Abstract
Changes in gene expression represent a major protective mechanism, and enforced overexpression of individual genes has been shown to protect cells. However, no large-scale comparison of genes involved in mammalian oxidative stress protection has yet been described. Using filter microarray and restriction fragment differential display technology, hydrogen peroxide (H2O2)-resistant variants of hamster HA-1 fibroblasts and human HL-60 promyelocytes were found to possess a surprising lack of commonality in specific modulated genes with the single exception of catalase, supporting the hypothesis that catalase overexpression is critical for resistance to H2O2. Comparison of two cell lines from the same species (hamster) selected with an exogenous oxidative stressing agent (H2O2) and an endogenous metabolic oxidative stressing agent (95% O2) also revealed little commonality in modulation of specific mRNAs with the exception of glutathione S-transferase enzymes and catalase. Acute oxidative stress in HL-60 led to the modulation of a limited subset of the genes associated with chronic oxidative stress resistance. Overall, these results suggest that mammalian resistance to oxidative and perhaps other stress does not require a significant number of common genes but rather only a limited number of key genes (e.g., catalase in our model systems) in combination with others that are cell type and stress agent specific.
Keywords: Oxidative stress, Hydrogen peroxide, Chronic resistance, HA-1, HL-60, Catalase, Oxygen toxicity, Microarray
RESISTANCE to oxidative stress-induced damage is an important part of cellular and organismal homeostasis. Both prokaryotic and eukaryotic organisms have evolved a plethora of defensive strategies to protect themselves against the destructive effects of both chronic and acute stress. Changes in gene expression are thought to represent a major protective mechanism involved in stress responses. Furthermore, enforced expression of individual genes has been shown to provide some protection, though usually not as effectively as the global responses seen naturally. Elevated levels of Grp78, Grp94, heat shock protein 70, and heme oxygenase have all been shown to protect cells against stress-induced damage (2,10,13,20,21). Knowledge gained from the study of these modulated genes not only lends insight into the basic mechanisms of stress resistance, but may also offer potential advantages as clinical targets in treating cancer and other diseases.
In addition to classical antioxidant enzymes (catalase, glutathione peroxidases, and superoxide dismutases), several novel potential mechanisms by which cells become resistant to oxidative stress have been suggested, including increased expression of redoxsensitive transcription factors (1,8) and increased expression of aldehyde-metabolizing enzymes (36). In addition, it is also highly likely that other as yet undiscovered alterations in gene expression could contribute to resistance of oxidative stress. To date, however, the number of genes assessed for cytoprotection is relatively small and limited to a few specific model systems. This is due, at least in part, to a lack of studies comparing the modulation of genes in oxidative stress-resistant cell lines. Those few studies that attempted a comprehensive analysis of gene expression in oxidative stress-resistant cells have concentrated on one resistant phenotype due, at least in part, to the labor-intensive nature of gene analysis methodology. This has changed, however, with the advent of gene microarray technology. This technology now allows for the analysis of thousands of genes in a single hybridization experiment.
In order to rapidly probe for gene-based mechanisms responsible for mammalian cellular resistance to oxidative stress, the current study utilized filter microarray technology as a global screening tool. Gene expression was compared in three model systems where resistance to oxidative stress in known to occur, with the long-term goal of obtaining a better understanding of the genetic basis for resistance to oxidative stress. These model systems include comparing two cell lines from different species (hamster and human) selected for resistance to the same agent (H2O2); comparing two cell lines from the same species (hamster) selected for resistance to two different forms of chronic oxidative stress (H2O2 and 95% O2); and comparing chronic to acute oxidative stress responses. In addition, restriction fragment differential display (RFDD) was also used to gain additional insight into the potential genetic mechanisms by which mammalian cells establish and maintain an oxidative stress-resistant phenotype.
MATERIALS AND METHODS
Derivation of Resistant HL-60 Cells
HL-60, HP50, and HP100 cells were derived from HL-60 cells by repeated exposure to 50 and 100 μM of hydrogen peroxide followed by the outgrowth of viable cells. HP50 and HP100 were approximately 40- and 340-fold more resistant to hydrogen peroxide than the HL-60 cells, respectively (14). These cells were routinely maintained in RPMI medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) in a humidified incubator atmosphere of 95% air and 5% CO2 at 37°C.
Derivation of Oxidative Stress-Resistant Hamster Fibroblasts
H2O2-resistant (OC14) and 95% O2-resistant (O2R95) cell lines were isolated following chronic exposure (>200 days) of parental HA-1 cells to progressively increasing concentrations of H2O2 or O2 as previously described (11,34,36). All hamster cell lines were maintained in Eagle’s minimum essential medium supplemented with Earle’s basic salts, 10% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) in a humidified 5% CO2, 95% air atmosphere at 37°C. Cells were seeded at a density of 2–5 × 105 cells per 100-mm tissue culture dish and grown exponentially to 75–85% confluence (4–5 × 106 cells per 100-mm dish) prior to experimental treatment and harvest. Plating efficiencies of both cell lines were 60–70%.
Acute Oxidative Stress
For acute stress studies, HL-60 cells were passed at a density of 100,000 cells/ml. After 2 days of incubation, the cells were divided into two groups, one receiving phosphate-buffered saline (PBS) only (controls) and the other 100 μM of hydrogen peroxide. Cells were then returned to the incubator for the designated period of time (4 h) and RNA extracted as described below. A stock solution of hydrogen peroxide (30%, w/w) was diluted in PBS for all cell treatment experiments.
RNA Isolation and Analysis
For HL-60 and HP100 cell lines, the cells were grown to equivalent cell densities. At this time, the cultures were centrifuged, washed with PBS, and total RNA extracted using RNA extractor according to the manufacturer (Genosys, The Woodlands, TX). The final RNA pellet was resuspended in diethylpyro-carbonate-treated distilled, deionized water. The same protocol was used to extract RNA from HL-60 control and peroxide-treated cells following incubation for 4 h (see above).
For HA-1, OC14, and O2R95 cell lines total RNA was isolated from 80% confluent cell monolayers directly from the 100-mm culture plate using 1 ml of Tri-Reagent (MRC, Cincinnati, OH) according to the manufacturer’s instructions. Extracted samples were stored in 100% EtOH at −80°C until analysis.
Microarray Hybridization
Human gene filter microarrays obtained from Research Genetics (Research Genetics, Huntsville, AL) were used for these studies. These arrays contain 5184 noncontrol cDNAs derived from various human tissues. cDNA was prepared from total RNA (6 μg each) from duplicate samples obtained from cells as described above, and the arrays probed and washed as described by the manufacturer (Research Genetics). A final wash stringency of 50°C and 0.5× SSC, 1% SDS was used for HA-1 sample probings, and of 55°C and 0.5× SSC, 1% SDS for HL-60 sample probings. The arrays were then phosphoimaged using a Storm 860 phosphoimager with ImageQuant software (Molecular Dynamics, Riverdale, CA).
Microarray Analysis
Arrays were compared for spot intensity variation using the Pathways 3.0 software (Research Genetics). Values were normalized to all data points. Duplicate arrays were averaged and those exhibiting a twofold or greater modulation, and of which both values were either up- or downmodulated, were reported. Of these, false positives originating from the shadow effects of adjacent strong expressors or nonspecific background signal were eliminated by visual examination using the Thumbnail image window of Pathways.
A strict data filtering criteria was employed specifying a minimum ratio of normalized intensity of 1.5 for either induced specie duplicate value, or maximum ratio of 0.67 for either reduced specie duplicate value.
RNA Analyses
Northern blot preparation and hybridization was carried out as previously described (3,4). Briefly, extracted RNA was electrophoresed in a 1.1% agarose/ formaldehyde gel, then transferred to Gene Screen (NEN Research Products, Boston, MA) by capillary transfer in 10× SSC and UV cross-linked with a UV Stratalinker 1800 (Stratagene Cloning Systems, La Jolla, CA). Blots were then prehybridized at least 2 h, then hybridized in 50 mM sodium phosphate buffer (pH 6.8), 1× SET, 5× Denhardts, 0.5% SDS, and 200 μg/ml denatured, fragmented salmon sperm DNA at 65°C in a Hybaid hybridization oven (Laboratory Product Sales, Rochester, NY). The hybridization solution contained random primed (Oligolabeling kit, Pharmacia, Piscataway, NJ) radiolabeled probe added to fresh prehybridization solution. After overnight hybridization, the blots washed two times for 10 min each at room temperature with 2× SET plus 0.2% SDS followed by two more washes for 20 min each at 65°C with 0.4× SET plus 0.2% SDS. The blots were then phosphoimaged.
Western Blot Analyses
HL-60 cells, pelleted by centrifugation, or HA-1 monolayer cells were washed in PBS and lysed with 1 ml of 15 mM CHAPS detergent, 1 mM EDTA, 20 mM Tris-HCl, pH 7.5, and protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN) per 107 cells. The lysates were then mixed with an equal volume of 2× Laemmli sample buffer (17) and 30 μg (per lane) electrophoresed on a 15% SDS-polyacryl-amide gel. After electroblotting to nitrocellulose membrane according to the manufacturer (Schleicher and Schuell, Keene, NH), immunoblots were hybridized to a 1:875 dilution of rabbit polyclonal antibody prepared against bovine ubiquitin (StressGen, Victoria, BC, Canada). This antibody detects free ubiquitin and ubiquinated proteins on immunoblots. Ubiquitin signal was detected using horse radish peroxidase-conjugated secondary antibody followed by signal development with ECL Plus according to the manufacturer (Amersham Pharmacia Biotech, Arlington Heights, IL) and signal quantitation with a Storm 860 phosphoimager loaded with ImageQuant software (Molecular Dynamics).
RESULTS
Filter Microarray Comparison of Hydrogen Peroxide-Resistant Cells From Two Different Cell Types
The human gene filter microarrays were hybridized to probes derived from the RNA of hydrogen peroxide-resistant HA-1 and HL-60 cells as described in Materials and Methods. An example of these results is shown in Figure 1 for HL-60 control compared with HP100-resistant cells. In the H2O2-resistant cell line derived from HA-1 following chronic exposure to H2O2 (OC14), 14 mRNAs were found to be modulated twofold or greater, 5 downregulated, and 9 upregulated. In H2O2-resistant cell line derived from HL-60 cells following chronic exposure to H2O2 (HP100), 5 mRNAs were downregulated and 16 were upregulated twofold or more. These results are shown in Tables 1 and 2, respectively. Comparison of Tables 1 and 2 revealed, surprisingly, that none of the upregulated or downregulated mRNA species were common between the resistant cell types. Although hybridizable catalase was not present on these arrays, it is important to note that both peroxide-resistant cells lines are known to dramatically overexpress catalase (11,15,31). Previously reported catalase activities for OC14 and HP100 cells relative to controls are approximately 20- and 18-fold, respectively (15,31) and this increased activity was confirmed in the cultures used during the current study.
Figure 1.
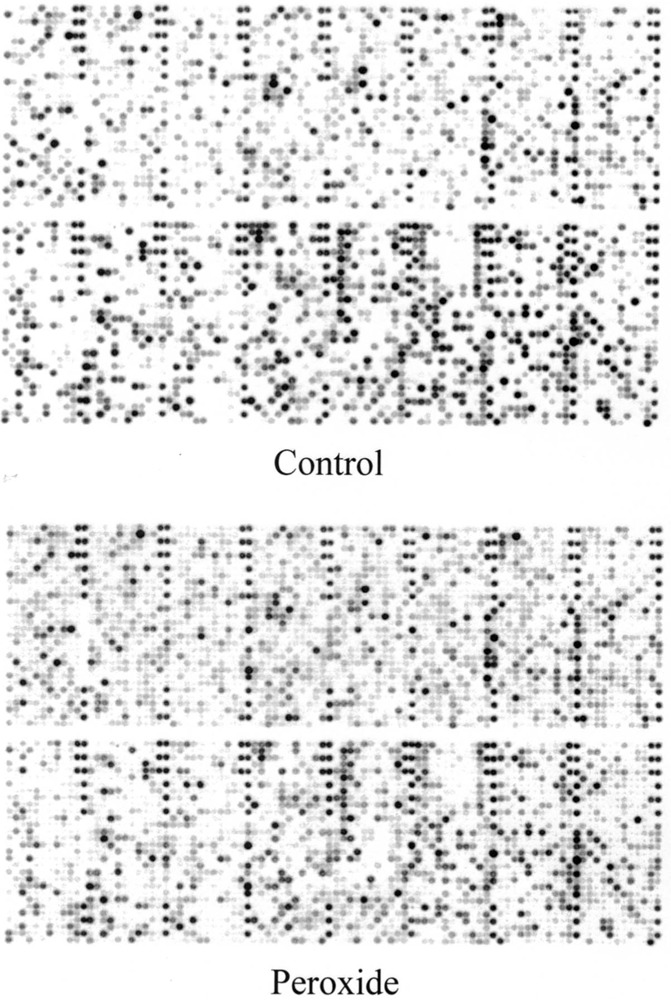
Example of microarray analysis. Human gene filter arrays containing 5184 noncontrol cDNAs were hybridized with probes obtained from HL-60 control and HP100-resistant cells. The cDNA probes were prepared from 6 μg of extracted total RNA in the presence of 33P radiolabel. After hybridization and washing, a final wash stringency of 55°C and 0.5× SSC, 1% SDS was employed. The arrays were then phosphoimaged using a Storm 860 phosphoimager with ImageQuant software.
TABLE 1.
MODULATED mRNA LEVELS IN HYDROGEN PEROXIDE-RESISTANT HA-1 CELLS DESIGNATED OC14
| Assession # | mRNA | Ratio |
|---|---|---|
| AA423944 | 37-kDa leucine-rich repeat (LRR) protein | 0.39 |
| R00707 | stearoyl-CoA desaturase (delta-9-desaturase) | 0.43 |
| AA293050 | mitogen-activated protein kinase kinase 4 | 0.47 |
| AA455316 | aryl hydrocarbon receptor-interacting protein | 0.48 |
| AA446108 | endoglin (Osler-Rendu-Weber syndrome 1) | 0.49 |
| AA456931 | cytochrome c oxidase subunit VIc | 2.03 |
| H50345 | succinate dehydrogenase complex, subunit A, flavoprotein | 2.03 |
| H57309 | slug (chicken homolog), zinc finger protein | 2.11 |
| H75599 | serum/glucocorticoid regulated kinase | 2.17 |
| T64625 | esterase D/formylglutathione hydrolase | 2.26 |
| R09815 | DKFZp586I1420 | 2.31 |
| W81684 | transcription elongation factor B (SIII), polypeptide 1 | 2.59 |
| T83829 | pim-1 oncogene | 2.92 |
| R33755 | glutathione S-transferase pi | 4.58 |
TABLE 2.
MODULATED mRNA LEVELS IN HYDROGEN PEROXIDE-RESISTANT HL-60 CELLS
| Assession # | mRNA | Ratio |
|---|---|---|
| R05886 | myeloperoxidase | 0.02 |
| AA431611 | thyroid hormone receptor interactor 7 | 0.24 |
| AA599178 | ribosomal protein L27a | 0.39 |
| AA410517 | protease inhibitor 6 (placental thrombin inhibitor) | 0.44 |
| AA424315 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 0.47 |
| R25823 | acetyl-Coenzyme A acetyltransferase 2 | 2.01 |
| AA188155 | actin related protein 2/3 complex, subunit 1B (41 kD) | 2.10 |
| T70122 | ATP-binding cassette, subfamily E (OABP), member 1 | 2.14 |
| N54914 | chromosome 15 open reading frame 3 | 2.20 |
| AA448396 | heat shock 10-kDa protein 1 (chaperonin 10) | 2.22 |
| AA465611 | ubiquitin specific protease 10 | 2.23 |
| R43325 | death associated protein 3 | 2.52 |
| AA598637 | chaperonin containing TCP1, subunit 4 (delta) | 3.38 |
| R61674 | protein tyrosine phosphatase type IVA, member 1 | 3.39 |
| H53340 | metallothionein 1G | 3.42 |
| R60317 | dihydrolipoamide dehydrogenase | 3.50 |
| N80129 | metallothionein 1L | 3.82 |
| H23075 | hydroxyacyl-coenzyme A dehydrogenase | 4.10 |
| H09590 | eukaryotic translation initiation factor 4A, isoform 1 | 4.37 |
| AA487812 | vimentin | 5.13 |
| AA458965 | natural killer cell transcript 4 | 13.24 |
Rows shown in bold also elevated by acute exposure to hydrogen peroxide.
The most dramatically modulated mRNA species identified by the array probings in the OC14 and HP100 H2O2-resistant cell lines were glutathione S-transferase (4.6-fold induction) and myeloperoxidase (49.5-fold reduction), respectively. Both observations are consistent with previous reports (15,35). Spitz et al. (33) have reported a fourfold increase in glutathione S-transferase enzymatic activity in OC14 cells using a nonisoform-specific substrate but the current finding shows this change in expression at the level of mRNA and indicates that the pi isoform is increased. Kasugai and Yamada (15) reported a dramatic decrease in myeloperoxidase enzymatic activity in HP100 cells that was not quantifiable due to a lack of detectable activity in the HP100 cells. In addition, several mRNAs coding for enzymes associated with oxidative energy metabolism, signal transduction, transcription, and proteolysis were also modulated in both the hamster and human H2O2-resistant cell lines, although the specific enzymes in each category were different between the two cell lines. There was also a strong induction of natural killer cell transcript 4 in the HP100 cell line (13.2-fold). However, the function of the protein product of this mRNA has not yet been reported.
Microarray Expression Profile Comparison of Hyperoxia-Resistant Cell Lines
Because the above results indicated a lack of specific common genes between different cell types from hamster and human origin with acquired resistance to peroxide, we also considered the comparative expression profiles of oxidant-resistant cells chronically adapted to a different oxidant but from the same cell type (hamster). The same filter microarrays were hybridized to probes derived from the RNA of O2R95 cells that were derived from HA-1 following chronic exposure to 95% O2. Hyperoxia is a known metabolic oxidative stress where the reactive species (superoxide and hydrogen peroxide) are believed to be derived from one-electron reductions of O2 from mitochondrial electron transport chains (11). Results indicated a surprising abundance of modulated mRNAs in O2R95 cells, relative to HA-1 (60 in total; see Table 3), especially those that are upregulated (44 in number). Also surprising was the nearly complete absence of commonly modulated mRNA species between the O2R95 cells and the OC14 hydrogen peroxide-resistant cells derived from the same parental cell line (compare with Table 1). The only exception was the elevated expression of glutathione S-transferase, albeit different isoforms, in both hyperoxia- and hydrogen peroxide-resistant HA-1 cells. Similar to the OC14 cells, the O2R95 cells have also been reported to have increased glutathione S-transferase activity (using the nonspecific substrate) and catalase activity and mRNA level (11,36).
TABLE 3.
MODULATED mRNA LEVELS IN HYPEROXIA-RESISTANT HA-1 CELLS
| Assession # | mRNA | Ratio |
|---|---|---|
| R43973 | eukaryotic translation elongation factor 1 gamma | 0.2 |
| H16958 | glyceraldehyde-3-phosphate dehydrogenase | 0.31 |
| R40850 | ARP1 (actin-related protein 1, yeast) homolog A (centractin alpha) | 0.32 |
| AA490047 | poly(rC)-binding protein 1 | 0.32 |
| R06311 | ESTs | 0.33 |
| H48420 | — | 0.34 |
| H77479 | EST | 0.27 |
| AA486626 | poly(A)-binding protein, cytoplasmic 1 | 0.35 |
| H65044 | ESTs | 0.35 |
| R54097 | RNA binding protein (autoantigenic) | 0.35 |
| AA115901 | cartilage linking protein 1 | 0.35 |
| H94236 | ESTs | 0.35 |
| AA428778 | ephrin-B1 | 0.37 |
| W03979 | ESTs | 0.37 |
| R00591 | — | 0.37 |
| AA031770 | pre-mRNA cleavage factor Im (25 kDa) | 0.38 |
| AA262988 | brain-derived neurotrophic factor | 2.12 |
| AAO18906 | transcription factor AP-2 beta | 2.17 |
| AA487452 | DNA fragmentation factor, 45 kDa, alpha subunit | 2.25 |
| AA490981 | CGI-28 protein | 2.44 |
| R52548 | superoxide dismutase 1, soluble ([myotrophic lateral sclerosis 1 (adult)] | 2.56 |
| H05580 | peptidylprolyl isomerase F (cyclophilin F) | 2.56 |
| AA448184 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 2.58 |
| AA504526 | low-density lipoprotein receptor defect C complementing | 2.59 |
| AA069414 | glial fibrillary acidic protein | 2.62 |
| AA490991 | heterogeneous nuclear ribonucleoprotein F | 2.76 |
| AA427472 | ATPase, H+ transporting, lysosomal noncatalytic accessory protein 1A | 2.97 |
| AA421977 | DR1-associated protein 1 (negative cofactor 2 alpha) | 3.01 |
| R63318 | hypothetical protein FLJ11137 | 3.01 |
| AA488233 | papillary renal cell carcinoma (translocation-associated) | 3.12 |
| AA453898 | sialyltransferase 4C (beta-galactosidase alpha-2,3-sialytransferase) | 3.24 |
| AA029041 | seven in absentia (Drosophila) homolog 2 | 3.30 |
| AA490493 | centaurin beta2 | 3.48 |
| W52803 | retinoblastoma-like 2 (pl30) | 3.92 |
| AA487034 | transforming growth factor, beta receptor II (70–80 kDa) | 3.94 |
| H53499 | ESTs, Weakly similar to Zn-finger-like protein (H. sapiens) | 3.96 |
| AA490124 | CGI-43 protein | 3.96 |
| AA290737 | glutathione S-transferase M4 | 4.19 |
| W17246 | peptidylprolyl isomerase E (cyclophilin E) | 4.30 |
| T72698 | splicing factor 3a, subunit 1, 120 kDa | 4.63 |
| AA452725 | nucleobindin 1 | 4.70 |
| AA456183 | CD151 antigen | 4.72 |
| N59851 | WAS protein family, member 1 | 4.74 |
| R78607 | deleted in oral cancer (mouse, homolog) 1 | 5.24 |
| AA404293 | triadin | 5.25 |
| AA281137 | KIAA0019 gene product | 5.34 |
| AA454743 | kallikrein 6 (neurosin, zyme) | 5.76 |
| AA455272 | ITBA1 gene | 5.85 |
| AA457199 | uncl 19 (C. elegans) homolog | 5.86 |
| AA397819 | DNA segment on chromosome 12 (unique) 2489 expressed sequence | 6.26 |
| W05628 | phosphoserine phosphatase-like | 6.37 |
| R91078 | cytochrome P450, subfamily IIIA, polypeptide 7 | 7.07 |
| T60223 | ribonuclease, RNase A family, 4 | 7.56 |
| R69153 | hypothetical protein FLJ20185 | 7.81 |
| R09561 | decay accelerating factor for complement (CD55) | 8.40 |
| T81764 | cell division cycle 27 | 8.46 |
| AA402960 | ring finger protein 5 | 8.64 |
| AA490771 | UV radiation resistance associated gene | 8.68 |
| T67474 | anaphase-promoting complex subunit 7 | 9.36 |
| R94775 | Homo sapiens clone 23596 mRNA sequence | 11.63 |
Other novel mRNA species modulated in the O2R95 cells, relative to the HA-1, of note include: Homo sapiens clone 23596 (11.6-fold), the protein product of which is homologous to endooligopeptidase A from human brain (9). This enzyme is thiol activated and thought to be involved in the maturation and degradation of neuropeptides. Anaphase-promoting complex (subunit 7 was elevated 9.4-fold) is a ubiquitin-protein ligase comprised of eight subunits that is important for progression through mitosis (7). UV radiation resistance associated gene (8.7-fold) partially complements the ultraviolet sensitivity of a xeroderma pigmentosa cell line (24) and is thought to be involved with DNA repair. Ring finger protein 5 contains a zinc-chelating domain thought to be involved in mediating protein–protein interactions (18). Also of interest is decay accelerating factor (8.4fold), a cell surface protein involved in cellular signaling and complement regulation, for which there is also evidence that it can act as a cytoprotectant (22); and Cu,Zn-superoxide dismutase (2.6-fold), another antioxidant gene in addition to glutathione S-transferase and catalase whose activity and immunoreactive protein have previously been reported to be elevated in O2R95 and OC14 (32,36). Finally, as was seen in the comparisons between the hamster and human H2O2-resistant cell lines, several mRNAs coding for proteins associated with oxidative energy metabolism, signal transduction, transcription, and the cell cycle were also modulated in the O2R95 cells, relative to HA-1 (Table 3), although the specific proteins in each category were different from those seen in the H2O2-resistant cell lines.
Although there were far fewer downregulated mRNAs (16) in O2R95 cells it is noteworthy that four of them (polyrC-binding protein 1, polyA-binding protein, RNA-binding protein, and pre-mRNA cleavage factor Im) are all nucleic acid binding proteins. The latter one appears to be involved in the processing of mRNA precursors (25).
Microarray Expression Profile Comparison: Acute Stress
We also performed microarray gene expression analysis on RNA extracted from HL-60 cells treated with a single acute dose of hydrogen peroxide (100 μM for 4 h). Comparison with the HP100 chronically adapted H2O2-resistant cells revealed three common modulations. These include heat shock chaperonin 10 (2.57-fold induction by acute peroxide), chromosome
15 open reading frame 3 (2.32-fold induction), and eukaryotic translation initiation factor 4A (2.02-fold induction). The three genes modulated by acute as well as chronic exposure to H2O2 in HL-60 cells are highlighted in Table 2.
Restriction Fragment Differential Display (RFDD)
Ubiquitin
Human RFDD was performed on HL-60 control and HP100 hydrogen peroxide-resistant RNAs extracted from log-phase unstressed cell cultures. Several mRNAs exhibited altered levels of expression, three of which were strongly modulated. One strongly modulated species was cloned and sequenced and found to encode ubiquitin. Ubiquitin RNA exists as three different forms: A, B, and C. The A form is further subdivided into ribosomal protein-conjugated sequences designated A-52 and A-80 (17). In HL-60 cells, all of these forms are expressed comparably. In the hydrogen peroxide-resistant HP50 and HP100 cell lines, however, there was little if any detectable signal in the B variant form compared with A and C (Fig. 2). Based on this dramatic reduction of ubiquitin B transcript in resistant HL-60 cells, we performed Western blot analysis to determine whether this reduction is also reflected in its translation product, a small 76-amino acid protein (17). As shown in Figure 3A (two separate analyses), no significant reduction in ubiquitin protein was observed. Our antibody also detects other ubiquinated proteins because these represent targets to which ubiquitin has attached. Comparison within lanes, and between the two Figure 3A experiments, were therefore also performed and revealed no obvious overall decrease in the ubiquination of other proteins in the lanes of the resistant phenotypes. Thus, the possible involvement of ubiquitin protein and target ubiquitination in peroxide resistance demonstrated by the cell lines is unlikely. More likely, a novel ubiquitin RNA effect is involved of yet unknown mechanism. The ubiquitin B-specific reduction presumably reflects the known variation in this transcript at the 5′ and 3′ untranslated region of ubiquitin B compared with A and C. Interestingly, distinct regulatory responses for the different ubiquitin genes have been reported previously (16,23).
Figure 2.
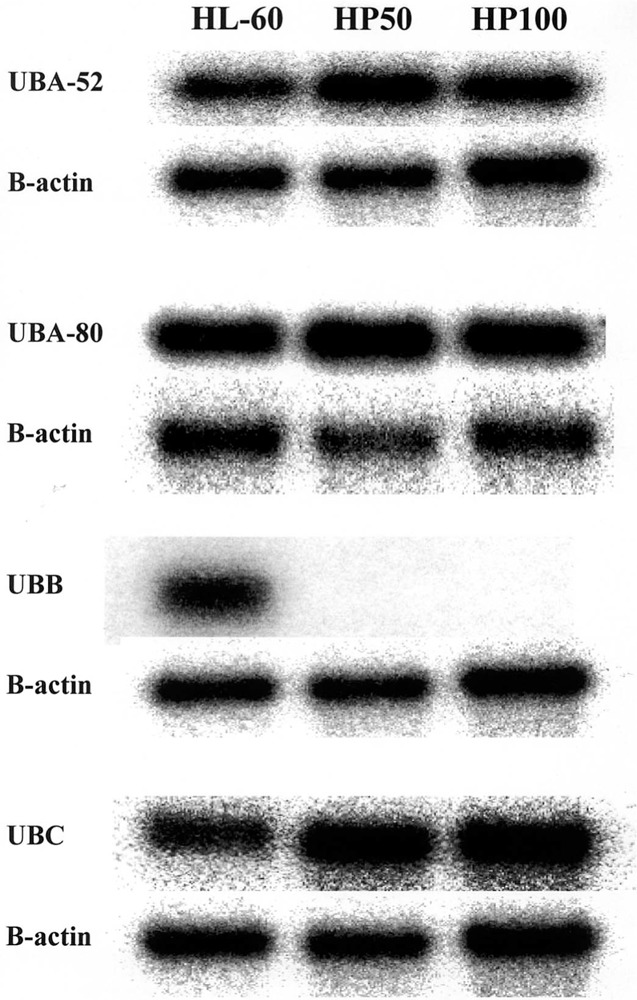
Expression of ubiquitin mRNA in HL-60 cells. Northern blot analysis of ubiquitin, identified by RFDD as a modulated mRNA. Extracted RNAs from HL-60 control, HP50, and HP100 log-phase unstressed cell cultures were electrophoresed, blotted to nylon paper, and then hybridized to radiolabeled ubiquitin cDNA probes specific for each ubiquitin variant indicated. β-Actin was used as a loading control.
Figure 3.
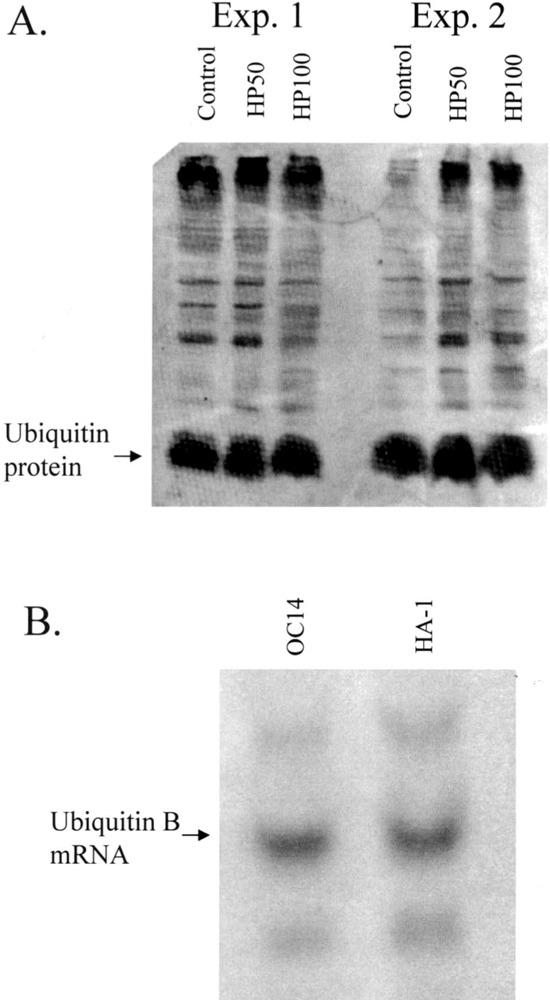
Expression of ubiquitin in oxidative stress-resistant human and hamster cells. (A) Western blot analysis of HL-60 versus resistant sublines. 30 μg of protein per lane was electrophoresed on a 15% SDS-polyacrylamide gel, electroblotted, and hybridized to a rabbit polyclonal antibody to bovine ubiquitin. Ubiquitin signal was detected using horse radish peroxidase-conjugated secondary antibody followed by signal development with ECL Plus and signal quantitation with a Storm 860 phosphoimager and ImageQuant software. Results are shown for two different experiments (Exp. 1 and Exp. 2). (B) Northern blot analysis of HA-1 versus OC14 subline. RNA extracted from HA-1 and OC14 cells was prepared, electrophoresed, blotted, and hybridized as above using radiolabeled ubiquitin as a probe.
To determine whether the observed downmodulation of ubiquitin mRNA level was specific to HL-60 cells, the RNA from peroxide-resistant OCl4 and control HA-1 cells was probed for ubiquitin. A similar strong downregulation of ubiquitin variant mRNAs was not observed between these two cell lines (Fig. 3B), indicating that this effect is not a general characteristic common to different oxidative stress-resistant phenotypes. Thus, as with genes identified by the above microarray analyses, the expression profile for RFDD identified ubiquitin as a gene modulated by H2O2 in oxidative stress-resistant HL-60 human cells, but this effect was not common to oxidative stress-resistant hamster cells.
Catalase and Myleoperoxidase
Two other strongly modulated mRNAs species were also identified in HP100 cells using RFDD. Cloning and sequencing revealed that the upregulated species was catalase and the downregulated was myeloperoxidase. Northern blot analysis (Fig. 4) confirmed these modulations, consistent with earlier reports that catalase and myeloperoxidase (MPO) activities in this cell line are significantly altered (15).
Figure 4.
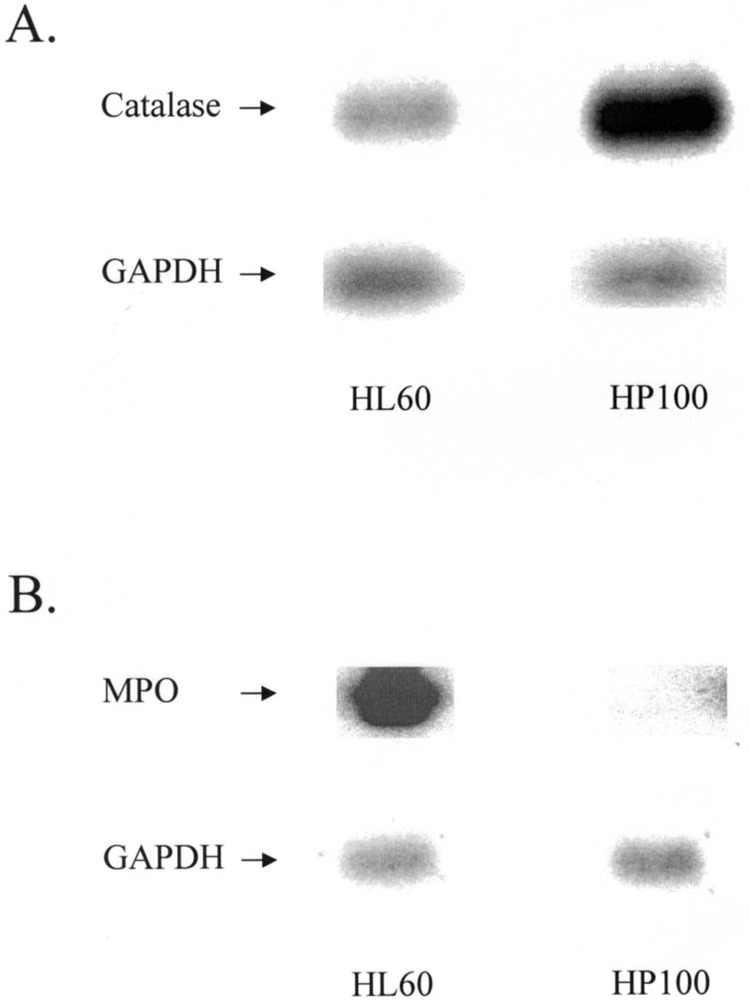
Northern blot confirmation of RFDD-identified catalase and myleoperoxidase. cDNAs to catalase and myleoperoxidase (MPO) were radiolabeled and used to probe Northern blots containing RNA from HL-60 and HP100 cells. GAPDH was used as a loading control.
DISCUSSION
In the current studies, the genetic basis of oxidative stress resistance was assessed in detail using micro-array technology. This study represents the first large-scale comparison of mammalian genes involved in oxidant stress resistance using these technologies. The most striking observation from these analyses is the relative paucity of specific mRNAs being modulated between the different resistant sublines. This surprising observation occurred despite the fact that catalase, myeloperoxidase, glutathione S-transferase, and superoxide dismutase were modulated in one cell line or another, all consistent with previous reports (1,8,15).
The implication of these results is that only a limited number of common key genes, and in our case only one (catalase), play a central role in chronic oxidative stress resistance, at least between these two cell types, and that other genes involved in resistance are more cell type and stress agent specific. Our data suggest that these latter genes include common activities such as oxidative energy metabolism, signal transduction, transcription, and proteolysis, although the specific enzymes in each category are different. In HA-1 cells, the upregulation of glutathione S-transferase in both OC14 and O2R95 cells suggests that this enzyme is important in resistance via the well-known ability of these enzymes to aid in the metabolic detoxification of cytotoxic byproducts of oxidative stress, such as aldehydes and hydroperoxides. This hypothesis has been supported by previous results using depletion of glutathione to sensitize these hamster cells to aldehydes and hydroperoxides implicated in oxidative stress (33,35). Aside from these examples, the lack of specific genes being modulated in a similar fashion when comparing OC14 with O2R95 hamster cell lines suggests that mechanisms of resistance are peculiar to the source of the oxidative stress (i.e., exogenous administration vs. endogenous metabolic oxidative stress). In HP100 human cells, the dramatic downregulations of myeloperoxidase and ubiquitin B RNA may be an indication that these genes are especially important in resistance to hydrogen peroxide in HL-60 cells and this has been confirmed for myeloperoxidase (38). It is also interesting to note that ubiquitin is known to be modulated by oxidative stress (12,29). Furthermore, a recent report demonstrates a new role for ubiquitin—activation of transcription—that is separate from protein degradation (27). Our data suggest yet another possible “new” role involving ubiquitin RNA and not its protein product in resistance.
Several models of stable oxidative stress-resistant cell lines have been developed following chronic exposure to oxidative stress. Most were developed as hydrogen peroxide-resistant lines, but others include hyperbaric oxygen-, redox quinone-, and excitotoxin-resistant lines as well (5). These lines were all selected following chronic exposure to oxidative stress and demonstrated stable resistant phenotypes. In addition to the OC14, O2R95, and HP100 cell lines, studies on these other model systems also support a central role for catalase in chronic resistance to oxidative stress. Chinese hamster cell line R-8, which exhibits a 10-fold resistance to hydrogen peroxide compared with control cells, exhibit a corresponding 10-fold increase in catalase activity but not superoxide dismutase, glutathione peroxidase, or glutathione reductase, suggesting that resistance is associated with the enhanced catalase activity (28). In neonatal cardiac myocytes, exposure to continuously generated hydrogen peroxide results in catalase mRNA induction but not superoxide dismutase or selenium-dependent glutathione peroxidase (19). rC18 cells are a PC12 pheochromocytoma neuronal-like cell line derivative developed by long-term exposure to the Alzheimer’s disease-derived amyloid beta protein (26). This protein apparently causes the generation of hydrogen peroxide in culture and, expectedly, the rC18 cells also become resistant to hydrogen peroxide and t-butyl hydroperoxide-mediated cell killing. These cells exhibit highly elevated levels of catalase and glutathione peroxidase but not the superoxide dismutases. Another neuronal model of oxidative stress resistance was developed by exposing mouse nerve cell line HT-22 to the excitotoxin glutamate, a neurotransmitter known to lead to the generation of reactive oxygen species in target cells (26). Again in this model system, catalase levels were elevated in the stress-resistant cells but not glutathione peroxidase or superoxide dismutase.
Another intriguing aspect of altered gene expression in response to oxidative stress is the differences between acute vs. chronic exposures. Modulations in gene expression that occur transiently in response to acute oxidative stress appear to involve a subset of the alterations that occur following chronic exposure, possibly indicating differences in the mechanisms by which mammalian cells adapt to acute vs. chronic oxidative stress. However, the relationship between transient and stable resistance has been largely ignored in mammalian cells. Here, we observe that three genes present in chronically hydrogen peroxide-resistant HP100 cells were also modulated by acute exposure to hydrogen peroxide, and in the same direction. These genes include heat shock chaperonin 10, chromosome 15 open reading frame 3, and eukaryotic translation initiation factor 4A. The induction of heat shock chaperonin 10, a mitochondrial matrix protein chaperone, could represent an important protective mechanism because, in combination with heat shock protein 60, chaperonin 10 has been demonstrated to confer protection against the oxidative stress produced by ischemia-induced injury in rat cardiomyoctes (19). Similar changes in gene expression have also been observed in control cells transiently exposed to menadione and long-term stable menadione-resistant cells (37). These results suggest that the acquisition of a stable oxidative stress-resistant phenotype may in part represent the stable integration of gene expression changes that normally occur transiently in cells exposed to acute episodes of oxidative stress. This slowly evolving stable integration of gene expression changes might also relate to the wide variety of changes in signal transduction-associated genes and genes associated with transcriptional regulation noted in the chronic oxidative stress-resistant cells analyzed in the current study and in previous studies (1,8). Perhaps changes in these signaling molecules and transcription factors are related to stably redirecting metabolic processes to limit the production of reactive species produced as by-products of oxidative energy metabolism as well as increasing the capacity of the cellular detoxification and repair processes to cope with the continuous accumulation of oxidative damage.
Overall, as expected, elevations in at least some anti-oxidant enzymes levels were observed in the stable oxidant-stress cell lines. These include glutathione S-transferase, superoxide dismutase, and catalase. Because they all act to detoxify reactive oxygen species, it would be expected that their induction would contribute to resistance against chronic oxidative stress as experienced by these cells. The very strong downregulation of myeloperoxidase (MPO) also would be expected to minimize the levels of toxic oxidized halogens produced by the oxidations catalyzed by this enzyme in the presence of hydrogen peroxide, and this hypothesis has been supported by recent observations (38). Also this downregulation of MPO represents another example of a metabolic biochemical pathway specific to the HL-60 cell lineage, which appears to be downregulated in order to limit the production of toxic by-products of oxidative metabolism during chronic oxidative stress.
Identification of oxidative stress-associated genes may be valuable not only in understanding basic mechanisms of resistance, but also in the identification of clinically relevant targets for manipulation of disease progression. Many human disease states are currently thought to involve chronic oxidative stress. Therefore, novel oxidative stress-associated gene regulation might be expected to contribute to cellular attempts to limit disease progression by 1) limiting the metabolic production of reactive species, 2) enhancing the detoxification of reactive species once formed, 3) enhancing the repair of oxidative damage to lipids, proteins, or nucleic acids, and/or 4) altering cellular signaling processes in order to reestablish a viable nonequilibrium steady state capable of supporting basic cellular processes in the face of chronic oxidative stress. In fact, oxidative stress-associated gene expression and the resulting protein products have been implicated in many clinically relevant conditions including ischemia–reperfusion, stroke, cardiovascular disease, cancer, inflammation, trauma, aging, autoimmunity, cystic fibrosis, diabetes, hearing loss, infectious disease, muscular dystrophy, periodontal disease, and hypertrophy (5,6,30). Gene expression changes involved in chronic oxidative stress-resistant phenotypes, such as those identified by the current study, may therefore represent potential useful clinical targets for manipulating pathological conditions that involve oxidative stress. Because ours is the first large-scale comparison of genes involved in mammalian oxidative stress protection, it will be interesting to determine whether future studies in other oxidative stress model systems lead to similar conclusions toward ultimately selecting the best genes for clinical targeting.
ACKNOWLEDGMENTS
The authors would like to thank Ms. Julia E. Sim for technical assistance in cell culture and RNA isolation. The studies were supported by NIH grants RO3HD35673 (D.R.C.) and RO1HL51469 (D.R.S.).
REFERENCES
- 1. Bradbury C. M.; Locke J. E.; Wei S. J.; Rene L. M.; Karimpour S.; Hunt C.; Spitz D. R.; Gius D. Increased activator protein 1 activity as well as resistance to heat-induced radiosensitization, hydrogen peroxide, and cisplatin are inhibited by indomethacin in oxidative stress-resistant cells. Cancer Res. 61:3486–3492; 2001. [PubMed] [Google Scholar]
- 2. Brostrom C. O.; Brostrom M. A. Regulation of translational initiation during cellular responses to stress. Prog. Nucleic Acid Res. Mol. Biol. 58:79–125; 1998. [DOI] [PubMed] [Google Scholar]
- 3. Crawford D. R.; Edbauer-Nechamen C. A.; Lowry C. V.; Salmon S. L.; Kim Y. K.; Davies J. M. S.; Davies K. J. A. Assessing gene-expression during oxidative stress. Methods Enzymol. 234:175–217; 1994. [DOI] [PubMed] [Google Scholar]
- 4. Crawford D. R.; Schools G. P.; Davies K. J. A. Oxidant-inducible adapt15 RNA is associated with growth arrest- and DNA damage-inducible gadd153 and gadd45. Arch. Biochem. Biophys. 329:137–144; 1996. [DOI] [PubMed] [Google Scholar]
- 5. Crawford D. R.; Suzuki S.; Davies K. J. A. Oxidant-modulated gene expression. In: Sen C. K.; Sies H.; Bauerle P. A. (eds.), Reactive oxygen species in biological systems: An interdisciplinary approach. San Diego, CA: Academic Press, 2000:21–45. [Google Scholar]
- 6. Floyd R. A. Role of oxygen free radicals in carcinogenesis and brain ischemia. FASEB J. 4:2587–2597; 1990. [PubMed] [Google Scholar]
- 7. Grossberger R.; Gieffers C.; Zachariae W.; Podtelejnikov A. V.; Schleiffer A.; Nasmyth K.; Mann M.; Peters J. M. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J. Biol. Chem. 274:14500–14507; 1999. [DOI] [PubMed] [Google Scholar]
- 8. Guyton K. Z.; Spitz D. R.; Holbrook N. J. Expression of stress response genes GADD153, c-jun, and heme oxygenase-1. Free Radic. Biol. Med. 20:735–741; 1996. [DOI] [PubMed] [Google Scholar]
- 9. Hayashi M. A.; Portaro F. C.; Tambourgi D. V.; Sucupira M.; Yamane T.; Fernandes B. L.; Ferro E. S.; Reboucas N. A.; de Camargo A. C. Molecular and immunochemical evidences demonstrate that endooligopeptidase A is the predominant cytosolic oligopeptidase of rabbit brain. Biochem. Biophys. Res. Commun. 269:7–13; 2000. [DOI] [PubMed] [Google Scholar]
- 10. Holwell T. A.; Schweitzer S. C.; Evans R. M. Tetra-cycline regulated expression of vimentin in fibroblasts derived from vimentin null mice. J. Cell Sci. 110:1947–1956; 1997. [DOI] [PubMed] [Google Scholar]
- 11. Hunt C. R.; Sim J. E.; Sullivan S. J.; Featherstone T.; Golden W.; Kapp-Herr C.; Hock R. A.; Gomez R. A.; Parsian A. J.; Spitz D. R. Genomic instability and catalase gene amplification induced by chronic exposure to oxidative stress. Cancer Res. 58:3986–3992; 1998. [PubMed] [Google Scholar]
- 12. Jahngen-Hodge J.; Obin M. S.; Gong X.; Shang F.; Nowell T. R. Jr.; Gong J.; Abasi H.; Blumberg J.; Taylor A. Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J. Biol. Chem. 272:28218–28226; 1997. [DOI] [PubMed] [Google Scholar]
- 13. Jamora C.; Dennert G.; Lee A. S. Inhibition of tumor progression by suppression of stress protein GRP78/BiP induction in fibrosarcoma B/C10ME. Proc. Natl. Acad. Sci. USA 93:7690–7694; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kasugai I.; Yamada M. Adaptation of human leukemia HL-60 cells to hydrogen peroxide as oxidative stress. Leuk. Res. 13:757–762; 1989. [DOI] [PubMed] [Google Scholar]
- 15. Kasugai I.; Yamada M. High production of catalase in hydrogen peroxide-resistant human leukemia HL-60 cell lines. Leuk. Res 16:173–179; 1992. [DOI] [PubMed] [Google Scholar]
- 16. Kemp L. M.; Latchman D. S. The herpes simplex virus type 1 immediate-early protein ICP4 specifically induces increased transcription of the human ubiquitin B gene without affecting the ubiquitin A and C genes. Virology 166:258–261; 1988. [DOI] [PubMed] [Google Scholar]
- 17. Kirschner L. S.; Stratakis C. A. Structure of the human ubiquitin fusion gene Uba80 (RPS27a) and one of its pseudogenes. Biochem. Biophys. Res Commun. 270:1106–1110; 2000. [DOI] [PubMed] [Google Scholar]
- 18. Kyushiki H.; Kuga Y.; Suzuki M.; Takahashi E.; Horie M. Cloning, expression and mapping of a novel RING-finger gene (RNF5), a human homologue of a putative zinc-finger gene from Caenorhabditis elegans . Cytogenet. Cell Genet. 79:114–117; 1997. [DOI] [PubMed] [Google Scholar]
- 19. Lai C. C.; Peng M.; Huang L.; Huang W. H.; Chiu T. H. Chronic exposure of neonatal cardiac myocytes to hydrogen peroxide enhances the expression of catalase. J. Mol. Cell. Cardiol. 28:1157–1163; 1996. [DOI] [PubMed] [Google Scholar]
- 20. Lee P. J.; Alam J.; Wiegand G. W.; Choi A. M. Overexpression of heme oxygenase-1 in human pulmonary epithelial cells results in cell growth arrest and increased resistance to hyperoxia. Proc. Natl. Acad. Sci. USA 93:10393–10398; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H.; Bowes R. C., 3; van de Water B.; Sillence C.; Nagelkerke J. F.; Stevens J. L. Endoplasmic reticulum chaperones GRP78 and calreticulin prevent oxidative stress, Ca2+ disturbances, and cell death in renal epithelial cells. J. Biol. Chem. 272:21751–21759; 1997. [DOI] [PubMed] [Google Scholar]
- 22. Mason J. C.; Yarwood H.; Sugars K.; Morgan B. P.; Davies K. A.; Haskard D. O. Induction of decay-accelerating factor by cytokines or the membrane-attack complex protects vascular endothelial cells against complement deposition. Blood 94:1673–1682; 1999. [PubMed] [Google Scholar]
- 23. Nenoi M.; Mita K.; Ichimura S.; Cartwright I. L.; Takahashi E.; Yamauchi M.; Tsuji H. Heterogeneous structure of the polyubiquitin gene UbC of HeLa S3 cells. Gene 175:179–185; 1996. [DOI] [PubMed] [Google Scholar]
- 24. Perelman B.; Dafni N.; Naiman T.; Eli D.; Yaakov M.; Feng T. L.; Sinha S.; Weber G.; Khodaei S.; Sancar A.; Dotan I.; Canaani D. Molecular cloning of a novel human gene encoding a 63-kDa protein and its sublocalization within the 11q13 locus. Genomics 41:397–405; 1997. [DOI] [PubMed] [Google Scholar]
- 25. Ruegsegger U.; Blank D.; Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1:243–253; 1998. [DOI] [PubMed] [Google Scholar]
- 26. Sagara Y.; Dargusch R.; Klier F. G.; Schubert D.; Behl C. Increased antioxidant enzyme activity in amyloid beta protein-resistant cells. J. Neurosci. 16:497–505; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salghetti S. E.; Caudy A. A.; Chenoweth J. G.; Tansey W. P. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651–1653; 2001. [DOI] [PubMed] [Google Scholar]
- 28. Sawada M.; Sofuni T.; Ishidate M. Jr. Induction of chromosomal aberrations in active oxygen-generating systems. II. A study with hydrogen peroxide-resistant cells in culture. Mutat. Res. 197:133–140; 1988. [DOI] [PubMed] [Google Scholar]
- 29. Shang F.; Gong X.; Taylor A. Activity of ubiquitin-dependent pathway in response to oxidative stress. Ubiquitin-activating enzyme is transiently upregulated. J. Biol. Chem. 272:23086–23093; 1997. [DOI] [PubMed] [Google Scholar]
- 30. Sies H. Oxidative stress: From basic research to clinical application. Am. J. Med. 91:31S–38S; 1991. [DOI] [PubMed] [Google Scholar]
- 31. Spitz D. R.; Adams D. T.; Sherman C. M.; Roberts R. J. Mechanisms of cellular resistance to hydrogen peroxide, hyperoxia, and 4-hydroxy-2-nonenal toxicity: The significance of increased catalase activity in H2O2-resistant fibroblasts. Arch. Biochem. Biophys. 292:221–227; 1992. [DOI] [PubMed] [Google Scholar]
- 32. Spitz D. R.; Elwell J. H.; Sun Y.; Oberley L. W.; Oberley T. D.; Sullivan S. J.; Roberts R. J. Oxygen toxicity in control and H2O2-resistant Chinese hamster fibroblast cell lines. Arch. Biochem. Biophys. 279:249–260; 1990. [DOI] [PubMed] [Google Scholar]
- 33. Spitz D. R.; Kinter M. T.; Roberts R. J. Contribution of increased glutathione content to mechanisms of oxidative stress resistance in hydrogen peroxide resistant hamster fibroblasts. J. Cell Physiol 165:600–609; 1995. [DOI] [PubMed] [Google Scholar]
- 34. Spitz D. R.; Li G. C.; McCormick M. L.; Sun Y.; Oberley L. W. Stable H2O2-resistant variants of Chinese hamster fibroblasts demonstrate increases in catalase activity. Radiat. Res. 114:114–124; 1988. [PubMed] [Google Scholar]
- 35. Spitz D. R.; Malcolm R. R.; Roberts R. J. Cytotoxicity and metabolism of 4-hydroxy-2-nonenal and 2-nonenal in H2O2-resistant cell lines. Do aldehydic by-products of lipid peroxidation contribute to oxidative stress? Biochem. J. 267:453–459; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sullivan S. J.; Oberley T. D.; Roberts R. J.; Spitz D. R. A stable O2-resistant cell line: Role of lipid peroxidation byproducts in O2-mediated injury. Am. J. Physiol. 262:L748–L756; 1992. [DOI] [PubMed] [Google Scholar]
- 37. Vallis K. A.; Wolf C. R. Relationship between the adaptive response to oxidants and stable menadione-resistance in Chinese hamster ovary cell lines. Carcinogenesis 17:649–654; 1996. [DOI] [PubMed] [Google Scholar]
- 38. Wagner B. A.; Buettner G. R.; Oberley L. W.; Darby C. J.; Burns C. P. Myeloperoxidase is involved in H2O2-induced apoptosis of HL-60 human leukemia cells. J. Biol. Chem. 275:22461–22469; 2000. [DOI] [PubMed] [Google Scholar]


