Abstract
Macrophage migration inhibitory factor (MIF) is deemed as an immunoregulatory and proinflammatory cytokine related to the progression of tuberculosis. A CATT short tandem repeat (STR) polymorphism at position −794 in the MIF gene promoter region is associated with the susceptibility to tuberculosis (TB). To investigate whether macrophage MIF gene mif CATT variants are associated with susceptibility to retreatment cases of TB and drug-resistant TB prevalence, genotyping of MIF −794 CATT polymorphism and quantifying of serum MIF were performed to associate MIF−794 CATT polymorphism with new patients and retreatment cases. Significant increases in MIF −794 CATT genotypes 7/8 and allele CATT 8 were observed in TB patients. Significant differences in the genotypic frequencies of MIF −794 CATT (5/X + 6/X vs 7/7 + 7/8) were demonstrated upon comparing the total cases and the new cases of TB with the controls. Significant differences in the allelic frequencies of MIF −794 CATT (5 + 6 vs 7 + 8) were observed in the total cases and new cases of TB. No differences in the genotypic frequencies of the MIF −794 CATT (5/X + 6/X vs 7/7 + 7/8) were observed between the retreatment cases and the controls or between the new cases and retreatment cases. In conclusion, the MIF −794 CATT genotypes 7/8 and allele CATT 8 were highly associated with TB; no differences in the genotypic frequencies of the MIF −794 CATT (5/X + 6/X vs 7/7 + 7/8) were observed between the new cases and retreatment cases.
Keywords: gene promoter; macrophage migration inhibitory factor; microsatellite polymorphism; Mycobacterium tuberculosis; tuberculosis, CATT polymorphism
Introduction
Tuberculosis (TB) is an infectious disease caused by the bacillus Mycobacterium tuberculosis and characterized by chronic lung inflammation leading to pulmonary destruction, loss of function, and disability. The disease is still a major global health problem, resulting in ill health for approximately 9 million people each year. It is estimated that about 2–3 billion individuals may have been infected with M. tuberculosis; however, only a relatively small proportion (5%–15%) among them will eventually develop TB during their lifetimes.1 Long-term surveillance of TB has indicated that different populations show different susceptibilities to M. tuberculosis infection. There is some evidence that genetic factors are close related to susceptibility to disease development.2
Macrophage migration inhibitory factor (MIF), which is coded by MIF gene as a proinflammatory cytokine, may act as a double-edged sword, as it can contribute to detrimental tissue inflammation but at the same time may be important for controlling infection. It is produced by T cells, macrophages, and pulmonary epithelial cells.3 There is a single MIF gene, MIF, in the human genome, which is located on chromosome 22q11.2, 0.8 kb long, and composed of three exons and two introns. The gene product MIF contains 115 amino acids with an approximate molecular weight of 12.5 kDa, and it is regarded as a proinflammatory cytokine that prohibits the migration and induces the recruitment of macrophages at the local inflammation or infection sites. Two types of polymorphisms with obvious functions have been confirmed in the MIF promoter region, namely, G/C single-nucleotide polymorphism at position −173 and −794 CATT5–8 (rs5844572) microsatellite polymorphism.4
The cytokine MIF has been shown to be involved in several diseases. Furthermore, polymorphisms in the human MIF promoter have been proved to be associated with the host’s susceptibility to infectious diseases.5 Several recent studies have suggested that both of these polymorphisms are associated with TB susceptibility.6,7
Following the World Health Organization (WHO) definition of TB cases in the Global Tuberculosis Report 2015, TB cases can be further divided into two types, new cases of TB and retreatment cases of TB.1 Globally, an estimated 3.3% of new cases and 20% of retreatment cases are multidrug-resistant tuberculosis (MDR-TB). In 2014, there were an estimated 480,000 new cases of MDR-TB worldwide, with approximately 190,000 deaths resulting from MDR-TB infection.1
The aim of this study was to further explore the potential association between the −794 CATT5–8 microsatellite polymorphism of the MIF gene and the susceptibility to new cases and retreatment cases of TB. This study was designed to systematically evaluate the correlation between functional CATT microsatellite polymorphisms in the MIF gene promoter and the susceptibility to new cases and retreatment cases of TB, which are one of the dominant drug-resistant TB pools.
Patients and methods
Study population
Our paired data study included 200 patients with pulmonary TB and 100 healthy controls. Pulmonary TB subjects were recruited from hospitalized patients at the Third Hospital of Kunming, who came from various cities and counties of Yunnan Province. The controls were healthy volunteers undertaking physical examinations at the Center for Disease Prevention and Control of Kunming, Yunnan, China. The objectives and procedures were explained to all potential participants before samples were collected. Individual data from participants and diagnostic results were kept strictly confidential. All subjects signed informed consent forms voluntarily and the research project was approved by the Medical Ethics Commission of Kunming Medical University.
Clinically diagnosed cases of pulmonary TB were further categorized into two types, new cases and retreatment cases based on the WHO definition of TB cases in the Global Tuberculosis Report 2015.1 New cases of TB refer to patients who have never been treated for TB or who have taken anti-TB drugs for less than 1 month. Retreatment cases of TB refer to patients who have been treated for at least 1 month with anti-TB drugs in the past. Retreatment cases were further classified by the outcome of their most recent course of treatment into four categories: (a) relapsed patients, who have previously been treated for TB, were declared cured or treatment completed at the end of their most recent course of treatment, and are now diagnosed with a recurrent episode of TB (either a true relapse or a new episode of TB caused by reinfection); (b) treatment after failure patients, who have previously been treated for TB and their most recent course of treatment failed, that is, they had a positive sputum smear or culture result at month 5 or later during treatment; (c) treatment after loss to follow-up patients, who have previously been treated for TB and were declared “lost to follow-up” at the end of their most recent course of treatment; and (d) other previously treated patients, who have previously been treated for TB but whose outcome after their most recent course of treatment is unknown or undocumented.1 The retreatment cases included in our study fell into categories (a) and (b). We excluded co-morbidity conditions like human immunodefiency virus (HIV) infection/AIDS, diabetes, chronic obstructive pulmonary disease (COPD) in included patients. The control population was chosen based on no history or clinical evidence of TB and other diseases.
Sample collection
The venous blood samples were collected from patients and healthy volunteers into tubes containing the anticoagulant ethylenediaminetetraacetic acid (EDTA). The blood samples were used to extract genomic DNA.
Total genomic DNA extraction
Genomic DNA samples were extracted from peripheral blood samples anti-coagulated with EDTA dipotassium salt (EDTA-K2) using a commercially available Whole-Blood DNA Extraction Kit (Tiangen Company, Beijing, China) according to the manufacturer’s instructions. The purified genomic DNA samples were kept at −20°C until further use.
Genotyping of MIF −794 microsatellite polymorphisms
A 346 bp fragment containing the MIF promoter microsatellite repeat sequence (GenBank No. rs5844572) was amplified by polymerase chain reaction (PCR). The forward primer sequence was 5′-TGCAGGA-ACCAATACCCATAGG-3′ and the reverse primer sequence was 5′-AATGGTAAACTCGGGGAC-3′. For genotyping MIF −794 microsatellite, a 50 µL reaction mixture, which contains reaction buffer, dNTPs, primers, Taq polymerase, and 100 ng DNA, was amplified in a MyGene MG96G PCR machine (Long Gene Co. Ltd., Hangzhou, China). Denaturation was performed at 95°C for 10 min and then followed by 35 amplification cycles at 95°C for 45 s, 53.8°C for 45 s, and 72°C for 45 s, with a final extension step at 72°C for 7 min.
The 346 bp PCR product of the target DNA was purified using a DNA Product Purification Kit (Tiangen Company, Beijing, China) and further confirmed by electrophoresis. The CATT microsatellite polymorphisms of the MIF gene promoter −794 site were determined by DNA sequencing of the 346 bp PCR product (GenScript Biotech Company, Nanjing, China).
Determination of serum MIF concentration by enzyme-linked immunosorbent assay
Serum samples were obtained from the peripheral blood of each patient and control and reserved at −80°C until use. The contents of the serum MIF protein were quantified using a commercially available enzyme-linked immunosorbent assay (ELISA) kit (R&D, USA) according to the manufacturer’s operating manual. Recombinant human MIF was chosen to make the standard curve.
Statistical analysis
All of the statistical analyses were performed using GraphPad PRISM version 6.0 for Windows (GraphPad Software, San Diego, CA, USA). The Hardy–Weinberg equilibrium was first examined for each group. The sequencing results of microsatellite polymorphism were analyzed using the Primer 6.0 software (http://www.primer-e.com). The chi-square test was chosen to compare the allele frequencies among the TB-affected and control individuals. The odds ratio (OR) with 95% confidence interval (CI) was calculated using the PRISM software, and the data are expressed as mean ± standard deviation (SD). The MIF concentrations were compared using Student’s t test, and the differences of MIF between groups were analyzed using the paired Student’s t test. Differences with P < 0.05 were deemed to be significant, and differences with P < 0.01 were deemed to be highly significant.
Ethics statement
This study was approved by the Ethical Review Committee of Kunming Medical University (no. 2012-019). All of the patients and controls voluntarily signed informed consent forms.
Results
Characteristics of study participants
The sex ratios (females/males) for the total cases, new cases, and retreatment cases of TB and the controls were 67/133, 42/58, 25/75, and 63/37, respectively. The mean ages ± SD for the total cases, new cases, and retreatment cases of TB and the controls were 40.56 ± 17.39, 40.64 ± 20.24, 40.52 ± 15.70, and 38.65 ± 9.15, respectively.
Genotype and Allele distributions of MIF −794 CATT polymorphisms
Both the genotype and allele distributions of MIF −794 CATT microsatellite polymorphisms in the total cases of TB and healthy controls were in accordance with the Hardy–Weinberg equilibrium (P = 0.065 in the total cases of TB and P = 0.076 in healthy controls). Statistically significant differences were observed in the distributions of both MIF genotypes −794 CATT 5/5 and CATT 7/8 between the TB patients and healthy controls. The distribution of MIF allele −794 CATT genotype 5/5 was significantly lower in the TB patients than in the controls (10.0% vs 23%, OR = 0.37, 95% CI = 0.19–0.72, P < 0.01). Moreover, a significant decrease in MIF allele −794 CATT 5 was observed in the TB patients compared with the controls (31.5% vs 43%, OR = 0.61, 95% CI = 0.43–0.87, P < 0.01). In contrast, significant increases in MIF −794 CATT, for both genotypes 7/8 (6.0% vs 1.0%, OR = 6.32, 95% CI = 0.81–49.31, P < 0.05) and alleles CATT 8 (3.0% vs 0.5%, OR = 6.15, 95% CI = 0.79–47.67, P < 0.05), were observed in the TB patients compared with the controls (Table 1).
Table 1.
Comparison of genotypic and allelic distributions of MIF −794 CATT polymorphisms between TB patients and controls.
| MIF −794 | Tuberculosis, n (%) | Controls, n (%) | OR (95% CI) | χ2 | P value |
|---|---|---|---|---|---|
| Genotype | |||||
| CATT 5/5 | 20 (10.0) | 23 (23.0) | 0.37 (0.19–0.72) | 9.18 | 0.002** |
| CATT 5/6 | 74 (37.0) | 34 (34.0) | 1.14 (0.69–1.89) | 0.26 | 0.61 |
| CATT 5/7 | 12 (6.0) | 6 (6.0) | 1.00 (0.36–2.75) | 0.00 | 1.00 |
| CATT 6/6 | 42 (21.0) | 20 (20.0) | 1.06 (0.59–1.93) | 0.04 | 0.84 |
| CATT 6/7 | 31 (15.5) | 15 (15.0) | 1.04 (0.53–2.03) | 0.01 | 0.91 |
| CATT 7/7 | 9 (4.5) | 1 (1.0) | 4.67 (0.58–37.35) | 2.53 | 0.11 |
| CATT 7/8 | 12 (6.0) | 1 (1.0) | 6.32 (0.81–49.31) | 4.02 | 0.045* |
| Allele | |||||
| CATT 5 | 126 (31.5) | 86 (43.0) | 0.61 (0.43–0.87) | 7.72 | 0.005** |
| CATT 6 | 189 (47.25) | 89 (44.5) | 1.18 (0.79–1.57) | 0.41 | 0.52 |
| CATT 7 | 73 (18.25) | 24 (12.0) | 1.64 (0.99–2.69) | 3.84 | 0.05 |
| CATT 8 | 12 (3.0) | 1 (0.5) | 6.15 (0.79–47.67) | 3.93 | 0.047* |
MIF: migration inhibitory factor; OR: odds ratio; CI: confidence interval.
P < 0.05, **P < 0.01.
Genotypic frequencies of MIF −794 CATT genotypes (5/X + 6/X vs 7/7 + 7/8) in total cases, new cases, and retreatment cases of TB
In order to determine the genotypic frequencies of the MIF −794 CATT genotypes in association with specific TB subgroups, the genotypic frequencies of the −794 CATT genotypes (5/X + 6/X vs 7/7 + 7/8) were calculated for the total cases (89.5% vs 10.5%), new cases (87.0% vs 13.0%), retreatment cases of TB (92.0% vs 8.0%), and the controls (98.0% vs 2.0%). Significant differences in the genotypic frequencies of MIF −794 CATT (5/X + 6/X vs 7/7 + 7/8) were demonstrated when comparing the total cases and new cases of TB with the controls (OR = 5.57, 95% CI = 1.32–25.03, P < 0.05, and OR = 7.32, 95% CI = 1.61–33.36, P < 0.01, respectively). In contrast, no differences in the genotypic frequencies of the MIF −794 CATT (5/X + 6/X vs 7/7 + 7/8) were observed between the retreatment cases and the controls or between the new cases and retreatment cases (Table 2). These results suggested that the MIF −794 CATT genotypes (7/7 + 7/8) were associated with new cases of TB and total cases of TB.
Table 2.
Comparison of genotypic frequencies of MIF −794 CATT genotypes (5/X + 6/X vs 7/7 + 7/8) between TB patients and healthy controls.
| Cases | 5/X + 6/X (%) | 7/7 + 7/8 (%) | OR (95% CI) | P value |
|---|---|---|---|---|
| Total cases of TB | 179 (89.5) | 21 (10.5) | 5.57 (1.32–25.03) | 0.01* |
| New cases | 87 (87.0) | 13 (13.0) | 7.32 (1.61–33.36) | 0.005** |
| Retreatment cases | 92 (92.0) | 8 (8.0) | 4.26 (0.88–20.59) | 0.101 |
| 0.58 (0.23–1.47) | 0.375a | |||
| Controls | 98 (98.0) | 2 (2.0) | 1.00 (reference) |
MIF: migration inhibitory factor; TB: tuberculosis; OR: odds ratio; CI: confidence interval.
For comparison between new cases of TB and retreatment cases of TB.
P < 0.05, **P < 0.01.
Allelic frequencies of MIF −794 CATT alleles (5 + 6 vs 7 + 8) in new cases, retreatment cases, and total cases of TB, and controls
To determine whether the allelic frequencies of the MIF −794 CATT alleles (5 + 6 vs 7 + 8) play a part in genetic susceptibility to TB, the allelic frequencies of the MIF −794 CATT alleles (5 + 6 vs 7 + 8) were analyzed for the total cases (78.7% vs 21.3%), new cases (77.0% vs 23.0%), and retreatment cases of TB (80.5% vs 19.5%), and the controls (87.5% vs 12.5%). Significant differences in the allelic frequencies of MIF −794 CATT (5 + 6 vs 7 + 8) were observed for the total cases of TB (OR = 0.53, CI = 0.33–0.86, P < 0.05) and new cases of TB (OR = 2.09, CI = 1.23–3.56, P < 0.01) when compared with the controls, respectively. In contrast, the allelic frequencies of MIF −794 CATT (5 + 6 vs 7 + 8) revealed no differences between the retreatment cases and the controls or between the new cases and the retreatment cases. These results suggested that the MIF −794 CATT alleles (7 + 8) were associated with both total cases and new cases of TB (Table 3).
Table 3.
Comparisons of allelic frequencies of MIF −794 CATT alleles (5 + 6 vs 7 + 8) between three TB patient groups and healthy controls.
| Cases | 5 + 6 (%) | 7 + 8 (%) | OR (95% CI) | P value |
|---|---|---|---|---|
| Total cases of TB | 315 (78.7) | 85 (21.3) | 0.53 (0.33–0.86) | 0.01* |
| New cases | 154 (77.0) | 46 (23.0) | 2.09 (1.23–3.56) | 0.009** |
| Retreatment cases | 161 (80.5) | 39 (19.5) | 0.59 (0.34–1.02) | 0.076 |
| 1.23 (0.76–1.99) | 0.463a | |||
| Controls | 175 (87.5) | 25 (12.5) | 1.00 (reference) |
MIF: migration inhibitory factor; TB: tuberculosis; OR: odds ratio; CI: confidence interval.
The allelic frequencies of MIF −794 CATT alleles (5 + 6 vs 7 + 8) of three TB patient groups were successively compared to that of healthy controls.
For comparison between new cases of TB and retreatment cases of TB.
P < 0.05, **P < 0.01.
Serum MIF levels in TB cases with MIF −794 genotypes (5/5 + 5/6 + 6/6) and (7/X + 8/X)
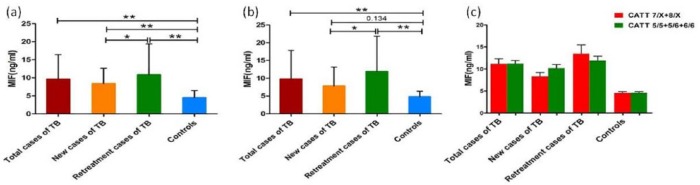
To investigate whether the MIF −794 CATT genotype is associated with the increase in serum MIF concentrations in TB cases, we quantified the MIF concentrations in different subgroups with the MIF −794 CATT genotypes (5/5 + 5/6 + 6/6) and (7/X + 8/X). The results demonstrated an increase in MIF concentrations for the new cases (8.38 ± 4.25 ng/mL) and retreatment cases of TB (10.85 ± 8.54 ng/mL) in TB patients with the MIF −794 CATT genotype (5/5 + 5/6 + 6/6), compared with the controls (4.50 ± 1.98 ng/mL). Furthermore, the serum MIF levels were significantly higher for the retreatment cases than for the new cases (P < 0.05, Figure 1(a)). Similarly, a significant increase in serum MIF concentrations was observed in TB patients with the MIF −794 CATT genotype (7/X + 8/X), with serum MIF concentrations of 9.78 ± 8.06 ng/mL and 11.92 ± 9.95 ng/mL being detected for the total cases and retreatment cases, respectively, compared with the control individuals (4.80 ± 1.59 ng/mL) (P < 0.01, Figure 1(b)). In addition, the serum MIF concentrations for the retreatment cases were significantly higher than those for the new cases (P < 0.05). In contrast, no significant increase in MIF expression was observed in the new cases of TB (7.84 ± 5.32 ng/mL), compared with the control group (P > 0.05, Figure 1(c)).
Figure 1.
Association comparisons of serum MIF concentrations with different MIF −794 CATT genotypes. (a) Comparison of serum MIF concentrations in MIF −794 CATT genotype (5/5 + 5/6 + 6/6) between TB cases and controls. *P < 0.05, **P < 0.01. (b) Comparison of serum MIF concentrations in MIF −794 CATT genotype (7/X + 8/X) between TB cases and controls. *P < 0.05, **P < 0.01. (c) Comparison of serum MIF concentrations between MIF −794 CATT genotypes (5/5 + 5/6 + 6/6) and (7/X + 8/X) for each TB group.
No differences were found between serum MIF concentrations in MIF −794 CATT genotypes (5/5 + 5/6 + 6/6) and (7/X + 8/X) cases within same TB group
To evaluate whether the MIF −794 CATT genotype plays an effect in MIF production in TB cases, we studied the serum MIF concentrations for the MIF −794 CATT genotype (5/5 + 5/6 + 6/6) and genotype (7/X + 8/X) cases within each group. No significant differences in MIF concentrations between genotype (5/5 + 5/6 + 6/6) and (7/X + 8/X) cases within the same group were observed (Figure 1(c)).
Discussion
Genetic factors may be related to an increased risk of patients developing active TB.8 Gene polymorphism could play an significant role in the occurrence and development of TB. Several TB-related innate immunity genes have been reported, including human leukocyte antigen (HLA), natural-resistance-associated macrophage protein-1 (Nramp1), mannose-binding lectin (MBL), vitamin D receptor (VDR), nitric oxide synthase 2A (NOS2A), SP110 nuclear body protein (SP110), and interferon genes (IFNGs).9 Therefore, the studies concentrating on the relationship between genetic polymorphisms and the susceptibility to TB are of great significance for both prevention and treatment of this bacterial infection.
Because of its wide expression in various cells, MIF is considered to be a multi-functional cytokine.10 As a proinflammatory cytokine, we believe that MIF may function as a double-edged sword which contributes to tissue-damaged inflammation but may also be key for mitigating infection.11 MIF has been proved to be a protective host factor against TB-causing bacteria. Oddo et al.8 demonstrated the growth inhibition of M. tuberculosis by MIF. Das et al. found that Mif-deficient mice succumbed much faster with higher numbers of the organisms burdened, increased lung pathology, and decreased innate cytokine production.6 Furthermore, Mif-deficient animals exhibited an increase in pulmonary neutrophil accumulation with a suppressed adaptive immune response.12 Therefore, MIF may provide a two-edged sword effect which induce an inflammation and protection against M. tuberculosis.
MIF −794 CATT5–8 microsatellite polymorphism has been preliminarily proved to be related to genetic susceptibility to active TB in the southwestern Han Chinese population.13 Similarly, a significant association was found between genotypes carrying MIF −794 CATT 7 or 8 and susceptibility to active lung TB. In contrast, no allele in the MIF −794 CATT polymorphism was involved in TB risk in the northwestern Colombian population.11 MIF −794 CATT5–8 microsatellite has been found to be linked with the transcription level alteration of MIF gene. The CATT repeat number regulates the activity of the MIF gene promoter; higher CATT repeat numbers lead to stronger activity of the promoter.14,15 Our current study has demonstrated a significant increase in MIF −794 CATT, for both genotypes 7/8 and alleles CATT 8, in TB patients compared with healthy controls. On the other hand, no significant difference for either CATT 5/5 or CATT 5 was found for the total TB cases compared with the control group. These results suggested that the genotypic frequencies of the MIF −794 CATT genotypes (7/7 + 7/8) and the allelic frequencies of the MIF −794 CATT alleles (7 + 8) were associated with the new cases and total cases of TB.
The MIF serum concentrations in TB cases with the MIF −794 genotype (5/5 + 5/6 + 6/6) and genotype (7/X + 8/X) were found to be greatly increased compared with the healthy controls. Moreover, the MIF expression levels in retreatment cases of TB were higher than those in new cases of TB. No differences were observed in the MIF concentrations between the MIF −794 CATT genotype (5/5 + 5/6 + 6/6) cases and genotype (7/X + 8/X) cases of TB within each individual group.
We speculate that the genotypes 7/8 and alleles CATT 8 at position −794 in the MIF gene promoter could be associated with high MIF expression, resulting in increased susceptibility to TB. Our results not only further confirm previous conclusions by other authors from studies on the Han Chinese population but also give a deeper and more extensive insight into the association between MIF −794 CATT5–8 microsatellite polymorphisms and susceptibility to active TB. Our focus on the investigation of new cases and retreatment cases of TB could confer a significant impact on prevention and control of drug-resistant TB.
Acknowledgments
A.L., S.P.V., and F.B. designed the research. A.L. and F.B. performed the research. A.L., S.P.V., and F.B. analyzed data. A.L., S.P.V., and F.B. wrote the paper.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This work was supported by grants from the National Natural Science Foundation of China (grant nos 81371835 and 31560051), the Scientific and Technological Development Project of Yunnan Province of China (grant no. 2017FE467-001), and a TRF Senior Research Scholarship from the Thailand Research Fund (grant no. RTA 5880005).
References
- 1. World Health Organization (WHO) (2016) Global Tuberculosis Report 2015. Geneva: WHO. [Google Scholar]
- 2. Casanova JL, Abel L. (2002) Genetic dissection of immunity to mycobacteria: The human model. Annual Review of Immunology 20: 581–620. [DOI] [PubMed] [Google Scholar]
- 3. Schindler L, Dickerhof N, Hampton MB, et al. (2018) Post-translational regulation of macrophage migration inhibitory factor: Basis for functional fine-tuning. Redox Biology 15: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gregersen PK, Bucala R. (2003) Macrophage migration inhibitory factor, MIF alleles, and the genetics of inflammatory disorders: Incorporating disease outcome into the definition of phenotype. Arthritis & Rheumatism 48: 1171–1176. [DOI] [PubMed] [Google Scholar]
- 5. Illescas O, Gomez-Verjan JC, García-Velázquez L, et al. (2018) Macrophage migration inhibitory factor −173 G/C polymorphism: A global meta-analysis across the disease spectrum. Frontiers in Genetics 9: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Das R, Koo MS, Kim BH, et al. (2013) Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. PNAS 110: E2997–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y, Yuan T, Lu W, et al. (2012) Association of tuberculosis and polymorphisms in the promoter region of macrophage migration inhibitory factor (MIF) in a Southwestern China Han population. Cytokine 60: 64–67. [DOI] [PubMed] [Google Scholar]
- 8. Oddo M, Calandra T, Bucala R, et al. (2005) Macrophage migration inhibitory factor reduces the growth of virulent Mycobacterium tuberculosis in human macrophages. Infection and Immunity 73: 3783–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y, Zeng Z, Deng S. (2012) Study of the relationship between human MIF level, MIF-794CATT 5-8 microsatellite polymorphism, and susceptibility of tuberculosis in Southwest China. Brazilian Journal of Infectious Diseases 16: 383–386. [DOI] [PubMed] [Google Scholar]
- 10. Kuai SG, Ou QF, You DH, et al. (2016) Functional polymorphisms in the gene encoding macrophage migration inhibitory factor (MIF) are associated with active pulmonary tuberculosis. Infectious Diseases 48: 222. [DOI] [PubMed] [Google Scholar]
- 11. Gómez LM, Sánchez E, Ruiznarvaez EA, et al. (2010) Macrophage migration inhibitory factor gene influences the risk of developing tuberculosis in northwestern Colombian population. Tissue Antigens 70: 28–33. [DOI] [PubMed] [Google Scholar]
- 12. Azad AK, Sadee W, Schlesinger LS. (2012). Innate immune gene polymorphisms in tuberculosis. Infection and Immunity 80: 3343–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Temple S, Cheong K, Price P, et al. (2008) The microsatellite, macrophage migration inhibitory factor-794, may influence gene expression in human mononuclear cells stimulated with E. coli or S. pneumoniae. International Journal of Immunogenetics 35: 309–316. [DOI] [PubMed] [Google Scholar]
- 14. Bloom J, Sun S, AI-Abed Y. (2016) MIF, a controversial cytokine: A review of structural features, challenges, and opportunities for drug development. Expert Opinion on Therapeutic Targets 20: 1463–1475. [DOI] [PubMed] [Google Scholar]
- 15. O’Garra A, Redford PS, Mcnab FW, et al. (2013) The immune response in tuberculosis. Annual Review of Immunology 31: 475. [DOI] [PubMed] [Google Scholar]