Abstract
Differential display (DD) is a well-established analytical tool for measuring gene expression that is still popular due to its documented success and ability to identify novel genes not yet available for analysis by more powerful microarray hybridization. For a comprehensive analysis of all mRNAs in a given cell, it is statistically predicted that at least 240 different DD primer combinations are required. This prediction, however, has never been empirically tested. Using far more primer combinations than that predicted to evaluate 90% of the mRNAs in a cell, plus other modifications, we identified and confirmed the induction of five mRNAs by hydrogen peroxide in HA-1 hamster cells. However, five other known oxidant-inducible mRNAs were not identified by DD. Filter microarray hybridization did not result in the identification of any additional species modulated twofold or greater but previous two-dimensional protein gel electrophoresis identified 15 induced protein species. We conclude that the current statistical prediction for comprehensive analysis of all the mRNAs in a given cell is inaccurate, at least in our hands, and further conclude that DD is a useful but less than comprehensive method for assessing changes in mRNA levels.
Keywords: Differential display, Gene expression, Hydrogen peroxide, Oxidative stress, Hamster fibroblasts
DIFFERENTIAL display (DD) is a PCR-based technique that is used to assess changes in gene modulation between samples at the level of mRNA. Its basic approach utilizes different combinations of PCR primers to generate subpopulations of DNA species that are then analyzed on a sequencing gel (1,4,13). Comparison of control and test sample lanes then allows identification of potential differentially modulated mRNAs. In this manner, both increased and decreased levels of individual mRNA species can be determined (8,13). Because DD is PCR-based, it is also a reasonably sensitive technique.
DD has been used extensively since its first report and continues to be a popular technology. Over the last 3 years alone, more than 1000 research articles have been published on this technique. These studies have included the use of DD to identify hundreds of novel genes; valuable insights regarding cellular responses under varied conditions such as acute stress or chronic disease; and the identification of certain mRNAs and their parent genes as potential diagnostic and therapeutic clinical targets [e.g., (10,11,21)]. Even with the advent of newer technologies such as microarrays and serial analysis of gene expression (SAGE) (14,16,17,20), and known drawbacks of DD such as a high number of false positives (18), DD continues to be a popular technique. This is due to its documented success, its ability to identify novel genes not yet available for analysis by microarray hybridization, and newly expanded uses for this technique (9,15,23).
Despite the past and present use of DD, an empirical determination of the comprehensiveness of this technique has never been reported. In other words, what percentage of all modulated mRNAs is successfully identified by a thorough DD analysis? To date, these considerations have relied on statistical predictions.
A comprehensive analysis of all mRNAs is based on the assumption that there are 15,000 different mRNA species per cell. It is statistically predicted that about 240 different DD primer combinations are required to assess 90% or more of these mRNAs in a given cell type (13). In this report, we decided to evaluate the validity of these statistical predictions using an oxidative stress cell culture model; namely, hamster HA-1 fibroblasts exposed to hydrogen peroxide. This approach has the advantage that oxidant-inducible genes have already been identified and can therefore be compared with our DD results.
MATERIALS AND METHODS
Cell Culture and Treatment Conditions
Hamster HA-1 cells, a Chinese hamster ovary fibroblast cell line (19), were maintained in Eagle’s minimal essential medium (MEM) supplemented with 15% heat-inactivated fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). The cultures were grown in a humidified incubator atmosphere of 95% air and 5% CO2 at 37°C.
Treatment of HA-1 Cells With Hydrogen Peroxide
HA-1 cells were trypsinized and plated at 200,000 cells per 60-mm plate. After 2 days of incubation, cells were divided into two groups, one receiving phosphate-buffered saline (PBS) only (controls) and the other 160 μM of hydrogen peroxide (H2O2). Cells were then returned to the incubator for the designated period of time.
RNA Isolation and Analysis
Total RNA was isolated by direct addition of RNA lysis buffer containing guanidinium isothiocyanate (RNAzol, Biotecz, Houston, TX and RNA Isolator, Genosys, Woodlands, TX) to PBS-washed cultures. The RNA was then extracted according to the manufacturer, and the final semidried pellet resuspended in diethylpyrocarbonate-treated distilled, deionized water. Electrophoresis, Northern blotting, and hybridization were performed as previously described (4,7). Final washed blots were exposed to X-ray film.
Differential Display
HA-1 cells treated with and without peroxide were extracted 30 min, 90 min, and 4 h after the initial peroxide exposure as described above. These RNAs were then used to prepare cDNA templates for DD PCR using three different approaches based on Gen-Hunter kits: the original GenHunter (Nashville, TN) kit; the “third-generation” GenHunter kit; and a modification of both kits.
The original GenHunter kit synthesizes 12 separate cDNA subgroups using four degenerate anchored oligo-dT primers (T12MA or 5′-TTTTTTTTTTTTMA-3′, T12MC, T12MG, and T12TM where M refers to either A, C, or G but not T). A comprehensive analysis for a given condition then involves using these primers for PCR in combination with 20 upstream “AP” 10mer primers, a total of 240 primer combinations, and was statistically predicted to cover all mRNA sequences (13). We synthesized an additional seven upstream primers for these analyses, bringing our actual total of possible primer combinations to 324. The “third-generation” GenHunter kit features three one-base-anchored oligo-dT primers in combination with 80 arbitrary upstream 13mer primers. This combination is predicted to detect about 90% or more of the possible mRNAs is a mammalian cell (12) when considering that the three one-base-anchored oligo-dT primers should generate cDNAs to all of the cellular mRNAs. We also used a third approach, which was to take some of the first- and third-generation kit primer combinations and include TaqStart Taq antibody (Clontech, Palo Alto, CA) in the PCR reaction. This is a hot start approach that was hoped would be especially advantageous for the low annealing temperature used in DD. This approach was used for approximately 50% and 20% of the possible primer combinations for the original and third-generation kits, respectively. DNAse treatment of the RNA, cDNA template synthesis, PCR amplification, sequencing gel electrophoresis, band excision, reamplification, amplicon cloning, and confirmation analyses were all carried out according to the GenHunter protocols.
Microarray Hybridization
Human gene filter microarrays obtained from Research Genetics (Research Genetics, Huntsville, AL) were used for these studies. These arrays contain 5184 noncontrol cDNAs derived from various human tissues. cDNA was prepared from total RNA (6 μg each) from duplicate samples obtained from HA-1 cells as described above (4-h peroxide treatment) and the arrays probed and washed as described by the manufacturer (Research Genetics). A final moderate wash stringency of 50°C and 0.5S SSC, 1% SDS was used to ensure sufficient hamster-to-human sequence hybridization. The arrays were then phosphoimaged using a Storm 860 phosphoimager with ImageQuant software (Molecular Dynamics) and duplicate modulations of twofold or greater determined using the Pathways software (Research Genetics) after normalizing to all data points.
Two-Dimensional (2D) Protein Gel Electrophoresis
The publication and detailed description of 2D protein gel electrophoresis was previously reported by Wiese et al. (24). In summary, HA-1 cells were exposed to 150 μM hydrogen peroxide or solvent (control). At various times (4, 8, 15, and 18 h) after exposure, cells were pulse-chased with [35S]methionine/cysteine and processed for 2D protein gel electrophoresis. Rates of protein synthesis were assessed by incorporation of the radiolabeled amino acids into acid-precipitable protein following peroxide treatment.
RESULTS
Example of Differential Display: Identification of an RNA Induced in HA-1 Cells by Hydrogen Peroxide
Total RNA was extracted from HA-1 cells treated for 30 min, 90 min, and 4 h with 160 μM hydrogen peroxide and from control cells, and DD analysis was performed as described in Materials and Methods. Using one particular combination of primers (5′-AGGTGACCGT-3′ upstream primer plus T12MT anchored primer), a band with elevated signal intensity was observed in the 4-h peroxide sample lane compared with control (Fig. 1A; this figure also shows a representative DD pattern). The upregulated band was then cut out of the gel, reamplified by PCR, cloned, and used to probe Northern blots containing RNA extracted from other HA-1 cultures also exposed to peroxide for 30 min, 90 min, and 4 h. A strong induction of an RNA species, designated adapt15, was observed on the Northern blot, confirming the original differential display result (Fig. 1B).
Figure 1.
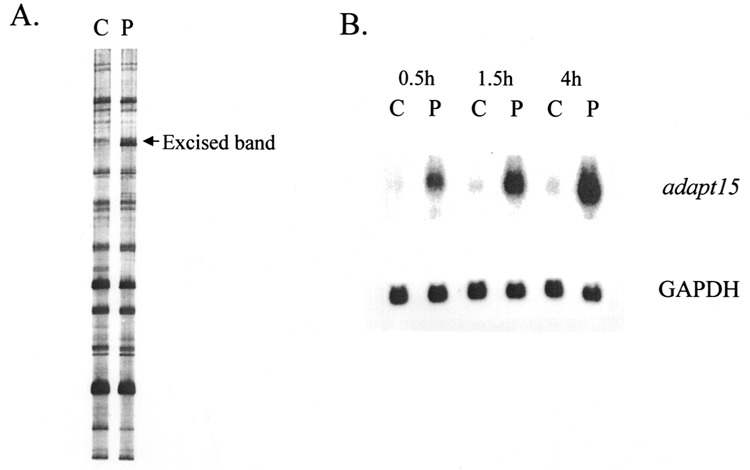
Example of using differential display to identify a modulated mRNA. (A) Representative differential display gel comparing the mRNA expression (as PCR DNA fragments) in control versus 4-h peroxide-treated HA-1 cells treated as described in Materials and Methods. C, control sample lane; P, peroxide-treated cell sample lane. Arrow denotes candidate band. (B) The candidate band denoted in (A) was excised, reamplified, cloned, radiolabeled, and used to probe a Northern blot containing RNA extracted from HA-1 hamster cells at multiple time points after peroxide exposure. The Northern blot was also probed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA as a loading control.
Other mRNA Species Were Also Induced by Hydrogen Peroxide
A comprehensive analysis of mRNA species induced by hydrogen peroxide was carried out using and combining the results of all three approaches described in Materials and Methods. These included using the complete combination of primers offered in both the original and the third-generation GenHunter DD kits as well as additional modifications. These additional modifications consisted of adding seven more upstream primers to the original kit analyses; of running parallel hot start PCR analyses with both kits; and of analyzing the PCR products at multiple earlier cycles besides the recommended 40 cycles to avoid plateau phase effects in one lane that might mute differences. In addition, an aggressive strategy was undertaken, excising and assessing confirmation on bands exhibiting only minor modulation on the DD gels. Combined, and including the additional modifications, these analyses should have assessed 90–100% of the total mRNA species in our HA-1 cells based on published statistical predictions (12,13). Overall, we identified, confirmed, sequenced, and reported the induction of five mRNAs by hydrogen peroxide: adapt15 (8), adapt33 (22), adapt66 (6), adapt73 (3), and adapt78 (5). The fold inductions for each of these adapt mRNAs are shown in Figure 2.
Figure 2.
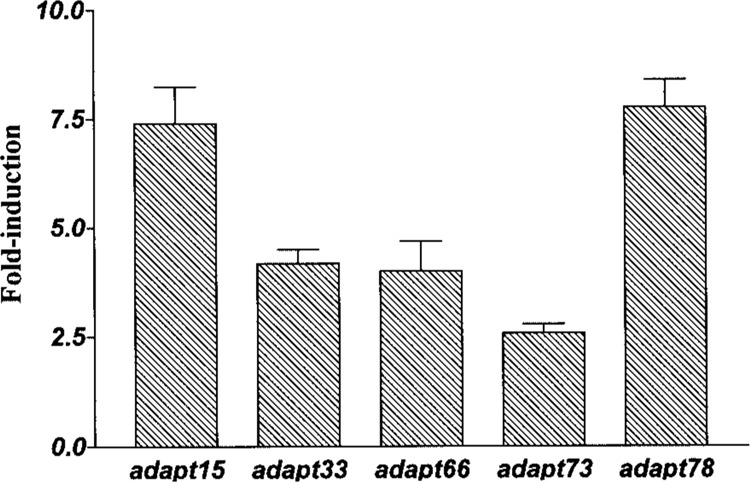
Fold inductions of adapt mRNA expression. Mean inductions for each individual adapt mRNA following exposure of HA-1 cells to 160 μM of hydrogen peroxide for 4 h. Data are expressed as the mean ± SEM (n = 4 or greater).
Other Peroxide-Induced mRNAs Were Not Identified by DD
Previous studies in a number of laboratories had identified a number of oxidant-inducible mRNAs using techniques other than DD (2). We used the cDNAs to six of these genes (heme oxygenase, gadd45, gadd153, c-fos, c-jun, and protein-tyrosine phosphatase CH134) to probe Northern blots containing the same RNA extracted and used for the above DD studies. We observed that five of these six mRNAs were significantly induced in the HA-1 cells by peroxide (Fig. 3). The fold inductions for each of these adapt mRNAs are shown in Figure 4. Thus, at best, only 5 out of 10 (and possibly more as yet undiscovered) or ∼50% of oxidant-inducible mRNAs were identified by DD, indicating that this technique is not comprehensive.
Figure 3.
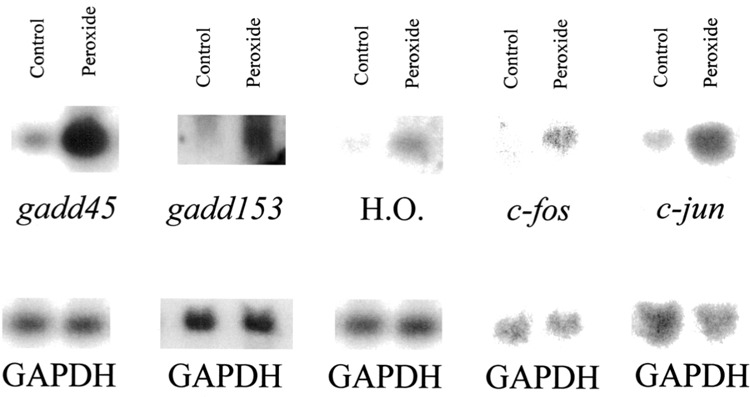
Other peroxide-induced mRNAs. HA-1 cells were treated with or without (control) 160 μM of hydrogen peroxide and RNA extracted at the appropriate time points. After electrophoresis and blotting, the nylon Northern blots were hybridized with cDNA probes to gadd45, gadd153, and heme oxygenase (H.O.), all to RNA extracted 4 h after initial peroxide exposure; with c-fos probe using RNA extracted 30 min after initial peroxide exposure; and with c-jun probe using RNA extracted 90 min after initial peroxide exposure. GAPDH was used as a loading control.
Figure 4.
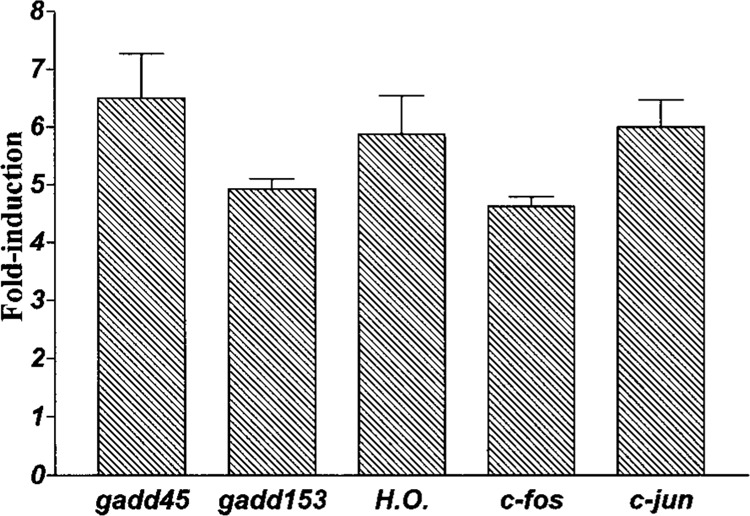
Fold inductions of other mRNAs. Mean inductions for each individual mRNA following exposure of HA-1 cells to 160 μM of hydrogen peroxide for the times indicated in Figure 3 legend. Data are expressed as the mean ± SEM (n = 3 or greater).
The unexpected absence of a DD identifiable and modulated mRNA was probably most prominent for gadd45 (Fig. 4), which is significantly induced and abundant in the peroxide-treated lane. We therefore designed two specific upstream primers for hamster gadd45 and performed DD. Still, no induced gadd45 band was observed, even when using the gadd45-specific primers in pair with any of the anchored primers.
Microarray Analysis
Because the above analyses indicated that DD is less comprehensive than previously thought, we decided to probe filter microarrays as a gauge of the extent of this discrepancy. Hybridization of human filter arrays containing over 5000 cDNAs was carried out using duplicate control and 4-h peroxide-treated HA-1 cell RNAs. A significant number of cDNAs cross hybridized, representing a wide cross section of intensity across the arrays as shown in Figure 5. We then analyzed for modulation of expression greater than twofold, which was the minimal fold modulation on DD gels that we used as criteria to subsequently pursue in follow-up studies (by Northern blot confirmation analysis), and also is less than the fold modulation we observed for all the adapt mRNAs and those mRNAs shown in Figure 3. No modulated species greater than twofold were observed by the array analyses.
Figure 5.
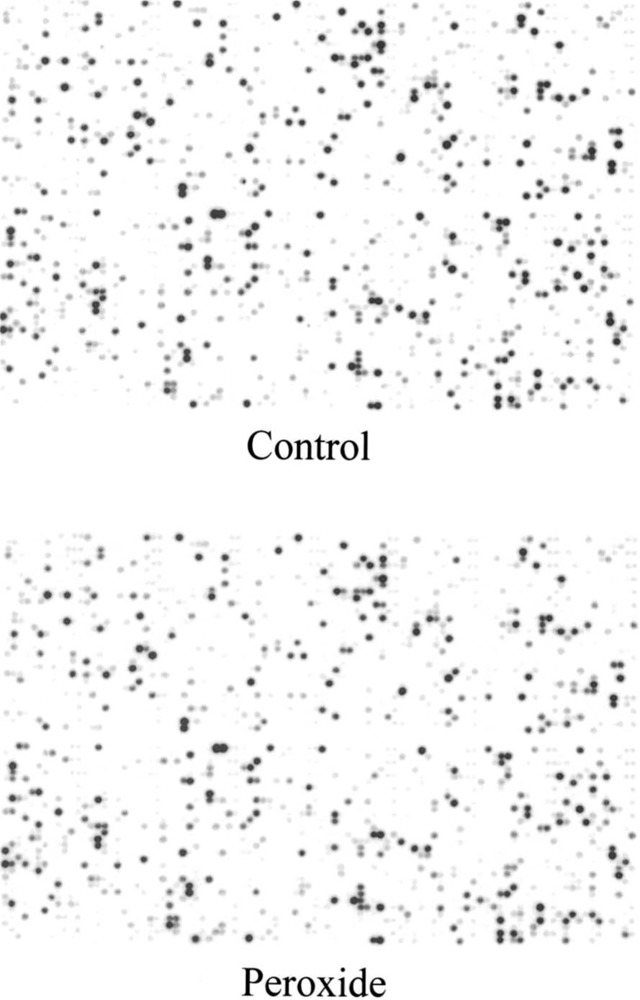
Microarray analysis. Human gene filter arrays containing 5184 noncontrol cDNAs were hybridized with control (A) and 4-h peroxide-treated (B) samples derived from HA-1 cells extracted as described. cDNA was prepared from 6 μg of RNA in the presence of 33P radiolabel and used to probe two separate human gene filter arrays. The figure images are representative array hybridization signals obtained following washing 2 × 20 min with 2× SSC plus 1% SDS at 50°C, then 0.5× SSC plus 1% SDS at 50°C for 30 min and phosphoimaging.
Previous Studies on Protein Synthesis
We have previously analyzed de novo protein synthesis following hydrogen peroxide using the same conditions as above (24). We observed that 15 pulse-labeled proteins were induced between 0 and 4 h post-hydrogen peroxide treatment (Fig. 6). Because this technique will only identify the more abundant species, this would represent a minimum. Thus, at least 15 new proteins and perhaps more were induced during the time period that the DD mRNA samples were obtained. We would therefore have expected to identify significantly more induced mRNA species by DD based on these protein expression results alone.
Figure 6.
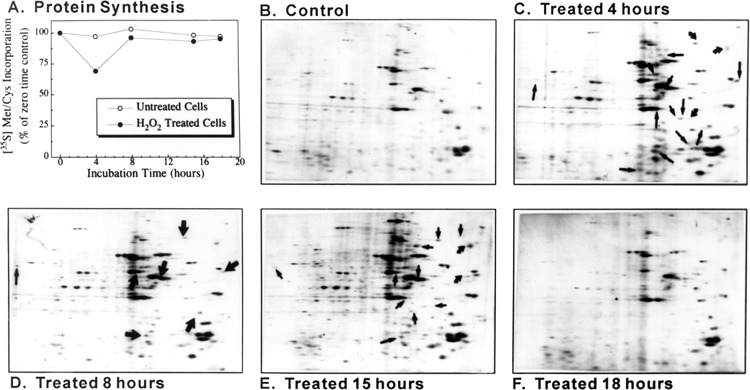
Two-dimensional gel electrophoresis. The publication and description of 2D protein gel electrophoresis was previously published by Wiese et al. (24). In summary, HA-1 cells were exposed to 150 μM hydrogen peroxide or solvent (control). At various times (4, 8, 15, and 18 h) after exposure, cells were pulse-chased with [35S]methionine/cysteine and processed for 2D protein gel electrophoresis. In (A), rates of protein synthesis were assessed by incorporation of the radiolabeled amino acids into acid-precipitable protein. (B–F) Representative 2D electrophoresis gels for control and peroxide-treated cells at various time points. Arrows in gels from peroxide-treated cells indicate some of the proteins whose synthesis increased following peroxide treatment.
DISCUSSION
No study testing the comprehensiveness of DD and the accuracy of statistical predictions has yet been reported. Here we empirically address this important question using the induction of oxidant-response genes as a model system. Based on the observed induction of most other oxidant-inducible mRNAs examined, our results, at least for this model system, demonstrate that the current statistical prediction of the number of DD primer combinations needed to achieve a comprehensive analysis of all the mRNAs in a given cell is inaccurate. However, this difference does not appear to be a gross underestimation based on our microarray analysis. That is, we did not observe any microarray spot (i.e., mRNA) exhibit a IP: 103.62.3 modulation of twofold or greater in response to peroxide, even though several thousand spots (representing expressing mRNAs) gave a hybridizable signal.
Our conclusion that DD is less than comprehensive is further supported by our previously published 2D gel electrophoresis results revealing the induction of synthesis of multiple proteins during this time. Here, we observed that at least 15 pulse-labeled proteins were induced between 0 and 4 h plus one new species (in addition to those observed 0–4 h) between 4 and 8 h post-hydrogen peroxide treatment (Fig. 6). We would therefore have expected to identify significantly more induced mRNA species by DD based on these protein expression results alone. There is a time delay between the time that elevation of an mRNA is observed and its subsequent protein synthesis. This might account for some of the discrepancy, although our DD analyses included multiple early time points. We expected elevated levels of the mRNAs corresponding to each protein spot would have existed at most of the time points during which mRNA sample was extracted for DD analysis.
The reasons for this lack of comprehensiveness are probably multiple and include considerations such as additional number of primer combinations. However, our results using gadd45-specific primers suggest that 100% comprehensive analysis is not possible regardless of the number of primer combinations used. Despite the successful use of this approach for identifying thymidine kinase cDNA by DD (13), the high level of gadd45 mRNA transcript in our peroxide-treated sample, and the use of two gadd45-specific primers, we were still unable to observe an induced gadd45 transcript by DD. This suggests that there is a selective resistance to the identification of certain sequences by DD, and may help explain the lack of a comprehensive mRNA detection.
Despite its drawbacks, DD remains popular due to its documented success and ability to identify novel genes not yet available for analysis by microarray hybridization. It will continue to be a valuable technique at least until, if not beyond, the point where high-throughput microarrays containing all known mRNAs are available. Our results demonstrate that comprehensive analyses using DD are limited, and that the current statistical prediction of the number of DD primer combinations needed to achieve a comprehensive analysis of all the mRNAs in a given cell is inaccurate, at least in our hands. We conclude that DD is a useful but less than comprehensive method for assessing changes in mRNA levels.
ACKNOWLEDGMENTS
We thank Drs. Al Fornace, Nikki Holbrook, Rex Tyrrell, and L. F. Lau for use of the gadd45, gadd153, heme oxygenase, and CH134 cDNA probes.
REFERENCES
- 1. Aittokallio T.; Ojala P.; Nevalainen T. J.; Nevalainen O. Automated detection of differently expressed fragments in mRNA differential display. Electrophoresis 22:1935–1945; 2001. [DOI] [PubMed] [Google Scholar]
- 2. Crawford D. R. Regulation of gene expression by reactive oxygen species. In: Gilbert D. L.; Colton C. A., eds. Reactive oxygen species in biological systems: An interdisciplinary approach. New York: Plenum Publishing; 1999:155–171. [Google Scholar]
- 3. Crawford D. R.; Davies K. J. A. Modulation of a cardiogenic shock-inducible RNA by chemical stress: adapt73/PigHep3. Surgery 121:581–587; 1997. [DOI] [PubMed] [Google Scholar]
- 4. Crawford D. R.; Edbauer-Nechamen C. A.; Lowry C. V.; Salmon S. L.; Kim Y. K.; Davies J. M. S.; Davies K. J. A. Assessing gene-expression during oxidative stress. Methods Enzymol. 234:175–217; 1994. [DOI] [PubMed] [Google Scholar]
- 5. Crawford D. R.; Leahy K. P.; Abramova N.; Lan L.; Wang Y.; Davies K. J. A. Hamster adapt78 mRNA is a Down syndrome critical region-homologue that is inducible by oxidative stress. Arch. Biochem. Biophys. 342:6–12; 1997. [DOI] [PubMed] [Google Scholar]
- 6. Crawford D. R.; Leahy K. P.; Wang Y.; Schools G. P.; Kochheiser J. C.; Davies K. J. Oxidative stress induces the levels of a MafG homolog in hamster HA-1 cells. Free Radic. Biol. Med. 21:521–525; 1996. [DOI] [PubMed] [Google Scholar]
- 7. Crawford D. R.; Schools G. P.; Davies K. J. A. Oxidant-inducible adapt15 RNA is associated with growth arrest- and DNA damage-inducible gadd153 and gadd45. Arch. Biochem. Biophys. 329:137–144; 1996. [DOI] [PubMed] [Google Scholar]
- 8. Crawford D. R.; Schools G. P.; Salmon S. L.; Davies K. J. A. Hydrogen peroxide induces the expression of adapt15, a novel RNA associated with polysomes in hamster HA-1 cells. Arch. Biochem. Biophys. 325:256–264; 1996. [DOI] [PubMed] [Google Scholar]
- 9. Fend F.; Raffeld M. Laser capture microdissection in pathology. J. Clin. Pathol. 53:666–672; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hua L. V.; Green M.; Warsh J. J.; Li P P. Molecular cloning of a novel isoform of diphosphoinositol poly-phosphate phosphohydrolase: A potential target of lithium therapy. Neuropsychopharmacology 24:640–651; 2001. [DOI] [PubMed] [Google Scholar]
- 11. Kroes R. A.; Jastrow A.; McLone M. G.; Yamamoto H.; Colley P.; Kersey D. S.; Yong V. W.; Mkrdichian E.; Cerullo L.; Leestma J.; Moskal J. R. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 156: 191–198; 2000. [DOI] [PubMed] [Google Scholar]
- 12. Liang P.; Bauer D.; Averboukh L.; Warthoe P.; Rohrwild M.; Muller H.; Strauss M.; Pardee A. B. Analysis of altered gene expression by differential display. Methods Enzymol. 254:304–321; 1995. [DOI] [PubMed] [Google Scholar]
- 13. Liang P.; Pardee A. B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967–971; 1992. [DOI] [PubMed] [Google Scholar]
- 14. Mills J. C.; Roth K. A.; Cagan R. L.; Gordon J. I. DNA microarrays and beyond: Completing the journey from tissue to cell. Nat. Cell Biol. 3:E175–E178; 2001. [DOI] [PubMed] [Google Scholar]
- 15. Pardinas J. R.; Combates N. J.; Prouty S. M.; Stenn K. S.; Parimoo S. Differential subtraction display: A unified approach for isolation of cDNAs from differentially expressed genes. Anal. Biochem. 257:161–168; 1998. [DOI] [PubMed] [Google Scholar]
- 16. Powell J. SAGE. The serial analysis of gene expression. Methods Mol. Biol. 99:297–319; 2000. [DOI] [PubMed] [Google Scholar]
- 17. Schulze A.; Downward J. Navigating gene expression using microarrays—a technology review. Nat. Cell Biol. 3:E190–E195; 2001. [DOI] [PubMed] [Google Scholar]
- 18. Sompayrac L.; Jane S.; Burn T. C.; Tenen D. G.; Danna K. J. Overcoming limitations of the mRNA differential display technique. Nucleic Acids Res. 23: 4738–4739; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spitz D. R.; Dewey W. C.; Li G. C. Hydrogen peroxide or heat shock induces resistance to hydrogen peroxide in Chinese hamster fibroblasts. J. Cell Physiol. 131:364–373; 1987. [DOI] [PubMed] [Google Scholar]
- 20. Velculescu V. E.; Zhang L.; Vogelstein B.; Kinzler K. W. Serial analysis of gene expression. Science 270: 484–487; 1995. [DOI] [PubMed] [Google Scholar]
- 21. Wang X.; Feuerstein G. Z. The use of mRNA differential display for discovery of novel therapeutic targets in cardiovascular disease. Cardiovasc. Res. 35:414–421; 1997. [DOI] [PubMed] [Google Scholar]
- 22. Wang Y.; Crawford D. R.; Davies K. J. A. adapt33, a novel oxidant-inducible RNA from hamster HA-1 cells. Arch. Biochem. Biophys. 332:255–260; 1996. [DOI] [PubMed] [Google Scholar]
- 23. Watson J. B.; Margulies J. E. Differential cDNA screening strategies to identify novel stage-specific proteins in the developing mammalian brain. Dev. Neurosci. 15:77–86; 1993. [DOI] [PubMed] [Google Scholar]
- 24. Wiese A. G.; Pacifici R. E.; Davies K. J. A. Transient adaptation to oxidative stress in mammalian cells. Arch. Biochem. Biophys. 318:231–240; 1995. [DOI] [PubMed] [Google Scholar]


