146b−/− mice were generated by deleting genomic sequence that encompasses the miR-146b precursor on mouse chromosome 19. The related animal experiments were approved by the Institutional Animal Care and Use Committee of City of Hope. For more detailed description see Supplementary Materials and Methods online.
Apoptosis, proliferation and luciferase assays
Proliferation was measured using either 3H thymidine incorporation or CellTiter-Glo® Luminescent cell viability assay (Promega, Madison, WI). Apoptosis was measured by means of flow cytometry after staining with 7-AAD and annexin V (Beckman Coulter, Brea, CA) as described before (Zimmermann et al., 2011). 3′UTR fragments of FERMT1 were inserted into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI) using the PCR primers containing NheI and SalI sites. For Luciferse assay, 30 nM of pre-miRNAs and 50 ng of the reporter plasmid were cotransfected into HEK293 cells. Luciferase activities were measured using Promega dual luciferase assay. For more detailed description see Supplementary Materials and Methods online.
Total RNA isolation and RT-qPCR
Total RNA was extracted using miRNeasy Mini Kit (Qiagen) or Direct-zol RNA MiniPrep kit (Zymo Research (Irvine, CA). For mRNA and miRNA RT-qPCR see Supplementary Materials and Methods online.
ISH and IF
ISH and IF were performed on 10 μm frozen skin sections. ISH was carried out using microRNA ISH Buffer and Controls Kit according to the manufacturer’s protocol (Exiqon, Vedbaek, Denmark). For immunofluorescence, anti-FERMT1 or KRT10 antibodies (Atlas antibodies, Stockholm, Sweden) were used. For additional information see Supplementary Materials and Methods online.
Statistics
Statistical analyses for RT-qPCR results, flow cytometry and proliferation assays were performed with GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). Student’s t-test or two-way ANOVA with Bonferroni correction were used. The results were considered significant at P<0.05 (*) and highly significant at P<0.01 (**). For genetic association analysis, SNPs from miR146a/b and 5 target genes (± 50 kb) were analyzed in silico using GWAS data of a total of 4,489 psoriasis cases and 8,240 controls. For additional information see Supplementary Materials and Methods online.
Supplementary Material
Acknowledgments
This work was supported by European Regional Fund with Archimedes Foundation, EU structural assistance grant SARMP12219T, Estonian Ministry of Education and Research grant REMARK (SARBS12096T), European Union through the European Regional Development Fund (Project No. 2014-2020.4.01.15-0012), institutional research grant IUT2-2 and personal research grants PUT214, PUT177, PUT1669 and PUT1465 from Estonian Research Council, Swiss National Science Foundation grants 320030-140772 and 31-30-156823 and the Christine Kühne-Center for Allergy Research and Education, Davos Switzerland (CK-CARE). This study makes use of genome-wide analysis data generated by the Wellcome Trust Case-Control Consortium (WTCCC). A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the WTCCC was provided by the Wellcome Trust under award 076113 and 085475. The KORA research platform (KORA, Cooperative Research in the Region of Augsburg) was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. The project received infrastructure support through the DFG Clusters of Excellence “Inflammation at Interfaces” (grants EXC306 and EXC306/2), and was by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (sysINFLAME, grant # 01ZX1306A). Support for the case-control psoriasis sample used for this study was provided by the National Institutes of Health (R01AR042742, R01AR050511, R01AR054966, R01AR062382, R01AR065183 to JTE). JTE is supported by the Ann Arbor Veterans Affairs Hospital.
Abbreviations
- AD
atopic dermatitis
- ALI
air-liquid interface
- CARD
caspase recruitment domain
- LNA
locked nucleic acid
- miRNA
microRNA
- STAT
signal transducer and activator of transcription
- Treg
regulatory T cell
- 3′UTR
3′ untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors state no conflict of interest.
References
- Ahn J, Lee H, Jung CH, Jeon TI, Ha TY. MicroRNA-146b promotes adipogenesis by suppressing the SIRT1-FOXO1 cascade. EMBO molecular medicine. 2013;5:1602–12. doi: 10.1002/emmm.201302647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdis M, Aab A, Altunbulakli C, Azkur K, Costa RA, Crameri R, et al. Interleukins (from IL-1 to IL-38), interferons, transforming growth factor beta, and TNF-alpha: Receptors, functions, and roles in diseases. The Journal of allergy and clinical immunology. 2016;138:984–1010. doi: 10.1016/j.jaci.2016.06.033. [DOI] [PubMed] [Google Scholar]
- Armstrong AW, Guerin A, Sundaram M, Wu EQ, Faust ES, Ionescu-Ittu R, et al. Psoriasis and risk of diabetes-associated microvascular and macrovascular complications. J Am Acad Dermatol. 2015;72:968–77 e2. doi: 10.1016/j.jaad.2015.02.1095. [DOI] [PubMed] [Google Scholar]
- Baurecht H, Hotze M, Brand S, Buning C, Cormican P, Corvin A, et al. Genome-wide comparative analysis of atopic dermatitis and psoriasis gives insight into opposing genetic mechanisms. Am J Hum Genet. 2015;96:104–20. doi: 10.1016/j.ajhg.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-kappaB. Immunol Rev. 2012;246:205–20. doi: 10.1111/j.1600-065X.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone SG, Jacobsen A, Federspiel B, Bardram L, Krogh A, Lund AH, et al. microRNA-146a inhibits G protein-coupled receptor-mediated activation of NF-kappaB by targeting CARD10 and COPS8 in gastric cancer. Mol Cancer. 2012;11:71. doi: 10.1186/1476-4598-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci U S A. 2013;110:11499–504. doi: 10.1073/pnas.1219852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duperret EK, Ridky TW. Kindler syndrome in mice and men. Cancer Biol Ther. 2014;15:1113–6. doi: 10.4161/cbt.29482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forloni M, Dogra SK, Dong Y, Conte D, Jr, Ou J, Zhu LJ, et al. miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. eLife. 2014;3:e01460. doi: 10.7554/eLife.01460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths CE, Reich K, Lebwohl M, van de Kerkhof P, Paul C, Menter A, et al. Comparison of ixekizumab with etanercept or placebo in moderate-to-severe psoriasis (UNCOVER-2 and UNCOVER-3): results from two phase 3 randomised trials. Lancet. 2015;386:541–51. doi: 10.1016/S0140-6736(15)60125-8. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part I: clinical and pathologic concepts. The Journal of allergy and clinical immunology. 2011a;127:1110–8. doi: 10.1016/j.jaci.2011.01.053. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part II: immune cell subsets and therapeutic concepts. The Journal of allergy and clinical immunology. 2011b;127:1420–32. doi: 10.1016/j.jaci.2011.01.054. [DOI] [PubMed] [Google Scholar]
- Herz C, Aumailley M, Schulte C, Schlotzer-Schrehardt U, Bruckner-Tuderman L, Has C. Kindlin-1 is a phosphoprotein involved in regulation of polarity, proliferation, and motility of epidermal keratinocytes. The Journal of biological chemistry. 2006;281:36082–90. doi: 10.1074/jbc.M606259200. [DOI] [PubMed] [Google Scholar]
- Hung PS, Liu CJ, Chou CS, Kao SY, Yang CC, Chang KW, et al. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PloS one. 2013;8:e79926. doi: 10.1371/journal.pone.0079926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E. Science. Vol. 349. New York, NY: 2015. GENE REGULATION. Breakers and blockers-miRNAs at work; pp. 380–2. [DOI] [PubMed] [Google Scholar]
- Kim J, Oh CH, Jeon J, Baek Y, Ahn J, Kim DJ, et al. Molecular Phenotyping Small (Asian) versus Large (Western) Plaque Psoriasis Shows Common Activation of IL-17 Pathway Genes, but Different Regulatory Gene Sets. J Invest Dermatol. 2015 doi: 10.1038/JID.2015.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger JG. Hiding under the skin: A welcome surprise in psoriasis. Nature medicine. 2012;18:1750–1. doi: 10.1038/nm.3025. [DOI] [PubMed] [Google Scholar]
- Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, Papp K, et al. Secukinumab in plaque psoriasis–results of two phase 3 trials. N Engl J Med. 2014;371:326–38. doi: 10.1056/NEJMoa1314258. [DOI] [PubMed] [Google Scholar]
- Li C, Fu W, Zhang Y, Zhou L, Mao Z, Lv W, et al. Meta-analysis of microRNA-146a rs2910164 G>C polymorphism association with autoimmune diseases susceptibility, an update based on 24 studies. PloS one. 2015;10:e0121918. doi: 10.1371/journal.pone.0121918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovendorf MB, Mitsui H, Zibert JR, Ropke MA, Hafner M, Dyring-Andersen B, et al. Laser capture microdissection followed by next-generation sequencing identifies disease-related microRNAs in psoriatic skin that reflect systemic microRNA changes in psoriasis. Experimental dermatology. 2015;24:187–93. doi: 10.1111/exd.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–29. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisgen F, Xu Landen N, Wang A, Rethi B, Bouez C, Zuccolo M, et al. MiR-146a negatively regulates TLR2-induced inflammatory responses in keratinocytes. J Invest Dermatol. 2014;134:1931–40. doi: 10.1038/jid.2014.89. [DOI] [PubMed] [Google Scholar]
- Papp KA, Leonardi C, Menter A, Ortonne JP, Krueger JG, Kricorian G, et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N Engl J Med. 2012a;366:1181–9. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- Papp KA, Reid C, Foley P, Sinclair R, Salinger DH, Williams G, et al. Anti-IL-17 receptor antibody AMG 827 leads to rapid clinical response in subjects with moderate to severe psoriasis: results from a phase I, randomized, placebo-controlled trial. J Invest Dermatol. 2012b;132:2466–9. doi: 10.1038/jid.2012.163. [DOI] [PubMed] [Google Scholar]
- Rebane A, Runnel T, Aab A, Maslovskaja J, Ruckert B, Zimmermann M, et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. The Journal of allergy and clinical immunology. 2014;134:836–47 e11. doi: 10.1016/j.jaci.2014.05.022. [DOI] [PubMed] [Google Scholar]
- Rognoni E, Widmaier M, Jakobson M, Ruppert R, Ussar S, Katsougkri D, et al. Kindlin-1 controls Wnt and TGF-beta availability to regulate cutaneous stem cell proliferation. Nature medicine. 2014;20:350–9. doi: 10.1038/nm.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahmatova L, Tankov S, Prans E, Aab A, Hermann H, Reemann P, et al. MicroRNA-155 is Dysregulated in the Skin of Patients with Vitiligo and Inhibits Melanogenesis-associated Genes in Melanocytes and Keratinocytes. Acta Derm Venereol. 2016 doi: 10.2340/00015555-2394. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PloS one. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Nikamo P, Lohcharoenkal W, Li D, Meisgen F, Xu Landen N, et al. MicroRNA-146a suppresses IL-17-mediated skin inflammation and is genetically associated with psoriasis. The Journal of allergy and clinical immunology. 2016 doi: 10.1016/j.jaci.2016.07.025. [DOI] [PubMed] [Google Scholar]
- Zhang W, Yi X, Guo S, Shi Q, Wei C, Li X, et al. A single-nucleotide polymorphism of miR-146a and psoriasis: an association and functional study. J Cell Mol Med. 2014;18:2225–34. doi: 10.1111/jcmm.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Koreck A, Meyer N, Basinski T, Meiler F, Simone B, et al. TNF-like weak inducer of apoptosis (TWEAK) and TNF-alpha cooperate in the induction of keratinocyte apoptosis. The Journal of allergy and clinical immunology. 2011;127:200–7. 7 e1–10. doi: 10.1016/j.jaci.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Ellinghaus E, Stuart PE, Capon F, Knight J, et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun. 2015;6:7001. doi: 10.1038/ncomms8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urgard E, Lorents A, Klaas M, Padari K, Viil J, Runnel T, et al. Pre-administration of PepFect6-microRNA-146a nanocomplexes inhibits inflammatory responses in keratinocytes and in a mouse model of irritant contact dermatitis. Journal of controlled release: official journal of the Controlled Release Society. 2016;235:195–204. doi: 10.1016/j.jconrel.2016.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


