Abstract
Growth and transcription factors provide important developmental cues to neural crest-derived precursors of enteric neurons. The basic helix–loop–helix transcription factors, HAND2 and HAND1, are expressed in the gastrointestinal tract, but neither the growth factors that induce their expression nor the cell types that express them in the gut are known. We show that transcripts encoding HAND2 are expressed in all segments of the developing gut while those encoding HAND1 are confined to the small intestine and colon. Using in situ hybridization combined with immunostaining using cell type-specific antigens, we demonstrate that transcripts encoding HAND2 are expressed in neurons of both the myenteric and submucoasl ganglia. Transcripts encoding HAND1 are expressed by cells in the epithelial lining of the small intestine and colon. The differential localization of HAND2 and HAND1 is reflected in nonoverlapping patterns of regulation by gut-derived factors. The expression of transcripts encoding HAND2 is increased in neural crest-derived cells when cocultured with E4 gut, suggesting a gut-derived factor regulates expression of HAND genes. Exposure of gut-derived neural crest-derived cells to BMP4 significantly increased the expression of HAND2 in all gut segments. In the esophagus and gizzard, where HAND1 is not normally expressed, treatment with BMP4 induced the expression of transcripts encoding HAND1 in nonneural crest-derived cells. GDNF failed to induce consistent expression of transcripts encoding HAND2 in neural crest cells but did support a modest increase in HAND2 expression in gut-derived crest cells obtained from the esophagus and colon. GDNF had no detectable effect on the expression of transcripts encoding HAND1. These results suggest; 1) that HAND2 has a function in the development of enteric neurons, and 2) that BMP and GDNF differentially regulate HAND2 and HAND1 gene expression in the developing gastrointestinal tract.
Keywords: HAND2, HAND1, Enteric nervous system, Neural crest, bHLH, Gut development
THE gut is a complex structure composed of epithelium (endoderm), mesenchyme (mesoderm), neurons and support cells (neural crest). During development, the gut changes from a simple tube to one divided into functionally and histologically distinct regions. Development along this axis is directed by epitheliel–mesenchymal interactions that involve sonic hedgehog (SHH), bone morphogenetic protein (BMP), glial-derived neurontrophic factor (GDNF), and other, as yet unknown, growth and transcription factors. As development proceeds, the mesenchyme becomes segregated into five distinct layers (from inner to outer): 1) epithelium, 2) lamina propria, 3) muscularis mucosae, 4) submucosa, and 5) circular and longitudinal smooth muscle (Fig. 1). The enteric nervous system, which is comprised of the myenteric and sub-mucosal ganglionic plexuses, is derived from the neural crest (11,13,14,24,25). Neural crest-derived cells arising from the vagal (somites 1–4), trunk (somites 5–7), and sacral (caudal to somite 28) levels of the neural axis contribute to different regions of the developing gut [for review see (16,53)]. The mechanisms directing how neural crest-derived cells reach their final sites of gangliogenesis to form the myenteric or submucosa plexuses remain unclear.
Figure 1.
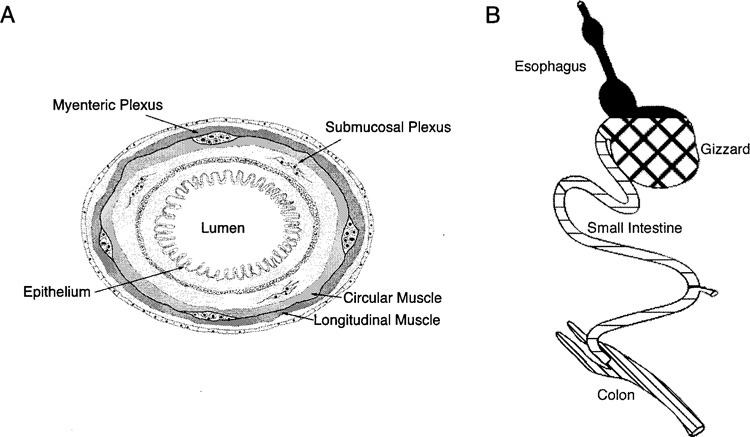
Schematic diagrams of gut morphology and functional domains. (A) The lumen of the gut is lined by an epithelium. The myenteric plexus is located between circular and longitudinal muscle. The submucosal plexus is located deep to the inner circular muscle in the submucosa. This schematic is presented in the same plane as tissue sections shown in Figures 3–5. (B) The gut tube differentiates into functionally and histologically distinct regions, as shown. Each region used in the current studies has been filled with patterns matching those in Figures 2, 7–10.
We have begun to investigate the mechanisms responsible for the patterning and differentiation of neurons in the enteric nervous system. To this end, we have concentrated on the basic helix–loop–helix (bHLH) DNA binding proteins HAND2 (dHAND) and HAND1 (eHAND). These genes are expressed in a restricted pattern in the periphery and have important roles in development of neural crest-derived sympathetic ganglia (22,23), limb bud (3,7), and heart (8,9,41,42,46). Additionally, HAND genes either regulate or are regulated by factors known to be important during early development of the gut, including SHH and BMP4.
In the chick, expression of SHH in the developing gut begins at Hamburger and Hamilton (HH) (17) stage 7 and by HH stage 10 is expressed in the anterior intestinal portal followed by expression in the posterior intestinal portal at HH stage 13 [(34), reviewed in (10)]. Expression of SHH is restricted to the endodermal epithelium and persists into adulthood (34). Intercellular signaling mediated by SHH functions, in part, by regulating expression of downstream target genes in the mesenchyme, BMP being the most notable for the current studies. The overall patterning of the gut is regulated by SHH by inhibiting formation of muscle and enteric ganglia while supporting differentiation of lamina propria and sub-mucosa [(43), reviewed in (10)]. Interaction of endoderm-derived SHH and adjacent mesoderm results in the differentiation of region-specific cell types, suggesting that SHH can affect the fate of cells resident in adjacent mesoderm (1,32,34). SHH is required for the proper formation of the limb and is expressed downstream of HAND2 (3,44). In the limb, SHH induces BMP, as it does in the gut, suggesting a possible interaction of these same regulatory molecules during gut morphogenesis.
BMP4 is expressed in a region-specific manner in the mesoderm of the developing gut, and appears to play multiple rules in gut morphogenesis (34,35,40,43). BMP4 represses differentiation of enteric neurons (43) and smooth muscle (40) as well as establishing the proper thickness of the mesoderm. This occurs in a region-specific manner through regulating apoptosis and cell proliferation (35,40). HAND2 is a downstream effector of BMP4 required for the differentiation of neural crest-derived noradrenergic neurons (22,23,51). This finding suggested a potential role of HAND2 and BMP in the differentiation of neural crest-derived enteric neurons.
Unlike SHH and BMP4, which appear to function primarily in patterning and differentiation of endodermal and mesodermal derivatives of the developing gut wall, GDNF appears to be primarily involved in the development of the enteric nervous system (28,37,39,45,50). In mice expressing a targeted null mutation of the GDNF gene, enteric neurons are lacking (26,30,36). The function of GDNF has been elucidated in tissue culture (4,18,19). GDNF supports the proliferation and survival of enteric neuron precursors as well as their differentiation into neurons (18,19). These results suggest a potential role for GDNF as a gut-derived factor that might regulate the expression of HAND genes.
Our goal in these initial studies was to describe the spatial and temporal expression patters of transcripts encoding HAND2 and HAND1 and identify growth factors that might regulate their expression. We show differential patterns of expression for transcripts encoding HAND2 and HAND1 as well as differential responsiveness of cells expressing these genes to GDNF and BMP4.
MATERIALS AND METHODS
In Situ Hybridization
In situ hybridization was performed according to published procedures (22,49). Embryos were staged according to Hamburger and Hamilton (17), removed from the egg, washed in PBS, and the gut was removed on ice. The whole gut was divided into the anatomical regions as shown in Figure 1 and fixed in 4% paraformaldehyde prepared fresh in PBS, pH. 7.4, at 4°C for 4 h [embryonic day (ED) 4/5] to overnight (ED8–18). Following fixation, the tissue was extensively washed (16 × 15 min) in PBS and stored in 30% sucrose in PBS at 4°C. Frozen sections were cut at 10 μm from tissues embedded in O.C.T. (Miles, Inc., Elkhart, IN) mounting medium and thaw mounted onto SuperFrost Plus (Fisher, Pittsburgh, PA) slides; sections were stored at −20°C until used. In situ hybridization was performed using digoxigenin-labeled antisense cRNA probes corresponding to the full-length chicken cDNAs encoding HAND1 or HAND2 (22,23). There was no visible signal in sections hybridized with sense strand control probe (data not shown). For in situ hybridization, sections were dried at 50°C for 15 min, postfixed with 4% paraformaldehyde for 20 min, washed in PBS, and then treated with 50 μg/ml of proteinase K (Sigma, St. Louis, MO) for 8 min. Tissue sections were fixed again in 4% paraformaldehyde for 15 min and washed in PBS. Sections were prehybridized in buffer containing 50% formamide, 5 × SSC, 0.3 mg/ml yeast tRNA, 50 μg/ml heparin, 1× Denhardt’s solution, 0.1% Tween 20, 0.1% CHAPS, and 5 mM EDTA, for 3 h at 65°C in a humid chamber containing 50% formamide and 5 × SSC. The sections were then hybridized with cRNA probes (1 μg/ml in pre-hybridization buffer) overnight at 65°C in a humidified chamber. Following hybridization, excess probe was removed by washing at 65°C, in 5× SSC, 1 × 15 min, and in 0.2× SSC, 2 × 30 min. To detect sites of hybridization, sections were washed with PBT (1× PBS, 2 mg/ml BSA, 0.1% Triton X-100) and incubated in blocking solution containing PBT and 20% heat-inactivated horse serum for 30 min at room temperature. The sections were then incubated with anti-digoxygenin antibody conjugated to alkaline phosphatase (1:2000; Roche, Indianapolis, IN) in the blocking solution at 4°C overnight. Sites of antibody binding were detected using BCIP/NBT according to manufacturer’s directions (Roche). Color development was carried out in the dark from 20 min to 2 h and stopped by washing in PBS. For combined in situ hybridization and immunostaining with HNK-1 to detect neural crest-derived cells, in situ hybridization was followed by immunocytochemistry, as described below.
Immunocytochemistry
To identify neurons, using TuJ1, immunocytochemistry was performed on serial sections of those used for combined in situ hybridization/immunocytochemistry, using our established procedures (21). Briefly, 10-μm frozen sections of the embryonic chick gut were rinsed with PBS and permeabilized in blocking buffer containing 0.3% Triton X-100 and 10% horse serum in PBS, for 30 min at room temperature. The sections were then incubated with primary antibody diluted in a buffer containing PBS, 0.3% Triton X-100, and 4% horse serum, overnight at 4°C. HNK1 (1:1000, Sigma) was used to identify neural crest-derived cells, and TuJ1 (1:2000, Covance, Richmond, CA) was used to identify neurons (neuron-specific β-tubulin). Unbound antibody was removed by washing 3 × 10 min in PBS containing 0.3% Triton X-100. Incubation in directly coupled species-specific secondary antibody (Kirkegaard and Perry, Gaithersburg, MD) was carried out at room temperature in the dark using fluorescein isothiocyanate (HNK-1) or tetramethylrhodamine (TuJ1).
Gut Explant Culture
The entire gut was removed from ED4 chick embryos and dissected in cold PBS. Segments of esophagus, gizzard, small intestine, and colon were dissected using the landmarks shown in Figure 1B. For each segment, three explants were plated on fibronectin (Life Technologies, Grand Island, NY)-coated culture dishes (24 μg/35-mm dish) and maintained in medium containing Eagle’s salts in minimal essential medium (MEM), supplemented with 10% 11-day chick embryo extract (CEE) (20) and 15% horse serum. Explants were maintained in the presence or absence of either 10 ng/ml BMP4 (R&D Systems, Minneapolis, MN) or 10 ng/ml GDNF (R&D Systems) for 2 days. Total cell RNA was subsequently extracted from the outgrowth of three explants using the Totally RNA Extraction Kit (Ambion, Austin, TX) according to the manufacturer’s instructions and RT-PCR performed as described below.
Gut-Derived Neural Crest-Derived Cell Culture
The whole gut was removed from ED4 chick embryos on ice in PBS and divided into segments of esophagus, gizzard, small intestine, and colon, as shown in Figure 1B. The segment explants were plated on fibronectin (24 μg/dish)-coated culture dishes and maintained in MEM supplemented with 2% CEE and 15% horse serum. Gut-derived neural crest-derived cells were allowed to migrate from each segment for 14–18 h, at which time the original ex-plant was removed using electrolytically sharpened tungsten needles. The neural crest-derived cells were grown for an additional 2 days in the presence or absence of either 10 ng/ml BMP4 (R&D Systems) or 10 ng/ml GDNF (R&D Systems). Total cell RNA was subsequently extracted from the outgrowth of six explants, as described for the outgrowth of whole gut segments, and RT-PCR was performed as described below.
Coculture of Neural Crest-derived Cells With Gut Explants
Neural crest cells were obtained from stage 14 (54) Japanese quail embryos (Coturnix coturnix japonica) as previously described (20,22,23) with minor modifications. Fertilized quail eggs (GQF, Savannah, GA) were incubated at 38°C for 43–47 h to obtain HH stage 14 embryos. To obtain neural crest-derived cells, the neural tube with associated somites was surgically removed from embryos using electrolytically sharpened tungsten needles and the neural tube fragments were incubated in 0.5% collagenase A (Boeringher Mannheim, Indianapolis, IN) for 12–15 min at room temperature. The neural tubes were released using gentle trituration with fire-polished Pasteur pipettes and then collected in fresh ice-cold growth medium and subsequently plated on 35-mm tissue culture plates coated with 24 μg/ml fibronectin (Life Technologies). Neural crest cells were allowed to migrate onto the dish for 14–16 h, at which time the neural tube explants were removed using tungsten needles. For these cocultures, a circle was scored in the fibronectin to divide the dish into two compartments. Neural crest explants were plated in the inner compartment.
Explants of ED4 chick gut were obtained, as described above, and plated in the outer compartment of the dish at the time that the neural crest explants were removed. Each dish contained 20 neural crest explants and three gut explants. The cells were maintained in MEM supplemented with 2% CEE and 15% horse serum for 7 days. Prior to the preparation of neural crest-derived total cellular RNA, gut explants were removed from the outer ring on the culture dish.
RT-PCR
RT-PCR was performed according to our published procedure (23,51) with minor modifications. Following DNase treatment, 0.5 μg total cellular RNA was used as template for first-strand synthesis in a 20-μl reaction containing 1 unit of SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA), 1 mM each dNTP, 3 mM MgCl2, 10 mM DTT, 2 units Rnasin (Promega, Madison, WI), 75 mM KCl, and 5 mM random hexanucleotides. The synthesis of cDNA was carried out at 37°C for 60 min. Equivalent aliquots of first-strand cDNA were amplified by PCR with primers specific for either chicken β-actin, HAND1, or HAND2. The final reaction contained 0.2 mM dNTPs, 0.4 mM of each primer, 2.2 mM MgCl2, and 1 U Platinum Taq polymerase (Invitrogen). The cycler protocol used for the amplification of HAND2 and β-actin was 94°C for 4 min, then 25 cycles of 94°C for 75 s, 57°C for 75 s, 72°C for 75 s, 72°C for 10 min, and hold at 4°C. Primers for β-actin were added to the reaction tube after 5 cycles. The cycler protocol for HAND1 and β-actin was 94°C for 4 min followed by 25 cycles of 94°C for 75 s, 55°C for 75 s, 72°C for 75 s, then 72°C for 10 min and hold at 4°C. The primers for β-actin were added to the reaction 9 cycles after the HAND1 PCR had begun. In pilot studies we established that amplification of neither β-actin, HAND1, nor HAND2 transcript is saturated and that our assays were done in the linear range of amplification. PCR products were fractionated in 1.5% agarose gels and the intensity of ethidium bromide-labeled PCR products quantified using a KODAK Gel Documentation System. The relative level of expression of transcripts encoding HAND1 or HAND2 was based on determining the product of the ratio of band intensity of HAND1 or HAND2 to β-actin.
RESULTS
Transcripts Encoding HAND Genes Are Expressed Differentially in the Developing Chick Gut
In our initial studies describing the cloning, expression pattern, and function of avian HAND2 (22), we reported expression of transcripts encoding HAND genes in the avian gut. Here we expand these studies with a thorough examination of the spatial and temporal expression pattern of transcripts encoding HAND2 and HAND1 in the developing avian gut. To determine relative levels of transcripts encoding HAND2 or HAND1, semiquantitative RT-PCR was used. Transcripts encoding HAND2 were expressed in all segments of the gut, albeit to varying degrees, beginning at ED4 and throughout ED18 (Fig. 2A). Expression of transcripts encoding HAND2 in the esophagus was significantly (p < 0.001) lower than in other more rostral regions of the gut. In the gizzard, small intestine, and colon, expression of transcripts encoding HAND2 remained relatively consistent both spatially and temporally from ED4 through ED18 (Fig. 2A).
Figure 2.
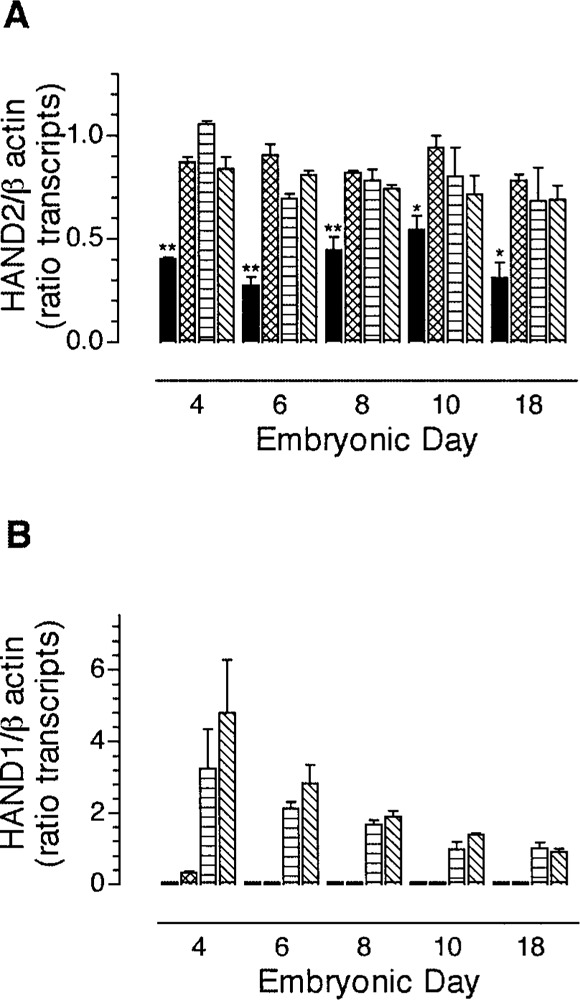
Transcripts encoding HAND2 and HAND1 are differentially expressed in the developing chick gut. RT-PCR was used to quantify the relative expression of transcripts encoding HAND2 (A) or HAND1 (B) in esophagus (filled bar), gizzard (hatched bar), small intestine (horizontal bar), and colon (diagonal bar) from ED4, ED6, ED8, ED10, and ED18 embryos. The segments of gut were dissected as shown in Figure 1B. The data are presented as the mean ± SEM of 3 determinations. The expression of transcripts encoding HAND2 was compared by one-way ANOVA followed by Bonferroni’s multiple comparison post hoc test. Student’s unpaired t-test was used for comparison of the level of expression of transcripts encoding HAND1 between stage 18 and other stages. *p < 0.05; **p < 0.01.
In contrast to the spatial distribution of transcripts encoding HAND2 (Fig. 2A), transcripts encoding HAND1 (Fig. 2B) were not detected in the esophagus from ED4 through ED18 and were only detected in the gizzard at ED4. Transcripts encoding HAND1 were expressed in the small intestine and colon throughout development (Fig. 2B). In contrast to HAND2, where the levels of expression of transcripts appeared constant with developmental age, the level of transcripts encoding HAND1 declined significantly with increasing developmental age (Fig. 2B). The data suggest differential regulation of HAND2 and HAND1 and raise the possibility that they are localized to different structures within the developing gut wall.
Transcripts Encoding HAND2, But Not HAND1, Are Expressed in Neurons Comprising Enteric Ganglia
As a first step toward understanding the function of HAND genes during development of the enteric nervous system, we used in situ hybridization to demonstrate in which structures of the gut wall transcripts encoding HAND genes are localized (Figs. 3–5). In situ hybridization was performed on esophagus, gizzard, small intestine, and colon from ED4/5, ED8, and ED18 segments of developing gut. In situ hybridization was combined with immunocytochemistry to allow identification of the cell type(s) expressing transcripts encoding HAND2 and HAND1 (Figs. 3–5, HAND + HNK-1).
Figure 3.
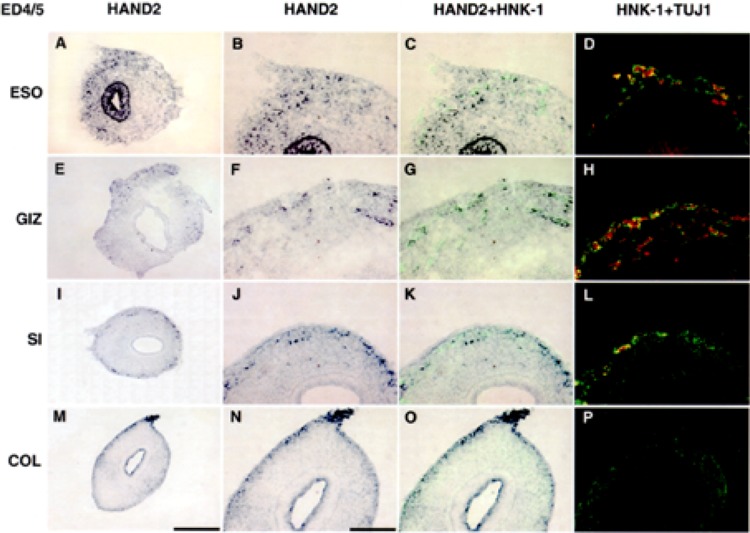
Transcripts encoding HAND2 are expressed in epithelium and neural crest-derived cells at ED4/5. In situ hybridization was used to localize transcripts encoding HAND2 in esophagus (A–D), gizzard (E–H), small intestine (I–L), and colon (M–P). Gut from ED4/5 embryos was dissected into the segments shown in Figure 1B and cut in cross section, as shown in Figure 1A. In situ hybridization was combined with immunocytochemistry (HAND2 + HNK-1, C, G, K, O) to simultaneously localize cells expressing transcripts encoding HAND2 (purple) and HNK-1 (green). HNK-1 is a marker for neural crest-derived cells (47). On serial sections, immunostaining using HNK-1 (green) and TUJ1 (red) (HNK-1 + TUJ) was used to demonstrate coexpression of HAND2, HNK-1, and TuJ1 in neural crest-derived neurons. At ED4/5, transcripts encoding HAND2 are expressed by cells scattered throughout the mesenchyme in all segments. In the esophagus (A) and colon (M), transcripts encoding HAND2 are also expressed in the epithelium. At this stage, neurons develop in the gizzard and esophagus, as demonstrated by the neuronal marker TuJ1. Low power (original magnification 20×), scale bar 200 μm (A, E, I, M). Higher magnification (original magnification 40×), scale bar 100 μm.
Figure 5.
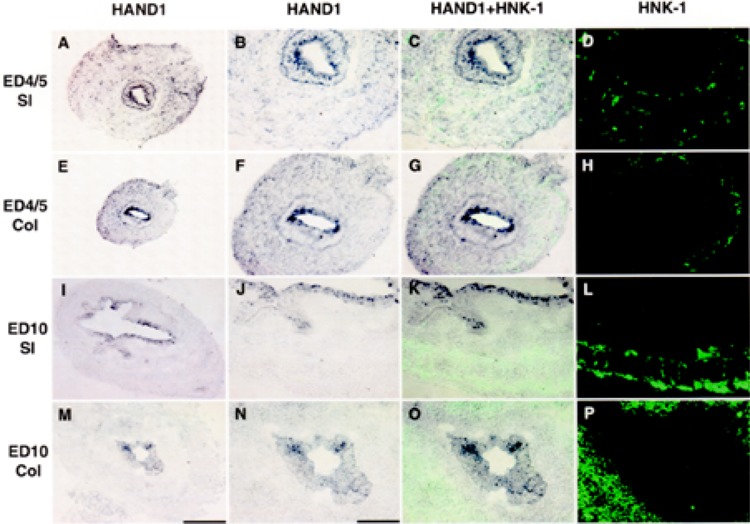
Transcripts encoding HAND1 are expressed by nonneural crest-derived cells. In situ hybridization was used to localize transcripts encoding HAND1 in small intestine (A–D, I–L) and colon (E–H, M–P) in ED5 (A–H) and ED10 (I–P) embryos. Segments of gut were dissected as shown (Fig. 1A, B). In situ hybridization was combined with immunocytochemistry (HAND2 + HNK-1, C, G, K, O) to simultaneously localize cells expressing transcripts encoding HAND1 (purple) and HNK-1 (green). The pattern of HNK-1 immunostaining is shown in serial sections (D, H, L, P). At ED4/5, cells expressing HAND1 are localized in the epithelium and scattered through the mesenchyme. At ED10, transcripts encoding HAND1 are found exclusively in the epithelium. Combined in situ hybridization with immunostaining for HNK-1 (C, G, K, O) demonstrates that neural crest-derived cells do not express transcripts encoding HAND1. Low power scale bar 200 μm (A, E, I, M). Higher magnification (original magnification 40×) scale bar 100 μm.
At ED4/5, transcripts encoding HAND2 were expressed in cells localized in the enteric mesenchyme in all segments (Fig. 3), as well as in the epithelium [esophagus (Fig. 3A) and colon (Fig. 3M)]. Based on the colocalization of transcripts encoding HAND2 and HNK-1, a marker of neural crest-derived cells, HAND2 is expressed primarily in neural crest-derived cells. The cells in the esophageal and colonic epithelium expressing transcripts encoding HAND2 have not yet been identified. At ED4/5, neurons had begun to differentiate in the esophagus (Fig. 3D) and gizzard (Fig. 3H). Based on co-immunolabeling with an antibody that recognizes neuron-specific β-tubulin (TuJ1) and HNK-1, transcripts encoding HAND2 were localized to neurons (Fig. 3D, H, L, P). Although in the small intestine at ED4/5, neural crest-derived cells (HNK-1) had migrated and localized in sites appropriate for neuronal differentiation (TuJ1), the expression of neuron-specific β-tubulin remained low at this level of the small intestine at this early developmental stage (Fig. 3L). In the colon, cells expressing transcripts encoding HAND2 were localized throughout the mesencyme.
At ED8, cells expressing transcripts encoding HAND2 had localized in myenteric and submucosal ganglia throughout the length of the gut (Fig. 4). Based on co-immunolabeling with TuJ1 and HNK-1, cells expressing transcripts encoding HAND2 differentiated into neurons (Fig. 4D, H, L, P). It should be noted that in the gizzard, a submucosal plexus did not develop even though at ED4/5, two separable groups of HAND2+, HNK-1+, TuJ1+ cells were easily identified (compare Figs. 3G, H and 4G, H); the myenteric plexus at this stage is a multilayered structure (47). At ED18, the adult pattern established by ED8 persisted (not shown) with neurons in both the sub-mucosal and myenteric ganglia of the small intestine and colon and myenteric ganglia in the gizzard and esophagus expressing transcripts encoding HAND2. Although not evident, based on RT-PCR (Fig. 2A), the level of expression of transcripts encoding HAND2 appeared to have decreased in the small intestine and colon.
Figure 4.
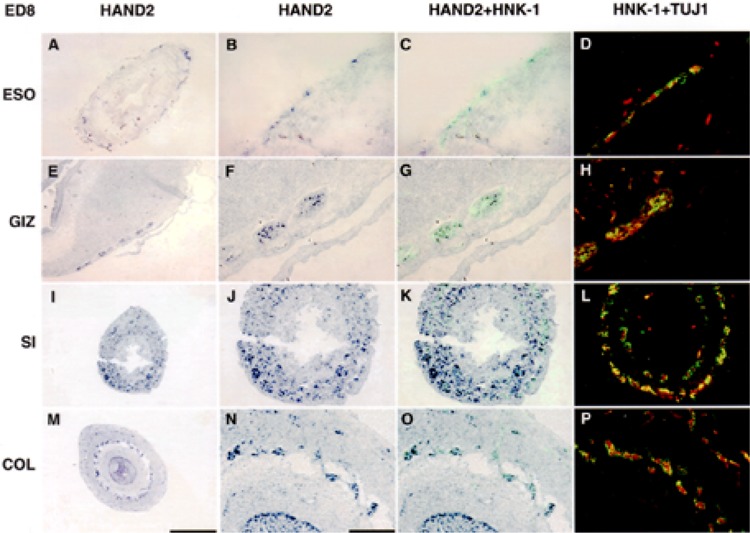
Transcripts encoding HAND2 are expressed by neural crest-derived neurons in both myenteric and submucosal ganglia at ED8. In situ hybridization was used to localize transcripts encoding HAND2 in esophagus (A–D), gizzard (E–H), small intestine (I–L), and colon (M–P) in ED8 segments of gut (Fig. 1A, B). In situ hybridization was combined with immunocytochemistry (HAND2 + HNK-1, C, G, K, O) to simultaneously localize cells expressing transcripts encoding HAND2 (purple) and HNK-1 (green). On serial sections, immunostaining using HNK-1 (green) and TUJ1 (red) (HNK-1 + TUJ) was used to demonstrate coexpression of HAND2, HNK-1, and TuJ1 in neural crest-derived neurons (D, H, L, P). By ED8, in all segments of gut, cells expressing transcripts encoding HAND2 and coexpressing HNK-1 and TuJ1 are found in myenteric and submucosal plexuses. In the epithelium, cells expressing transcripts encoding HAND2 remain only in the colon (M–P). Low power scale bar 400 μm (A, E, I M). Higher magnification (original magnification 40×) scale bar 100 μm.
Although transcripts encoding HAND2 were preferentially localized to neural crest-derived cells that will differentiate into neurons, transcripts encoding HAND1 were expressed in cells scattered throughout the gut wall (Fig. 5). Based on the results obtained with RT-PCR, where transcripts encoding HAND1 were found in small intestine and colon, in situ hybridization was used to identify cells expressing transcripts encoding HAND1 in small intestine and colon from ED4/5 (Fig. 5A–H) and ED10 colon (Fig. 5I–P). These embryonic ages represent times at which HAND1 transcript levels were high (ED4/5) and low (ED10). Transcripts encoding HAND1 were found in cells scattered throughout the gut mesenchyme as well as in the epithelium of both the small intestine and colon. The expression of HAND1 in the epithelium was maintained while that in the mesenchyme was reduced with increasing developmental age (Fig. 5). We were unable to demonstrate colocalization of neural crest-derived cells expressing HNK-1 with cells scattered in the mesenhcyme expressing transcripts encoding HAND1. This pattern of expression suggests that cells expressing HAND1 in the gut are not derived from the neural crest and thus do not contribute to the enteric nervous system.
We were unable to demonstrate, either spatially or temporally, overlapping patterns of transcripts encoding HAND1 and HAND2. The expression of transcripts encoding HAND2 by neural crest-derived cells that differentiate into neurons suggested that growth factors influencing the development of enteric neurons might affect the transcription or function of HAND2.
The Microenvironment of the Developing Gut Induces Expression of HAND2, But Not HAND1, in Neural Crest-Derived Cells
The localization of transcripts encoding HAND2 to neurons in enteric ganglia suggested the possibility that gut-derived soluble factors might regulate expression of HAND2 within neural crest-derived cells of the gut wall. To address this issue, we first tested whether cells within the microenvironment of the gut wall produce soluble factors that induce expression of HAND genes in neural crest-derived cells. We used a coculture paradigm in which neural crest-derived cells were grown with ED4 gut explants in a nonper-missive growth factor environment for HAND2 expression (23).
In the absence of gut-derived soluble factors, transcripts encoding HAND2 were expressed in 50% of explants (Fig. 6A) but HAND1 was not expressed (Fig. 6B). This result suggested that some factor in the gut microenvironment could differentially influence the expression of HAND genes. We tested this possibility by determining if explants of gut secrete soluble factors that could influence expression of transcripts encoding HAND genes in neural crest-derived cells. Coculture of neural crest-derived cells with chick gut significantly increased the expression of transcripts encoding HAND2 in neural crest-derived cells (n = 8, p < 0.01) (Fig. 6A), but also induced a small increase in the expression of transcripts encoding HAND1 from these same cells (Fig. 6B). The data support the hypothesis that HAND gene expression is regulated in neural crest-derived cells by gut-derived soluble factors.
Figure 6.
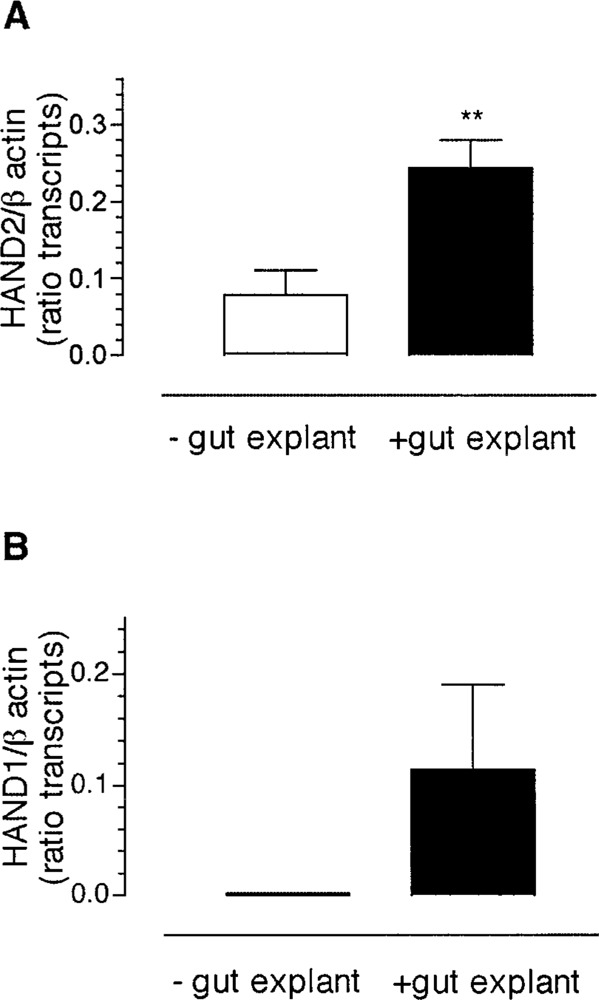
The microenvironment of the developing gut induces expression of HAND2, but not HAND1, in neural crest-derived cells. Quail neural crest-derived cells were grown in coculture with ED4 chick gut explants. Semiquantitative RT-PCR was used to detect the expression of transcripts encoding HAND2 or HAND1 in neural crest-derived cells after 2 days of exposure to gut-derived soluble factors. The level of expression of transcripts encoding HAND genes is normalized with that of β-actin and presented as the ratio of band intensity of HAND2 (A) or HAND1 (B) to β-actin. The data are presented as mean ± SEM, Statistical significance was determined using Student’s unpaired t-test (HAND2) or one-sample -test (HAND1). **p < 0.01.
BMP4 Induces Expression of Transcripts Encoding HAND2 in Gut-Derived Neural Crest-Derived Cells
The finding that there were soluble gut-derived factors that can influence transcription of HAND2 in neural crest-derived cells suggested BMP4 as one likely candidate. BMP4 has been shown to induce expression of HAND2 in neural crest-derived cells, which differentiate into noradrenergic neurons (23,33,38,51) and BMP4 is present in the gut beginning from ED2 (34). To determine if BMP4 induces expression of transcripts encoding HAND2 and/or HAND1 in neural crest-derived cells that have migrated and localized within the developing gut, gut-derived neural crest-derived cells (Fig. 7A) or whole gut explants obtained from E4 chick (Fig.7B) were treated with BMP4 for 2 days in nonpermissive growth factor conditions. Exposure of neural crest-derived cells obtained from esophagus, gizzard, small intestine, or colon to BMP4 was sufficient to increase the expression of transcripts encoding HAND2 (Fig. 7A) in all segments of the gut (n = 5, p < 0.05). In explants of gut from the same anatomical domains, however, exposure to BMP4 caused an increase in expression of transcripts encoding HAND2 only in explants of the gizzard (n = 3, p < 0.05) but not other segments of the gut (Fig. 7B).
Figure 7.
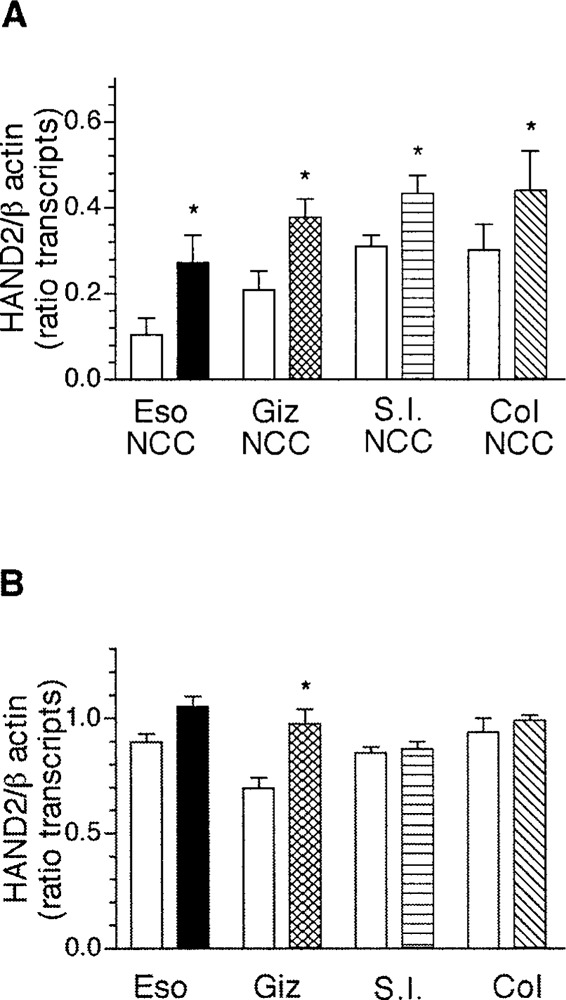
BMP4 induces expression of transcripts encoding HAND2 in gut-derived neural crest-derived cells. Gut-derived neural crest-derived cells (A) or whole gut explants (B) obtained from ED4 chick were treated with BMP4 and the expression of transcripts encoding HAND2 was determined using semiquantitative RT-PCR. The data are presented as mean ± SEM of the ratio of band intensity for HAND2 and β-actin from 3–5 determinations. For all comparisons between control and BMP4-treated samples, statistical significance was determined using Student’s unpaired t-test, except for the colon-derived neural crest-derived cells (Col NCC) where paired t-test was used. *p < 0.05.
In contradistinction to the results obtained for expression of transcripts encoding HAND2, transcripts encoding HAND1 were not expressed in gut-derived neural crest-derived cells, in culture, from any segment of the developing gut, with the exception of the small intestine (Fig. 8A). Further, in contrast to the results obtained with transcripts encoding HAND2, the addition of BMP4 did not induce significant expression of transcripts encoding HAND1 in gut-derived neural crest-derived cells. This result supports our previous findings that cells in the gut mesenchyme and not neural crest-derived cells express transcripts encoding HAND1 (Fig. 5). In gut explants, BMP4 robustly induced expression of transcripts encoding HAND1 in the esophagus (n = 3) and the gizzard (n = 4) (Fig. 8B), suggesting that some cells other than neural crest-derived cells express HAND1 in response to BMP4. In the small intestine and the colon, growth in the presence of added BMP4 did not affect the endogenous expression of transcripts encoding HAND1 (Fig. 8). Our data suggest there is differential regulation of transcripts encoding HAND1 between rostral and caudal gut.
Figure 8.
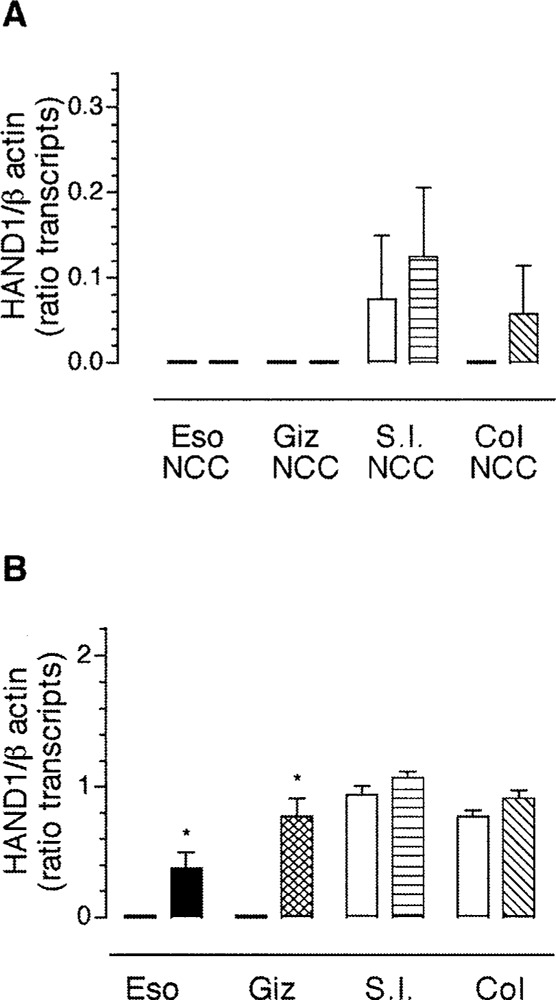
BMP4 induces expression of transcripts encoding HAND1 in the foregut. Gut-derived neural crest-derived cells (A) or whole gut explants (B) from ED4 chick were treated with BMP4 and the expression of transcripts encoding HAND1 was determined using semiquantitative RT-PCR. The expression of transcripts encoding HAND1 was normalized to β-actin. The data are presented as mean ± SEM of the product of the ratio of HAND1/ β-actin (minimum of 3 determinations). To determine statistical significance, Student’s unpaired t-test was used for comparison between control and BMP treatment for all groups except the esophagus (ESO) and gizzard (Giz) explants and the colon-derived neural crest-derived cells (Col NCC) where one-sample t-test was used. *p < 0.05.
GDNF Differentially Induces Expression of Transcripts Encoding HAND Genes in Functionally Distinct Domains of the Gut
GDNF is a gut-derived neurotrophic factor required for the development of the enteric nervous system. GDNF regulates the migration, proliferation, and differentiation of neural crest-derived cells in the gut and the differentiation of enteric neurons (4,18,52). To determine if GDNF regulates the expression of transcripts encoding HAND genes in the gut, gut-derived neural crest cells or whole gut explants obtained from E4 chick were treated with GDNF for 2 days in culture and the expression of transcripts encoding HAND1 and HAND2 was assessed by semi-quantitative RT-PCR. Exposure to GDNF resulted in increased expression of transcripts encoding HAND2 from neural crest-derived cells obtained from the esophagus (n = 3, p < 0.01) and the colon (n = 3, p < 0.05), but not from that of the gizzard or the small intestine (Fig. 9A). With the exception of the esophagus (n = 3, p < 0.05), treatment of gut explants with GDNF had no effect on the expression of transcripts encoding HAND2 (Fig. 9B).
Figure 9.
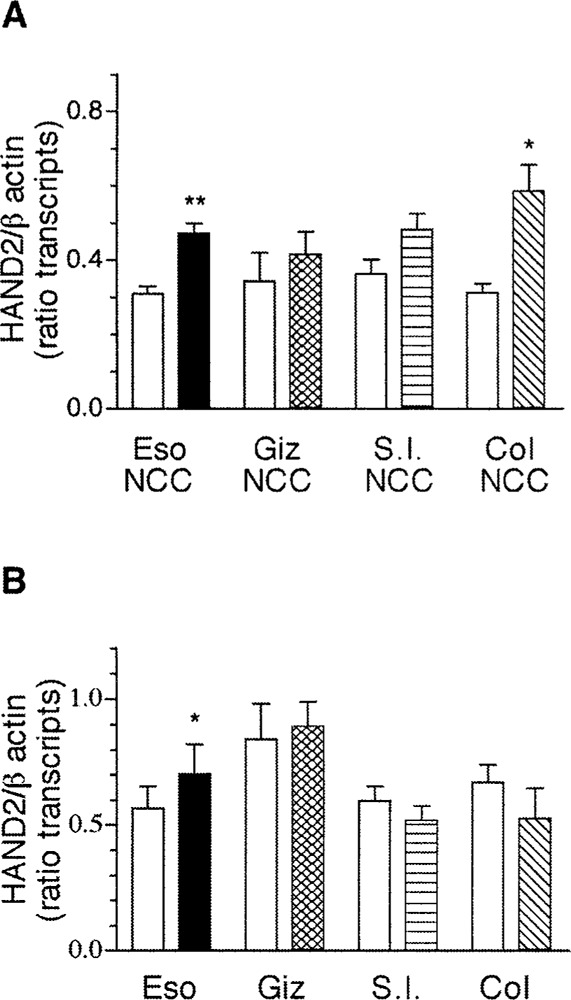
GDNF induces expression of transcripts encoding HAND2 in esophagus and colon. ED4 chick gut-derived neural crest-derived cells (A) or whole gut explants (B) were treated with GDNF and the expression of transcripts encoding HAND2 was assessed by semiquantitative RT-PCR. The expression of transcripts encoding HAND2 was normalized to the expression of transcripts encoding β-actin. The data are presented as mean ± SEM of the product of the ratio of HAND2/β-actin (3–4 determinations). Statistical significance was determined using Student’s unpaired t-test for results reported in (A) and paired t-test was used for results reported in (B). *p < 0.05; **p < 0.01.
Expression of transcripts encoding HAND1 was not detected in neural crest-derived cells from any region of the gut and GDNF failed to induce expression of transcripts encoding HAND1 in these cells (Fig. 10). Although transcripts encoding HAND1 were detected in explants of the gizzard, small intestine, and colon, treatment with GDNF had no appreciable effect on the levels of transcript encoding HAND1 (Fig. 10B). The data suggest that expression of HAND1 and HAND2 is differentially regulated by GDNF in a region-specific manner.
Figure 10.
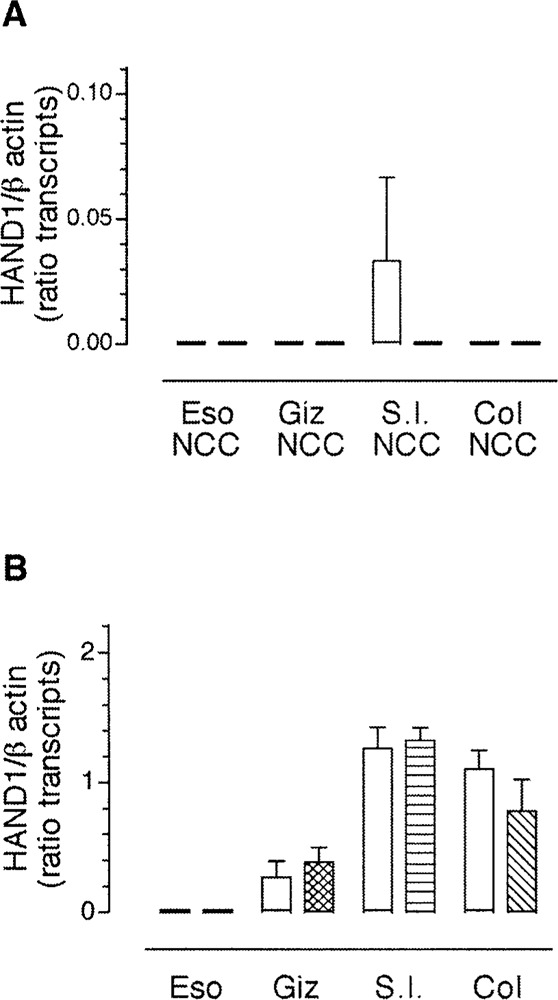
GDNF does not affect the expression of transcripts encoding HAND1 in the gut. Gut-derived neural crest-derived cells (A) or whole gut explants (B) obtained from ED4 chick were treated with GDNF and the expression of transcripts encoding HAND1 was assessed by semiquantitative RT-PCR. The data are presented as mean ± SEM of the product of HAND1/β-actin, from at least 3 determinations. Statistical significance was determined using Student’s unpaired t-test.
DISCUSSION
Growth and transcription factors provide important developmental cues to neural crest-derived cells as they differentiate into enteric neurons in the microenvironment of the gut. The bHLH transcription factors HAND2 and HAND1 are expressed in developing gut but neither the growth factors that induce their expression nor their role in the gut are known. As a first step toward understanding the function of HAND gene products in morphogenesis of the gastrointestinal tract, we sought to determine the spatial and temporal expression patterns of transcripts encoding HAND genes in the developing enteric nervous system (ENS) and to identify gut-derived growth factors that influence their expression.
We have defined a dynamic expression pattern of transcripts encoding HAND genes during development of the chick gastrointestinal tract. The spatial pattern of expression coincides with two lineages of neural crest-derived cells that colonize different regions of the gastrointestinal tract and that also give rise to sympathetic chain ganglia [(3,6), for review see (12,27)]. The overall pattern suggests that HAND2 is not a lineage marker in the ENS as it is expressed at all levels of the ENS.
Transcripts encoding HAND2 are expressed throughout the gut from ED4 through ED18 but the level of expression is consistently lower in the esophagus than in more caudal regions. It is established that neural crest-derived cells that colonize the esophagus are derived from the neural crest adjacent to somites 6–7 (6). The decreased level of expression in this domain of the gut may be due to the lack of a submucosal plexus reflecting a smaller number of neurons, although this is not apparent in the gizzard, which also lacks a submucosal plexus. However, in the gizzard the myenteric plexus consists of a multilayered gan-glionic plexus most likely containing more neurons than in the esophagus (47). In more caudal regions of the gut, HAND2 gene expression is uniformly distributed both spatially and temporally. In all anatomical domains of the ENS, expression of transcripts encoding HAND2 is primarily confined to neurons.
Using neuron-specific β-tubulin as a neuronal marker, our combined in situ hybridization/immuno-cytochemistry demonstrates that neural crest-derived neurons expressing HAND2 contribute to both myenteric and submucosal ganglia. Although we have not found transcripts encoding HAND2 in premigratory or migrating neural crest-derived cells (22), it appears that, as HNK-1+ cells enter the gut microenvironment, a subgroup of these cells also expressed transcripts encoding HAND2, prior to expression of the cell type-specific marker, TuJ1. For example, at ED8 in the small intestine, there are cells expressing transcripts encoding HAND2 located between the myenteric and submucosal ganglia. It is possible that these cells are migrating from the myenteric to the submucosal ganglia at this time in development (15,29). This conclusion is further supported by the finding that these cells are no longer visible by ED18; at this time, based on in situ hybridization, all cells expressing transcripts encoding HAND2 are located in myentric or submucosal ganglia (Howard, data not shown). It is also interesting to note that, in the ED8 colon, the submucosal ganglia appear to develop prior to the myenteric ganglia and show an increased density of cells expressing transcripts encoding HAND2 [(2), M. Epstein, personal communication]. The expression of HAND2 in neural crest-derived enteric neurons coincides with its expression in neural crest-derived sympathetic ganglion neurons (22,23), both of which share a common lineage. Our data are in agreement with those of White and Anderson (48), suggesting that neural crest-derived cells make the decision to contribute to autonomic or enteric lineages prior to their lineage segregation into neurons or glial cells.
Expression of HAND1, in the developing gastrointestinal tract, is confined to nonneural crest-derived cells. This conclusion is based on our results showing no apparent overlap between cells expressing HNK-1 and those expressing transcripts encoding HAND1. This is a somewhat unexpected finding because the expression of transcripts encoding HAND1 appears dependent upon the expression of HAND2 in neural crest-derived cells that differentiate into sympathetic ganglion neurons (23). At later stages of development, cells expressing transcripts encoding HAND1 are confined to the epithelium in the small intestine and colon; expression of HAND1 in the mesenchyme is lost by ED10. Differences in the spatial and temporal pattern of transcript expression suggest different functional roles for HAND2 and HAND1 in the development of components of the gut wall. HAND2 appears to be required for the specification/differentiation of neural crest-derived enteric neurons. The function of HAND1 in the developing gut remains unknown. The expression of HAND2 in neurons and HAND1 in the epithelium suggests that different factors within the gut microenvironment regulate their expression Signals from the microenvironment of the developing gut are required for the differentiation of neural crest-derived enteric neurons as well as for the proper patterning and differentiation of the gut wall [(12,26 30,35,40,43); for review see (10)]. Our results indicate that as neural crest-derived cells enter the gut microenvironment they express transcripts encoding HAND2; this is temporally separated from the subsequent expression of neuron-specific β-tubulin. We speculated that a gut-derived factor induces the expression of transcripts encoding HAND2 in neural crest-derived cells as they enter the gut. To determine if the microenvironment of the gut induces expression of HAND genes, we used primary cultures of avian neural crest-derived cells in coculture with explants of ED4 gut and assessed the expression of transcripts encoding HAND2 and HAND1. We found that expression of transcripts encoding HAND2, but not HAND1, was significantly increased in neural crest-derived cells in response to gut-derived factors. This supports our conclusion that HAND2 expression is induced in neural crest-derived cells once they enter the gut microenvironment and that HAND1 is not expressed by neural crest-derived cells in the gut. Two gut-derived factors, BMP4 and GDNF, were then tested as potential candidate factors that might regulate expression of HAND genes in the gut.
We chose to test GDNF and BMP because each of these factors is required for normal development of the gut. Targeted deletion of the GDNF gene results in loss of enteric neurons in all gut regions caudal to the stomach (26,30,36). Once neural crest-derived cells enter the gut, GDNF increases the proliferation and survival of neural precursors as well as their differentiation into neurons (4,18,53). To determine if GDNF regulates the expression of transcripts encoding HAND genes in the gut, gut-derived neural crest cells or whole gut explants obtained from E4 chick were treated with GDNF and the expression of transcripts encoding HAND genes was assessed by semi-quantitative RT-PCR. Exposure of gut-derived neural crest-derived cells to GDNF significantly increased expression of transcripts encoding HAND2 in the esophagus and colon but not in the small intestine or gizzard. The expression of transcripts encoding HAND1 was not affected by exposure to GDNF. This result supports our data showing that HAND1 does not appear to be expressed by neural crest-derived cells in the gut.
Because targeted knock-out of GDNF results in regional loss of neurons, it is not unexpected to find differential response of cells to GDNF in different regions of the gut [(26,30 36), reviewed in (53)]. Although enteric neurons differentiate in the esophagus in the absence of GDNF, we suggest that if GDNF is an endogenous factor that influences HAND2 expression in the esophagus, then expression of HAND2 is not sufficient for the differentiation of enteric neurons in the esophagus.
It has been previously reported that neural crest cells selected from the gut at ED4.7 respond to GDNF by increasing proliferation and neurogenesis (18), suggesting that GDNF may support expression of HAND2 as neural crest-derived cells reach and enter the gut microenvironemnt caudal to the esophagus. If this is the case, our results further suggest that there is also an endogenous inhibitor that prevents induction of additional transcripts encoding HAND2. Alternatively, our data can be interpreted to suggest that GDNF does not regulate HAND gene expression distal to the esophagus. It is possible that additional factors resident within the gut wall regulate expression of HAND genes. BMP4 was a likely candidate, because it has multiple functions in gut morphogenesis.
BMP4 is required for specification of the mesoderm (1,34) but regulates differentiation of smooth muscle as well (40). In addition, BMP4 induces a cascade of transcription factors, including HAND2, which are required for the differentiation of neural crest-derived noradrenergic sympathetic ganglion neurons (23).
Neural crest-derived cells isolated from all regions of the developing gastrointestinal tract responded to BMP4 by significantly increasing expression of transcripts encoding HAND2. It is not unexpected that added BMP4 can influence the expression of transcripts encoding HAND2 in gut-derived neural crest-derived cells. BMP4 is an endogenous factor that normally influences HAND2 gene expression in neural crest-derived cells as they localize along the dorsal aorta during formation of sympathetic chain ganglia (23,33,38).
In the context of the gut explants, addition of BMP4 significantly increased transcripts encoding HAND2 only in the gizzard. BMP4 is known to be expressed by ED4 throughout the midgut and hindgut but it is not present in the gizzard at ED4 (35). It is likely that BMP4 had an effect on the gizzard because these cells have the capacity to respond to it, although they are not exposed to it in situ. The data suggest either that BMP 4 is not a gut-derived factor that influences the expression of HAND2 in the gut microenvironment, or that there is an additional inhibitory factor in the gut that normally prevents BMP4 from interacting with neural crest-derived cells at this early stage of their differentiation within the gut wall.
The ability of BMP4 to support an increase in transcripts encoding HAND1 in the esophagus and gizzard suggests that BMP4 may be a mesodermally derived signal that functions to regulate HAND1 in the gut wall. However, it also seems likely that other factors within the gut wall regulate HAND1 expression because in those regions of the gut where BMP4 is expressed, added BMP4 did not affect a change in endogenous levels of HAND1. Inasmuch as HAND1 is not expressed in neural crest-derived cell types within the gut wall, the fact that there is no significant influence on levels of transcript encoding HAND1 in neural crest-derived cells once they have entered the gut wall is not unexpected.
As a whole, our studies implicate HAND2 as an important regulator of enteric neuron differentiation and further suggest different roles for BMP and GDNF in the regulation of HAND gene expression in the developing ENS. Our studies suggest the presence of additional factors that regulate HAND gene expression in the gut microenvironment.
ACKNOWLEDGMENTS
The authors wish to thank Drs. Miles Epstein and Phyllis Pugh for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health grants DK57237 and NS40644 to M.J.H.
REFERENCES
- 1. Apelqvist A.; Ahlgren U.; Edlund H. Sonic hedgehog directs specialized mesoderm differentiation in the intestine and pancreas. Curr. Biol. 7:801–804; 1997. [DOI] [PubMed] [Google Scholar]
- 2. Burns A. J.; Le Douarin N. M. The sacral neural crest contributes neurons and glia to the post-umbilical gut: Spatiotemporal analysis of the development of the enteric nervous system. Development 125:4335–4347; 1998. [DOI] [PubMed] [Google Scholar]
- 3. Carnahan J. F.; Anderson D. J.; Patterson P. H. Evidence that enteric neurons may derive from the sympathoadrenal lineage. Dev. Biol. 148:552–561; 1991. [DOI] [PubMed] [Google Scholar]
- 4. Chalazonitis A.; Rothman T. P.; Chen J.; Gershon M. D. Age-dependent differences in the effects of GDNF and NT-3 on the development of neurons and glia from neural crest-derived precursors immunoselected from the fetal rat gut: Expression of GFRalpha-1 in vitro and in vivo. Dev. Biol. 204:385–406; 1998. [DOI] [PubMed] [Google Scholar]
- 5. Charite J.; McFadden D. G.; Olson E. N. The bHLH transcription factor dHAND controls sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development 127:2461–2470; 2000. [DOI] [PubMed] [Google Scholar]
- 6. Durbec P. L.; Larsson-Blomberg L. B.; Schuchardt A.; Costantini F.; Pachnis V. Common origin and developmental dependence on c-ret of subsets of enteric and sympathetic neuroblasts. Development 122:349–358; 1996. [DOI] [PubMed] [Google Scholar]
- 7. Fernandez-Teran M.; Piedra M. E.; Kathiriya I. S.; Srivastava D.; Rodriguez-Rey J. C.; Ros M. A. Role of dHAND in the anterior–posterior polarization of the limb bud: Implications for the sonic hedgehog pathway. Development 127: 2133–2142; 2000. [DOI] [PubMed] [Google Scholar]
- 8. Firulli A. B.; McFadden D. G.; Lin Q.; Srivastava D.; Olson E. N. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 18:266–270; 1998. [DOI] [PubMed] [Google Scholar]
- 9. Firulli A. B.; Thattaliyath B. D. Transcription factors in cardiogenesis: The combinations that unlock the mysteries of the heart. Int. Rev. Cytol. 214:1–62; 2002. [DOI] [PubMed] [Google Scholar]
- 10. Fukuda K.; Yasugi S. Versatile roles for sonic hedgehog in gut development. J. Gastroenerol. 37:239–246; 2002. [DOI] [PubMed] [Google Scholar]
- 11. Furness J. B.; Costa M. The enteric nervous system. New York: Churchill Livingstone; 1987. [Google Scholar]
- 12. Gershon M. D. Genes and lineages in the formation of the enteric nervous system. Curr. Opin. Neurobiol. 7:101–109; 1997. [DOI] [PubMed] [Google Scholar]
- 13. Gershon M. D. The enteric nervous system. Ann. Rev. Neurosci. 4:227–272; 1981. [DOI] [PubMed] [Google Scholar]
- 14. Gershon M. D. Lessons from genetically engineered animal models II. Disorders of enteric neuronal development: Insights from transgenic mice. Am. J. Physiol. 277:G262–267; 1999. [DOI] [PubMed] [Google Scholar]
- 15. Gershon M. D.; Epstein M. L.; Hegstrand L. Colonization of the chick gut by progenitors of enteric serotonergic neurons: Distribution, differentiation, and maturation within the gut. Dev. Biol. 77:41–51; 1980. [DOI] [PubMed] [Google Scholar]
- 16. Gershon M. D.; Chalazonitis A.; Rothman T. P. From neural crest to bowel: Development of the enteric nervous system. J. Neurobiol. 24:199–214; 1993. [DOI] [PubMed] [Google Scholar]
- 17. Hamburger V.; Hamilton H. L. A series of normal stages in the development of the chick embryo. J. Morphol. 88:49–92; 1951. [PubMed] [Google Scholar]
- 18. Hearn C. J.; Murphy M.; Newgreen D. GDNF and ET-3 differentially modulate the numbers of avian enteric neural crest cells and enteric neurons in vitro. Dev. Biol. 197:93–105; 1998. [DOI] [PubMed] [Google Scholar]
- 19. Heuckeroth R. O.; Lampe P. A.; Johnson E. M.; Milbrandt J. Neurturin and GDNF promote proliferation and survival of enteric neuron and glial progenitors in vitro. Dev. Biol. 200:116–129; 1998. [DOI] [PubMed] [Google Scholar]
- 20. Howard M. J.; Bronner-Fraser M. The influence of neural tube-derived factors on differentiation of neural crest cells in vitro. I. Histochemical study on the appearance of adrenergic cells. J. Neurosci. 5:3302–3309; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howard M. J.; Gershon M. D. Role of growth factors in catecholaminergic expression by neural crest cells: In vitro effects of transforming growth factor beta 1. Dev. Dyn. 196:1–10; 1993. [DOI] [PubMed] [Google Scholar]
- 22. Howard M.; Foster D. N.; Cserjesi P. Expression of HAND gene products may be sufficient for the differentiation of avian neural crest-derived cells into catecholaminergic neurons in culture. Dev. Biol. 215:62–77; 1999. [DOI] [PubMed] [Google Scholar]
- 23. Howard M. J.; Stanke M.; Schneider C.; Wu X.; Rohrer H. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development 127:4073–4081; 2000. [DOI] [PubMed] [Google Scholar]
- 24. Le Douarin N. The neural crest. Cambridge: Cambridge University Press; 1982. [Google Scholar]
- 25. Le Douarin N. M.; Teillet M. A. The migration of neural crest cells to the wall of the digestive tract in avian embryo. J. Embryol. Exp. Morphol. 30:31–48; 1973. [PubMed] [Google Scholar]
- 26. Moore M. W.; Klein R. D.; Farinas I.; Sauer H.; Armanini M.; Phillips H.; Reichardt L. F.; Ryan A. M.; Carver-Moore K.; Rosenthal A. Renal and neuronal abnormalities in mice lacking GDNF. Nature 382:76–79; 1996. [DOI] [PubMed] [Google Scholar]
- 27. Newgreen D.; Young H. M.. Enteric nervous system: Development and developmental disturbances—Part 1. Pediatr. Dev. Pathol. 5:224–247; 2002. [DOI] [PubMed] [Google Scholar]
- 28. Pachnis V.; Mankoo B.; Costantini F. Expression of the c-ret proto-oncogene during mouse morphogenesis. Development 119:1005–1017; 1993. [DOI] [PubMed] [Google Scholar]
- 29. Payette R. F.; Tennyson V. M.; Pham T. D.; Mawe G. M.; Pomeranz H. D.; Rothman T. P.; Gershon M. D. Origin and morphology of nerve fibers in the aganglionic colon of the lethal spotted (ls/ls) mutant mouse. J. Comp. Neurol. 257:237–252; 1987. [DOI] [PubMed] [Google Scholar]
- 30. Pichel J. G.; Shen L.; Sheng H. Z.; Granholm A. C.; Drago J.; Grinberg A.; Lee E. J.; Huang S. P.; Saarma M.; Hoffer B. J.; Sariola H.; Westphal H. Defects in enteric innervation and kidney development in mice lacking GDNF. Nature 382:73–76; 1996. [DOI] [PubMed] [Google Scholar]
- 31. Pisano J. M.; Birren S. J.. Restriction of developmental potential during divergence of the enteric and sympathetic neuronal lineages. Development 126:2855–2868; 1999. [DOI] [PubMed] [Google Scholar]
- 32. Ramalho-Santos M.; Melton D. A.; McMahon A. P. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development 127:2763–2772; 2000. [DOI] [PubMed] [Google Scholar]
- 33. Reissmann E.; Ernsberger U.; Francis-West P. H.; Rueger D.; Brickell P. M.; Rohrer H. Involvement of bone morphogenetic protein-4 and bone morphogenetic protein-7 in the differentiation of the adrenergic phenotype in developing sympathetic neurons. Development 122:2079–2088; 1996. [DOI] [PubMed] [Google Scholar]
- 34. Roberts D. J.; Johnson R. L.; Burke A. C.; Nelson C. E.; Morgan B. A.; Tabin C. Sonic hedgehog is an endodermal signal inducing Bmp-4 and Hox genes during induction and regionalization of the chick hindgut. Development 121:3163–3174; 1995. [DOI] [PubMed] [Google Scholar]
- 35. Roberts D. J.; Smith D. M.; Goff D. J.; Tabin C. J. Epithelial-mesenchymal signaling during the regionalization of the chick gut. Development 125:2791–2801; 1998. [DOI] [PubMed] [Google Scholar]
- 36. Sanchez M. P.; Silos-Santiago I.; Frisen J.; He B.; Lira S. A.; Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382:70–73; 1996. [DOI] [PubMed] [Google Scholar]
- 37. Schiltz C. A.; Benjamin J.; Epstein M. L. Expression of the GDNF receptors ret and GFR alpha1 in the developing avian enteric nervous system. J. Comp. Neurol. 414:193–211; 1999. [PubMed] [Google Scholar]
- 38. Schneider C.; Wicht H.; Enderich J.; Wegner M.; Rohrer H. Bone morphogenetic proteins are required in vivo for the generation of sympathetic neurons. Neuron 24:861–870; 1999. [DOI] [PubMed] [Google Scholar]
- 39. Schuchardt A.; D’Agati V.; Larsson-Blomberg L.; Costantini F.; Pachnis V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367:380–383; 1994. [DOI] [PubMed] [Google Scholar]
- 40. Smith D. M.; Nielsen C.; Tabin C. J.; Roberts D. J. Roles of BMP signaling and Nkx2.5 in patterning at the chick midgut–foregut boundary. Development 127:3671–3681; 2000. [DOI] [PubMed] [Google Scholar]
- 41. Srivastava D.; Cserjesi P.; Olson E. N. A subclass of bHLH proteins required for cardiac morphogenesis. Science 270:1995–1999; 1995. [DOI] [PubMed] [Google Scholar]
- 42. Srivastava D.; Thomas T.; Lin Q.; Kirby M. L.; Brown D.; Olson E. N. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 16:154–160; 1997. [DOI] [PubMed] [Google Scholar]
- 43. Sukegawa A.; Narita T.; Kameda T.; Saitoh K.; Nohno T.; Iba H.; Yasugi S.; Fukuda K. The concentric structure of the developing gut is regulated by Sonic hedgehog derived from endodermal epithelium. Development 127:1971–1980; 2000. [DOI] [PubMed] [Google Scholar]
- 44. te Welscher P.; Fernandez-Teran M.; Ros M. A.; Zeller R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 16:421–426; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taraviras S.; Pachnis V. Development of the mammalian enteric nervous system. Curr. Opin. Gene. Dev. 9:321–327; 1999. [DOI] [PubMed] [Google Scholar]
- 46. Thomas T.; Kurihara H.; Yamagishi H.; Kurihara Y.; Yazaki Y.; Olson E. N.; Srivastava D. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development 125:3005–3014; 1998. [DOI] [PubMed] [Google Scholar]
- 47. Tucker G. C.; Ciment G.; Thiery J. P. Pathways of avian neural crest cell migration in the developing gut. Dev. Biol. 116:439–450; 1986. [DOI] [PubMed] [Google Scholar]
- 48. White P. M.; Anderson D. J. In vivo transplantation of mammalian neural crest cells into chick hosts reveals a new autonomic sublineage restriction. Development 126:4351–4363; 1999. [DOI] [PubMed] [Google Scholar]
- 49. Wilkinson D. G. In situ hybridization: A practical approach. Oxford: Oxford University Press; 1998. [Google Scholar]
- 50. Worley D. S.; Pisano J. M.; Choi E. D.; Walus L.; Hesion C. A.; Cate R. L.; Sanicola M.; Birren S. Developmental regulation of GDNF resposne and receptor expression in the enteric nervous system. Development 127:4383–4393; 2000. [DOI] [PubMed] [Google Scholar]
- 51. Wu X.; Howard M. J. Two signal transduction pathways involved in the catecholaminergic differentiation of avian neural crest-derived cells in vitro. Mol. Cell. Neurosci. 18:394–406; 2001. [DOI] [PubMed] [Google Scholar]
- 52. Young H.M.; Hearn C. J.; Farlie P. G.; Canty A. J.; Thomas P. Q.; Newgreen D.F. GDNF is a chemoattractant for enteric neural cells. Dev. Biol. 229:503–516; 2001. [DOI] [PubMed] [Google Scholar]
- 53. Young H. M.; Newgreen D. Enteric neural crest-derived cells: Origin, identification, migration, and differentiation. Anat. Rec. 262:1–15; 2001. [DOI] [PubMed] [Google Scholar]
- 54. Zacchei A. M. Lo sviluppo embrionale della quaffia giaponese (Coturnix coturnix japonica). Arc. Ital. Anat. Embriol. 66:36–72; 1961. [PubMed] [Google Scholar]


