Abstract
This review focuses on the spliced leader (SL) RNA and uridylic acid-rich small nuclear RNAs (U-snRNAs) involved in pre-mRNA processing in trypanosomatid protozoa, with particular emphasis on the mechanism of transcription and cap formation. The SL RNA plays a central role in mRNA biogenesis by providing the unique cap 4 structure to the 5′ end of all mRNAs by trans-splicing. The trimethylguanosine capped U-snRNAs, on the other hand, represent an unusual example among eukaryotic snRNAs in that they are transcribed by RNA polymerase III. This implies the existence of a distinctive mechanism for capping enzyme selection by the transcriptional machinery. Furthermore, the transcription units of U-snRNA genes offer yet another example of the variety of choices that have been established during eukaryotic evolution, namely that an upstream tRNA gene or tRNA-like gene provides extragenic promoter elements for a downstream small RNA gene.
Keywords: Spliced leader RNA, trans-Splicing, Cap formation, Cap 4 structure, Trimethylguanosine cap, U-snRNAs, RNA polymerase III transcription, tRNA genes
HISTORICALLY, flagellate protozoa of the family Trypanosomatidae have attracted the interest of the scientific community as well as the public at large, because they include various members of the genera Leishmania and Trypanosoma that cause debilitating diseases primarily in developing countries. However, more recently these organisms have become particularly valuable in the study of fundamentally important biological phenomena. Trypanosomatidae are among the most ancient eukaryotes and as such have retained many distinctive features. For instance, the survey of RNA metabolism in these organisms has been instrumental in the discovery of new concepts in eukaryotic RNA biology, such as polycistronic transcription (38), trans-splicing of pre-mRNAs (1,55,81), and mitochondrial RNA editing (3,28,74).
To date, one of the most intriguing aspects of trypanosomatid biology remains the regulation of gene expression. Because it appears that in general these organisms do not to use transcription initiation as a regulatory step to control the abundance of mRNA, the research effort in this field has concentrated on the elucidation of potential regulatory mechanisms at the posttranscriptional level. One area that has attracted considerable interest is the maturation of the pre-mRNA by RNA processing reactions and the identity and role of small nuclear RNAs (snRNAs) and small nuclear ribonucleoprotein particles (snRNPs) that participate in this process. In this review, we will concentrate on two peculiar aspects that have surfaced in the course of these studies, namely the promoter architecture of snRNA genes and the mode of cap formation.
A FEW FUNDAMENTALS OF GENE EXPRESSION IN TRYPANOSOMATIDS
We thought it would be helpful to begin this review by highlighting a few unique features of gene expression in trypanosomatids. One trait is that the almost universal eukaryotic rule “one gene = one promoter” does not apply to these organisms. Protein coding genes are organized as polycistronic rather than monocistronic transcription units; thus, the primary transcript is a polycistronic pre-mRNA. As a consequence, the 5′ end of all mature mRNAs is formed by trans-splicing, an RNA processing reaction, rather than by transcription initiation as in most eukaryotic organisms. The partners in trans-splicing are the polycistronic pre-mRNA, which contains several mRNA coding regions (3′ exons) preceded by a 3′ splice site, and the spliced leader (SL) RNA, which provides the 39-nt-long SL sequence (5′ exon) and the 5′ splice site.
Another curiosity in trypanosomatids is the apparent lack of promoters for RNA polymerase II, the enzyme typically responsible for the transcription of protein coding genes and of the U-snRNA genes, with the exception of the U6 snRNA gene. Although transcription is carried out by three RNA polymerases with α-aminitin sensitivities similar to higher eukaryotic RNA polymerase I, II, and III, and promoters for RNA polymerase I and III transcription units have been described, so far it has not been possible to map transcription initiation sites for protein coding genes transcribed by RNA polymerase II. [In Trypanosoma brucei there are two examples of developmentally regulated protein coding genes that are transcribed by RNA polymerase I (38).] The characterization of RNA polymerase II promoters was further delayed, when it was realized that the U-snRNA genes in Trypanosomatidae are transcribed by RNA polymerase III (17). Finally, the study of snRNAs has exposed quite a number of unusual properties, which will be described in more detail in this review.
A COMPLETE SET OF SPLICEOSOMAL U-snRNAs
The first uridylic acid-rich small nuclear RNA (U-snRNA) isolated from Trypanosomatidae was the T. brucei U2 snRNA (78), and over the years all five spliceosomal U-snRNAs (U1, U2, U4, U5, and U6 snRNA) have been identified in a variety of organisms (Table 1). Originally, most of the work employed the classical T. brucei system, but several investigators also turned to the Leptomonas system, which offers some experimental advantages, like efficient in vivo expression of snRNAs. Because the U2, U4, and U6 snRNAs were first described over 10 years ago (50,77,78), we will concentrate in the following sections on the more recent identification of the U1 and U5 snRNAs.
TABLE 1.
PROPERTIES OF TRYPANOSOMATID SPLICEOSOMAL U-snRNAs
| Synthesis | Cap Structure | |
|---|---|---|
| U1 snRNA | RNA polymerase III* | TMG |
| U2 snRNA | RNA polymerase III | TMG |
| U4 snRNA | RNA polymerase III | TMG |
| U5 snRNA | RNA polymerase III* | ?† |
| U6 snRNA | RNA polymerase III | ? |
Predicted from promoter structure.
Possibly a 5′ phosphate end.
The U5 snRNA has eluded detection in Trypanosomatidae for quite some time. One major reason is that the U5 snRNA does not have a trimethylguanosine (TMG) cap at its 5′ end (16,85). Thus, the classical selection of U-snRNAs by anti-TMG immunoprecipitations did not enrich for the U5 snRNA in T. brucei (50). The fact that the primary sequence of U5 snRNAs, apart from the 11-nucleotide 5′ loop sequence (5′-YGCCUUUYAYY-3′), is poorly conserved in different organisms, further complicated the identification of a trypanosomatid U5 homologue. Thus, a few years ago, three different laboratories independently employed alternative methods and succeeded in detecting the U5 snRNA in T. brucei (16,40) and L. collosoma (85).
One approach was to use in vivo cross-linking with the bifunctional reagent psoralen to search for small RNAs that interact with the SL RNA (16,82). Initially, these studies identified a novel small RNA, termed spliced leader-associated RNA or SLA RNA, that was proposed to be the T. brucei U5 snRNA (82). However, a phylogenetic analysis did not corroborate this hypothesis (60). Subsequent characterization of a second abundant RNA cross-linked to the SL RNA revealed a 62-nt “U5-like RNA” possessing the canonical 5′ loop sequence present in all known spliceosomal U5 snRNAs (16). A second approach to identify the U5 snRNA was to isolate the evolutionarily highly conserved U5 snRNP-specific protein PRP8/p220 in T. brucei and then look for an associated small RNA (40). This strategy generated an antibody against the C-terminal domain of the T. brucei PRP8 homologue, which coimmunoprecipitated the U5 snRNA. And finally, fractionation analysis of abundant snRNPs in L. collosoma revealed a small RNA of 80 nt that has all the characteristic structural features of a U5 snRNA (85). Subsequent to the initial identification of the U5 snRNA in T. brucei and L. collosoma, the corresponding sequences were isolated from L. seymouri (6) and Crithidia fasciculata (68).
The cross-link of the T. brucei U5 snRNA with the SL RNA was mapped between the 5′ loop sequence and the free SL exon, an intermediate in trans-splicing (16). This interaction is analogous to what happens in cis-splicing between the U5 snRNA loop and the free first exon splicing intermediate (54). Furthermore, based on the location of the cross-links, it was proposed that the U5 snRNA engages in extensive base pairing with both exon and intron sequences across the SL RNA 5′ splice site (16). This interaction is phylogenetically conserved in all trypanosomatids analyzed so far (6,85) and is supported by genetic experiments in L. collosoma (86).
The most recent spliceosomal snRNA to be identified in Trypanosomatidae was the U1 snRNA, which plays a crucial role in the initial recognition of the 5′ splice site of intervening sequences (4). Ever since the first evidence for trans-splicing was reported in 1982 (9), the notion developed over the years that trypanosomatids lack intervening sequences and therefore the machinery to carry out cis-splicing. This belief was based on negative observations such as that the genes characterized so far did not have introns and that trypanosomatids appeared to lack a homologue of the U1 snRNA. Furthermore, experiments in nematodes showed that, in contrast to cis-splicing, trans-splicing does not require the U1 snRNA (47), thus supporting the apparent absence of the U1 snRNA in trypanosomatids. However, quite frequently convictions that are not substantiated by experimental data do not last forever and in 1999 a report appeared describing the identification of a candidate U1 snRNA in two trypanosomatids, namely C. fasciculata and Leishmania tarentolae (66). This was followed soon afterward by the discovery of intervening sequences in the T. brucei and T. cruzi poly(A) polymerase gene (45) and the description of a candidate U1 snRNA in T. brucei (15).
In several respects the trypanosomatid U1 snRNAs exhibit features typical of other U1 snRNAs. First, the 5′ end is capped with a TMG structure and followed by additional modifications (15,66). Second, the three trypanosomatid U1 snRNAs contain a putative binding site for spliceosomal core proteins, and immunoprecipitations with anti-common protein antibodies revealed that the T. brucei U1 snRNA indeed forms of a core ribonucleoprotein particle (15). Third, there is the potential to form a U1-like stem–loop I structure, which in mammalian systems binds the U1-specific protein U1-70K (73). However, the three available sequences do not make a strong case for the existence of these intramolecular base pairing interactions, because very few compensatory base changes occur in the potential helical region. And finally, primary sequence conservation has been noted between the 5′ terminus of the trypanosome U1 snRNAs and other U1 snRNAs, which could be involved in interactions with sequences at the 5′ splice site (66).
On the other hand, one peculiar feature of the trypanosomatid U1 snRNAs is the absence of the highly conserved stem–loop II. Per se, this fits the pattern of other U-snRNAs in trypanosomatids, which in comparison with their counterparts in other eukaryotes are generally smaller in size and as a consequence harbor structural differences. However, in the case of the U1 snRNA the missing stem–loop II provides the binding site for the U1A protein in other organisms (73). U1A not only functions in splicing, but plays a role in the linkage between splicing and polyadenylation (12,44). The suggestion has been made that the spliced leader-associated (SLA) RNA harbors a U1-like stem–loop II structure (67), but experimental data are needed to validate this notion.
In cis-splicing sequences at the 5′ splice site of the intron interact through base pairing with the 5′ terminal sequence of the U1 snRNA (4). Indeed, examination of the nucleotide sequences of the T. brucei, C. fasciculata, and L. tarentolae U1 snRNA revealed a potentially extensive sequence complementarity of the 5′ end with the boundaries of the 5′ splice site of the T. brucei and T. cruzi poly(A) polymerase introns across 11 nucleotides (45). It is worth pointing out that it is quite unexpected to find 11 conserved nucleotides at the 5′ splice sites of such divergent species as T. brucei and T. cruzi. As a first step toward understanding the signals and interactions required for intron removal, mutations were introduced in the 5′ splice site region of the T. brucei intron (45). Substitutions of nucleotides 1–10 of the conserved sequence abolished cis-splicing in vivo, whereas mutations in nucleotides not conserved between the T. brucei and T. cruzi intron did not detectably affect intron removal. Although these results support a possible interaction of the U1 snRNA with the 5′ splice site, definite proof will come once compensatory mutations in the U1 snRNA are able to suppress the splicing defect.
Sm PROTEIN HOMOLOGUES CONSTITUTE THE COMMON PROTEINS OF U-snRNPS AND OF THE SL RNP
One characteristic feature of most U-snRNAs is the highly conserved Sm site with the sequence 5′-PuA(U)nGPu-3′. The Sm site forms the binding site for the common Sm proteins, which are recognized by anti-Sm antibodies. Our knowledge on the assembly of the seven canonical Sm polypeptides (SmB/B′, -D1, -D2, -D3, -E, -F, and -G) and their interactions is mainly based on studies in the mammalian and yeast systems. Crystallographic models are consistent with electronmicroscopic images (37,62) to view the mammalian Sm proteins as a heptameric, ring-like structure (36). The task towards the characterization of the core proteins associated with trypanosomal snRNPs has been difficult and time consuming, but the effort was rewarded very recently by the cloning of seven core proteins. Originally, affinity purification procedures allowed the identification of at least five polypeptides of 8.5, 10, 12.5, 14, and 15 kDa, which were associated with the core of the SL RNP, the U2 snRNP, the U4/U6 snRNP, and the U5 snRNP (40,58). These common proteins, as expected, localized predominantly in the nucleus (57) and bound to a region in the snRNA that resembled the Sm site (23). However, there were questions whether these common proteins were indeed Sm protein homologues. First, trypanosomatid snRNPs do not cross-react with Sm antibodies. Second, there is no highly conserved sequence analogous to the Sm site, and the putative binding site for common proteins in U-snRNAs is fairly degenerate. Third, in contrast to other eukaryotes, there appeared to be no correlation between the binding of core proteins and the formation of a trimethylated cap structure (see below). Nevertheless, two recent publications have shed some light on this issue by demonstrating that the seven canonical Sm proteins are present in Trypanosomatidae (21,59). Each of the Sm polypeptides carries the bipartite Sm motif (motif 1 and 2), but also several deviations in highly conserved positions, some of which might explain the lack of recognition of the trypanosome snRNPs by Sm antibodies. Despite the divergence at the primary structure level, in vitro interactions strongly argue that the seven trypanosome polypeptides also form a heptameric ring-like structure with the same arrangement as in the mammalian Sm core (59). This apparent structural conservation is unexpected at first glance, but in reality seems sensible considering that these RNPs are involved in the fundamental process of RNA splicing, which mechanistically is extremely conserved throughout evolution. Furthermore, because one of the trypanosome Sm protein homologues (SmG) was able to functionally complement a corresponding mutation in yeast (59), this indicates that at least some of the functional aspects are also conserved.
GENOMIC ORGANIZATION OF U-snRNA GENES
All trypanosomatid U-snRNA genes isolated to date are single copy, with the only exception being the U6 snRNA gene in C. fasciculata, which is encoded by two copies (84). One common feature of U-snRNA gene organization in these organisms is the presence of a divergently oriented tRNA gene or a tRNA-like gene in the immediate 5′ flanking region (Fig. 1). An intriguing hallmark of this genomic arrangement is the highly conserved distance between the tRNA gene or tRNA-like sequence and the associated U-snRNA gene (between 94 and 99 base pairs). This arrangement has been the topic of a previous review (53) and is schematically summarized in Figure 1. The recently described U1 and U5 snRNA genes from a variety of trypanosomatids show the same organization as that described for the U2 and U6 snRNA genes. The L. tarentolae U1 snRNA gene has a tRNAArg gene upstream (70), whereas a tRNA-like sequence is found associated with the T. brucei U1 snRNA gene (15). On the other hand, in the L. collosoma (85), L. seymouri (6,) and C. fasciculata (68) U5 snRNA genes a tRNACys gene is located 98, 95, and 98 bp upstream, respectively, whereas in T. brucei there is a tRNA-like sequence at the same location (6). Taken together, the available information indicates that the genomic organization of U-snRNA genes in T. brucei is unique. For instance, the U6 snRNA gene in T. brucei has a tRNAThr upstream (51), whereas a tRNAGln is present in Crithidia (51,84), Leishmania (51,83), Leptomonas (19,84), and Phytomonas (83). Similarly, the associated small RNA gene for the U1 and U5 snRNA genes in T. brucei is distinct from that in other organisms (see above). These observations are consistent with the available evidence indicating that Crithidia, Leishmania, Leptomonas, and Phytomonas are more closely related to each other than each of them is to T. brucei.
Figure 1.
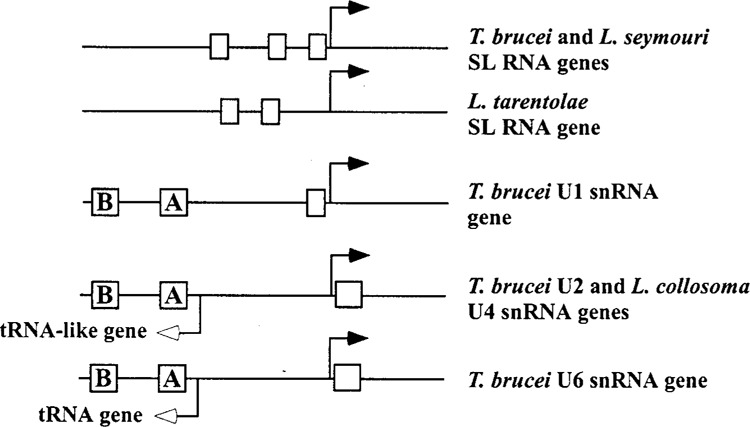
Promoter architecture of SL RNA and U-snRNA genes in trypanosomatids. Solid arrows indicate the direction of transcription of SL RNA and U-snRNA genes, whereas the open arrows indicate the direction of transcription of tRNA-like and tRNA genes associated with U-snRNA genes. Open boxes depict mapped regulatory elements. Only representative examples are shown and the drawing is not to scale.
How the association between tRNA and small RNA genes originated and was maintained, at least partially in some cases, in the family Trypanosomatidae is at present only a matter of speculation. However, the facts that to date several examples have been described where the associated tRNA-like sequences cannot be folded into a convincing cloverleaf structure and that the A and B boxes are nevertheless partially conserved in sequence and position strongly suggest that, apart from the A and B boxes, the surrounding sequences do not play a functional role in this association (see below). The major evolutionary and functional constraint in this genomic organization resides in the spacing between the genes, whereas the primary sequence separating the U-snRNA and its upstream tRNA or tRNA-like gene is not conserved. This is clearly illustrated by the two copies of the U6 snRNA gene in C. fasciculata, where several base changes have accumulated in the region between the U6 snRNA and the upstream tRNAGln gene (84).
PROMOTER ARCHITECTURE OF snRNA GENES
The study of U-snRNA gene expression in trypanosomatid protozoa has uncovered some remarkable features. First, it was shown experimentally that the T. brucei U2 snRNA represents a unique example among eukaryotic small nuclear RNAs: the gene is transcribed by RNA polymerase III, but the RNA possesses a trimethylguanosine (TMG) cap (17), which is the hallmark of RNA polymerase II-transcribed U-snRNA genes in other eukaryotic organisms (32). Similarly, the promoter structure of the U1, U4, and U5 snRNA genes in a variety of trypanosomatids suggests that they are also transcribed by RNA polymerase III, whereas these U-snRNA genes are transcribed by RNA polymerase II in all other eukaryotes studied to date (32). Second, the transcription units of trypanosomatid U-snRNA genes offer yet another example of the variety of promoter architectures that have been established during eukaryotic evolution. An upstream tRNA gene or a tRNA-like gene provides extragenic promoter elements for a downstream small RNA gene and these upstream elements coincide with the A and B boxes of the respective tRNA or tRNA-like gene (tRNA genes are transcribed by RNA polymerase III and are endowed with almost exclusively intragenic cis-acting elements consisting of two conserved sequence motifs called the A and B boxes).
Initially, in vivo expression studies with the T. brucei U2 snRNA gene showed a requirement for two upstream sequence motifs, which conformed to the complement of the A and B box consensus sequences (17). Similarly, expression of the L. collosoma U4 snRNA gene was dependent on an upstream tRNA-like B box, but in this case the six nucleotide changes introduced in the A box did not affect expression (39). In addition, the T. brucei U2 coding region contains an intragenic element close to the transcription initiation site, which is involved in positioning the RNA polymerase at the correct start site (17). A comparable situation was uncovered by surveying the T. brucei U6 snRNA gene using saturation mutagenesis (51,52). Three regulatory elements were identified to be essential and sufficient: the A and B boxes of the upstream tRNAThr gene and sequences in the U6 snRNA coding region close the 5′ end. The intragenic sequences of the U2 and U6 snRNA genes share a common motif (ANCYYYTCGG) that is fairly well conserved in other organisms. However, it has also become apparent from the analysis of the T. brucei U1 snRNA gene that intragenic regulatory elements are not necessarily the rule for the expression of U-snRNA genes in trypanosomatids (15). Although this gene has upstream regulatory elements that are reminiscent of the T. brucei U2 (17) and L. collosoma U4 snRNA gene (39), U1 snRNA expression is not dependent on intragenic sequences. Instead, there appears to be a third element in the 5′ flanking region immediately upstream of the transcription start site (our unpublished data).
The detailed mutational analysis of the T. brucei U6 snRNA gene also tested whether the observed high conservation of spacing between the U-snRNA genes and the upstream A and B boxes was of functional importance (52). This revealed that the B box can be moved, but that the spacing between the A box and the gene internal element is crucial for expression. Furthermore, in vitro studies uncovered that the A and B boxes play distinct roles in U6 snRNA gene expression. Whereas the A box is required for both in vivo and in vitro expression, the B box is dispensable for in vitro expression.
The identification of regulatory elements required for expression of U-snRNA genes in associated tRNA or tRNA-like sequences raises the question whether these elements carry out a dual function (i.e., are these elements active for transcription of both genes?). Northern blot analysis of total T. brucei RNA with a probe encompassing the A and B box of the U2 gene revealed a small RNA of 90 nucleotides, which is consistent with it being initiated upstream of the A box and terminated at a string of T residues that serves as transcription terminator for RNA polymerase III (53). Similarly, primer extension analysis suggested the synthesis of a transcript from the tRNA-like gene upstream of the L. collosoma U4 snRNA gene (39). Finally, for the T. brucei U6 snRNA gene locus in vivo expression studies demonstrated that the associated tRNAThr gene is a true gene (51) and that the A and B box motifs are required for expression of the tRNAThr gene both in vivo and in a heterologous in vitro system (51).
Despite the revelation of rather remarkable transcription units for trypanosomatid U-snRNA genes and the subsequent establishment of T. brucei (25) and L. seymouri (34) cell-free extracts competent for accurate transcription of the respective U2 snRNA gene, there have been no further reports to provide insights into the underlying mechanism. At this point it remains doubtful whether the assembly of transcription complexes on U-snRNA genes will become a major focus of trypanosomatid research, considering that it will most likely reveal a variation to the well-studied RNA polymerase III systems in other organisms. What has attracted recent attention, however, is the expression of the SL RNA and the mechanism of cap formation, which will be discussed in the following sections.
SL RNA GENE TRANSCRIPTION
The SL RNA plays a key role in the biogenesis of mRNA in trypanosomatids by providing the capped SL sequence for the 5′ end of every mRNA. SL RNAs are ubiquitously found in trypanosomatid organisms (81). The SL portion of the RNA is highly conserved in length (39 nucleotides) and sequence, whereas the intronic sequence is of variable length and much less conserved (81). Towards the 3′ end of the molecule there is a sequence motif, RAY3–8GR, which is reminiscent of the Sm binding site. Indeed, the SL RNA forms a core RNP with Sm protein homologues that are shared among all spliceosomal snRNPs (58).
Thus, considering the central role of the SL RNA in RNA metabolism, the expression and maturation of this RNA has become the focus of several investigations. In this section, we will review the current knowledge on the transcription mechanism of the SL RNA gene. With regard to the RNA polymerase responsible for SL RNA synthesis, there has been a long-lasting debate between RNA polymerase II and III, with most researchers leaning towards RNA polymerase II (10). Indeed, immunodepletion experiments with an antibody specific for the C-terminal extension of the largest subunit of RNA polymerase II resulted in the loss of SL RNA transcription in a L. seymouri cell-free system, whereas RNA polymerase III transcription of the U6 snRNA gene was not affected (Gilinger and V. Bellofatto, personal communication). However, because it is not possible to restore transcription activity by adding back recombinant protein(s) in this particular experiment, the possibility remains that the antibody removed another essential factor(s). Thus, to resolve this issue to everybody’s satisfaction, it will be important to confirm this result by additional methods, like capturing the RNA polymerase on immobilized DNA templates or chromatin immunoprecipitations.
Despite the above uncertainty, the promoter architecture of the SL RNA gene has been dissected in T. brucei (26), T. cruzi (56), L. tarentolae (65,87,88), L. amazonensis (2), L. collosoma (20), and L. seymouri (14,31,34,41) using a combination of in vitro and in vivo expression systems. These studies uncovered two or three regulatory elements upstream of the transcription start site in most organisms. Saturation mutagenesis on the L. tarentolae upstream region localized two regions, GN3CCC at −39 to −33 and GACN5G at −66 to −58 with respect to the transcription start site, to be required for efficient SL RNA gene transcription (88). The position of the two elements with respect to each other, as well as with respect to the transcription start site, appeared to be important for expression and correct transcription initiation. Because the core nucleotide sequences are only conserved in some Trypanosomatidae SL RNA genes, the SL RNA gene promoter is most likely composed of species-specific regulatory elements. Indeed, in T. brucei (our unpublished results) and L. seymouri (43) a third element, located immediately upstream of the initiation nucleotide, contributes in correctly positioning the RNA polymerase. Furthermore, SL RNA genes from T. brucei and L. amazonensis cannot be expressed in L. collosoma, a more distantly related trypanosomatid (18,20).
The identification and cloning of SL RNA gene-specific transcription factors is still at the very early stages. Proteins binding specifically to mapped regulatory elements have been detected in L. tarentolae (88) and L. seymouri (42,43). In particular, in Leptomonas two factors (PBP-1 and PBP-2) have been characterized in more detail (43). By DNase I foot-printing, PBP-1 binds within the −60 to −70 element, whereas PBP-2 was shown to interact with the DNA-bound PBP-1 and the protected region extended from −28 to −82, thus including both the −60 to −70 and −30 to −40 regulatory element. PBP-1 is a heterotrimer and two subunits of 41 and 57 kDa have been cloned (V. Bellofatto, personal communication). In addition, there is evidence for a third factor that binds to the initiator element close to the transcription start site (43). However, despite these encouraging developments, there has not yet been an effort to carry out a classical biochemical fractionation of a cell-free extract to characterize transcription factors other than DNA binding proteins.
MODIFICATIONS PRESENT ON U-snRNAS AND THE SL RNA
Similar to other eukaryotes, the trypanosomatid U1, U2, and U4 snRNAs contain a 2,2,7-trimethyl guanosine cap (15,50,78), whereas the U6 snRNA most likely carries a γ-monomethylphosphate cap (Table 1). In contrast, the 5′ end of the U5 snRNA appears to lack a TMG or m7G cap structure and most likely contains a phosphate terminus, but the exact structure remains to be determined (16,85). As a side note, there appears to be a slight inconsistency about the position of the U5 snRNA 5′ end and/or modifications present in this region. Whereas one study reports that the four most 5′ end nucleotides are modified in L. collosoma (85), as judged by primer extension analysis, another interpretation was that primer extension actually detected a longer form of the U5 snRNA (68).
Very little information is available about additional modifications present in trypanosomatid U-snRNAs. In L. collosoma 2′-O-methyl modifications were mapped in the U4 snRNA at residues A63 and A72, and in the U6 snRNA at residues G42, G47, U50, C53, and U56 (39). These modifications are in canonical positions, but overall there appear to be fewer modifications than in the mammalian homologues. For the U5 snRNA, the available data indicate that 2′-O-methyl modifications are absent in the invariant 5′ loop and that pseudouridine residues are not detectable in the RNA (8,16,68). Although these modifications, including the 5′ terminal TMG cap, are highly conserved in other eukaryotic organisms, there is also evidence that the cap structure and 2′-O-methyl and pseudouridine modifications are not essential for U5 snRNA function (69). One curious finding was that the first transcribed nucleotide of the U1 snRNA was identified as a hypermethylated adenosine residue with the structure N6,N6,2′-O-trimethyladenosine that is also present at the 5′ end of the SL RNA (66). This hypermethylated A residue appears to be restricted to the U1 snRNA and SL RNA and is not found at the 5′ end of the U2 or U4 snRNA (66). As pointed out by others, this specificity of methylation might be brought about by the fact that the 5′ terminal sequence of the U1 and SL RNA share six out of seven nucleotides (66).
The cap structure of the trypanosomatid SL RNA, which is identical to that of the mature mRNA, is unique in the eukaryotic kingdom in terms of its high content of modified nucleotides (Fig. 2). In higher eukaryotes the distinctive 5′ terminal structure of mRNAs consists of 7-methylguanosine residue linked to the first transcribed nucleotide by a 5′ to 5′ triphosphate bridge (m7GpppN or cap 0), and very often the first and second transcribed nucleotide are further modified by the addition of 2′-O-methyl groups leading to cap 1 and cap 2 structures. In contrast, four nucleotides adjacent to the m7G cap are methylated at the SL RNA 5′ end to generate a cap 4 structure: m7guanosine-ppp-N6,N6,2′-O-trimethyladenosine-p-2′-O-methyladenosine-p-2′-O-methylcytosine-p-N3,2′-O-methyluridine (5). Importantly, this structure contains two nucleosides, namely N6,N6,2′-O-trimethyladenosine and N3,2′-O-dimethyluridine, which are specific to trypanosomes and to date have not been identified in any other organism. Hence, it is anticipated that the enzymes, and more specifically the RNA methyltransferases, that catalyze these modifications are unique to trypanosomatids. In addition, it is likely that the enzymes catalyzing cap 4 formation, including the capping enzyme, play a pivotal role in regulating the availability of trans-splicing “competent” SL RNA and, therefore, these enzymes might be major players in regulating trans-splicing activity as a whole.
Figure 2.
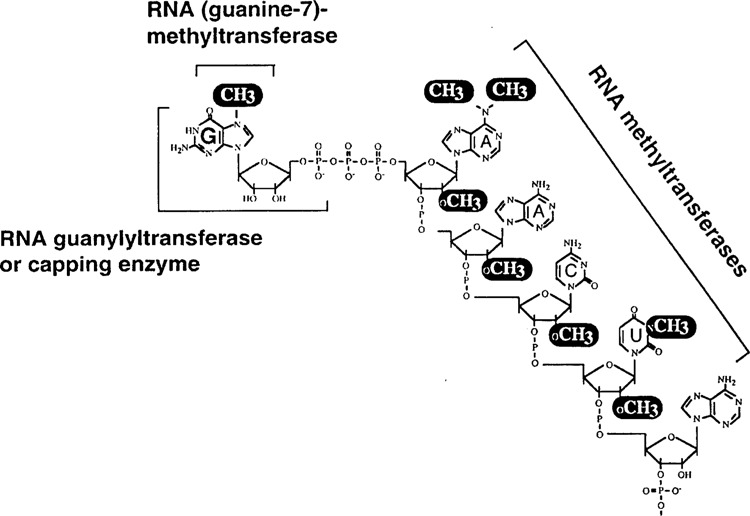
Enzymes involved in the synthesis of the trypanosomatid cap 4 structure.
The elaborate SL RNA cap structure has been conserved throughout evolution of the family Trypanosomatidae (5), thus suggesting an important functional role. Although proper modification of this cap structure is essential for utilization of the SL RNA in trans-splicing in T. brucei, T. cruzi, and L. amazonensis cells (49,80), the precise function of the cap 4 structure remains to be determined. Thus far, it is established that the extensive modifications are not essential for (i) the stability of the SL RNA (80); (ii) the folding of the SL RNA into a particular secondary structure (30); and (iii) the assembly into a core SL RNP (80).
m7G CAP FORMATION: A PROBLEM WITH SPECIFICITY
In all eukaryotic cells analyzed so far, addition of the m7G cap is the earliest event in the modification of the 5′ terminus of RNA polymerase II transcripts (13,27,35,63). Capping occurs cotranscriptionally when the RNA chain has achieved a chain length of about 30 nt, which is most likely as soon as the 5′ end of the nascent RNA is extruded from the elongating RNA polymerase molecule. Capping is mediated by a series of three consecutive reactions; namely, a 5′ RNA triphosphatase makes a diphosphate terminus, which in turn is used by an RNA guanylyltransferase or capping enzyme to add a guanosine residue in a 5′–5′ triphosphate bridge and then the N7 position of guanine is methylated by a RNA (guanine-7) methyltransferase (71). In most eukaryotes the capping reaction adds an m7G residue exclusively to RNAs produced by RNA polymerase II, namely pre-mRNA and the RNA polymerase II-transcribed U-snRNAs. This selective behavior in vivo is not reproduced in vitro, because capping accepts as a substrate any triphosphate or diphosphate terminated RNA. Recent data showed that capping is linked to the RNA polymerase II transcriptional complex through direct interaction of the capping enzyme with the carboxy-terminal domain (CTD) of RNA polymerase II, thus providing an elegant explanation for the specificity of the capping reaction (11,33,48,89).
In trypanosomatids the capping enzyme acts upon the SL RNA, which is most likely transcribed by RNA polymerase II (see above), and upon a specific subset of RNA polymerase III transcripts, namely U1, U2, U3, and U4 snRNAs. In contrast, the most abundant RNA polymerase III transcripts (tRNAs, 5S rRNA, and 7SL RNA) are not capped. Thus, in principle the trypanosome capping enzyme is capable of an even greater selectivity than its higher eukaryotic counterpart, in that it discriminates between transcripts synthesized by the same RNA polymerase. Once the RNA polymerase responsible for transcription of the SL RNA is unequivocally identified, it will become clear whether the trypanosome capping enzyme can cap RNAs other than RNA polymerase III transcripts. Notwithstanding this uncertainty, one can speculate that in the case of the U-snRNAs it is unlikely that the specificity of the trypanosome capping enzyme can solely be accounted for by an interaction with the large subunit of RNA polymerase III. There are several alternate possibilities that come to mind. For instance, it is possible that interaction between the capping enzyme and the RNA polymerase III transcriptional machinery is mediated by factors that are gene specific and that assemble on gene-specific promoter elements. Indeed, the analysis of RNA polymerase III promoters of the U2 and U6 snRNA genes has demonstrated the existence of extragenic and intragenic control regions (see above). Whereas the extragenic control regions coincide with the A and B boxes of tRNA or tRNA-like gene promoters for the U6 and U2 genes, respectively, the intragenic promoter elements are clearly different in sequence between the two genes and might provide information for the assembly of gene-specific, and possibly capping enzyme-specific, transcription complexes. If this were the case, capping would still occur cotranscriptionally, similar to the capping mechanism in higher eukaryotes. However, the finding that U1 snRNA gene expression does not depend on an intragenic element casts some doubt on this hypothesis. Another possibility is that the selection of RNA transcripts to be capped is based upon RNA determinants, which might be provided either by nascent or perhaps full-length transcripts. The recognition of an RNA determinant by the trypanosome capping enzyme would be an unprecedented mechanism of substrate selection for this class of enzymes.
TMG CAP FORMATION ON RNA POLYMERASE III TRANSCRIPTS
The maturation pathway of trypanosomatid U-snRNAs, including the determinants for cap trimethylation, has not yet been studied in any detail. The biogenesis of the TMG cap has been dissected in Xenopus oocytes (64). Briefly, these snRNAs are initially capped with a m7G cap and, after export to the cytoplasm and assembly with the core Sm proteins, the cap is further modified in the cytoplasm to produce the characteristic 2,2,7 trimethyl guanosine cap. Nuclear import of the matured U-snRNAs then takes place.
In T. brucei U-snRNAs and the SL RNA bind a set of common proteins that are analogous to the Sm proteins (58,59). However, inspection of the available T. brucei sequences does not reveal a highly conserved sequence motif similar to the Sm site that serves as the binding site for the Sm proteins. Furthermore, in vitro reconstitution experiments with the T. brucei U2 snRNA showed that the Sm analogous region is not essential for core snRNP assembly (23,24). Consistent with these observations, the analysis of mutant U2 snRNAs in T. brucei cells revealed that binding of the common proteins is not dependent on the region analogous to the Sm site (22). Although these results deviate from the well-characterized mechanism of U-snRNP biogenesis in higher eukaryotes, it is important to point out that experiments in Leptomonas, a distantly related trypanosomatid, have shown that the Sm site of the U4 and U5 snRNA is essential for core RNP assembly (6,7). Thus, it appears that there is going to be a certain degree of variation in the maturation pathways of U-snRNAs in different trypanosomatids. Nevertheless, for the T. brucei U2 snRNA it was also shown that the Sm analogous site is not required for cap trimethylation. Actually, excluding the 5′ terminal 24 nucleotides of the promoter, the T. brucei U2 snRNA sequences do not harbor determinants for TMG cap formation. In addition, binding of common proteins is not a prerequisite for TMG cap formation. These results are intriguing and clearly require further investigations. In particular, it will be interesting to characterize the interaction of the U2 snRNA 5′ end with the snRNA-(guanosine-N2)-methyltransferase, which converts m7G to TMG, because there is evidence that in mammals the core protein complex provides a binding site for this methyltransferase (61). We should also keep in mind that TMG-capped trypanosomatid U-snRNAs are synthesized by RNA polymerase III and that as a consequence the capping pathway, including the conversion to TMG, will most likely display several unexpected nuances to the well-understood mechanism in yeast and mammals.
FORMATION OF THE SL RNA CAP 4 STRUCTURE
As mentioned earlier, one of the essential steps of gene expression in trypanosomes involves the maturation of pre-mRNAs by trans-splicing. This process entails the transfer of the first 39 nucleotides of the SL RNA, a small capped RNA of about 90–140 nucleotides depending on the species, to the 5′ end of all mRNAs. Thus, the primary role of trans-splicing is to create 5′ ends for individual mRNAs derived from polycistronic pre-mRNAs, and in doing so trans-splicing fulfills the important function of providing mRNAs with a cap structure. Although the structure of the polycistronic pre-mRNA 5′ end is not known, it is established that the cap structure of the mature mRNA is derived from the SL RNA and is not further modified after trans-splicing.
The biogenesis of the cap 4 structure can be subdivided into two major steps: a “classical” capping reaction and subsequent modification reactions carried out by SL RNA-specific methyltransferases (Fig. 2). In this scheme, the SL RNA primary transcript is first capped by the addition of GMP by RNA guanylyltransferase, or capping enzyme, and then RNA (guanine-7-) methyltransferase acts to form the monomethylated cap 0 structure. Finally, the nucleotides adjacent to the m7G cap are modified by the addition of seven methyl groups: each nucleotide is methylated at the 2′-O position of the ribose and the first and fourth nucleotide are also modified by the addition of methyl groups to the base. This two-step mechanism is supported by inhibition studies using S-adenosyl-l-homocysteine (80).
What have we learned so far about the actual mechanism of cap 4 biosynthesis? The first significant step towards a biochemical characterization of the enzymatic activities involved in cap 4 modification was the development of a cell-free system (79). The substrate in this assay consisted of radiolabeled SL RNP, synthesized in detergent-permeabilized T. brucei cells in the presence of adenosyl-l-homocysteine to prevent methylation of the SL RNA 5′ end. Under these conditions, approximately half of the SL RNA in the resulting RNP preparation carried a m7G cap, whereas the remainder was capped with an unmodified guanosine residue. Nevertheless, no modifications were detectable on the four nucleotides after the cap. Incubation of this “undermodified” substrate with whole cell extracts from T. brucei resulted in accurate addition of the seven methyl groups characteristic of the cap 4 structure. Furthermore, consistent with the conservation of the cap 4 structure between such divergent genera as Trypanosoma and Crithidia (5), trypanosomatid protozoa appear to share a common machinery for SL RNA cap formation (79).
Preliminary fractionation experiments with T. brucei and C. fasciculata cell-free extracts revealed that a macromolecular complex of approximately 300 kDa is involved in modification of the cap 4 structure and work is in progress to identify and clone the constituents of this machinery (our unpublished results). Independently, the guanylyltransferase, which adds the capping guanosine residue, was purified from C. fasciculata and the corresponding genes were cloned from C. fasciculata and T. brucei (72). More recently, a genomic clone encoding the L. major guanylyltransferase was deposited in the NCBI database (Accession number AL499622). All three polypeptides arecomposed of two distinct domains: whereas the carboxy-terminal half harbors the six signature motifs characteristic of eukaryotic and viral capping enzymes, the function of the amino-terminal domain needs to be determined.
To address the sequence requirements in the SL RNA for trans-splicing and cap 4 formation, in vivo functional assays have been developed in L. seymouri (41) and L. tarentolae (75,76). Somewhat surprisingly, the results emerging from these two studies are contradictory. In particular, whereas severe defects in cap 4 modification, due to mutations in the SL exon sequences, were accompanied by very low or undetectable trans-splicing in the Leptomonas system (41), the opposite outcome was reported in the Leishmania system (76). Furthermore, the primary sequence of the L. seymouri SL exon played a role in trans-splicing, but was not critical in the L. tarentolae counterpart. At present, the results obtained in the Leptomonas system are consistent with previous studies, and it is difficult to rationalize the contrasting Leishmania results, recalling that (i) the cap 4 structure is essential for trans-splicing in T. brucei, T. cruzi, and L. amazonensis cells (49,80), and (ii) the trypanosomatid SL sequence is highly conserved (81) and, unlike the Ascaris SL (29), is not part of the SL RNA gene promoter (see above). There are a number of experimental differences between the two studies, including the method for generating the mutants (linker substitution versus changing the sequence to the complementary one), and the trans-splicing assay (PCR versus primer extension), which could contribute to these contrasting results. Finally, one severe limitation in both studies was to rely exclusively on primer extension analysis for assaying the modification status of the SL RNA 5′ end. This led to the bizarre result in the Leishmania system, where substitution of nucleotides 1 through 9 did not significantly affect cap 4 formation of this mutant SL RNA (76). Thus, to clarify the issue of whether cap 4 determinants are present in the SL sequence will require a direct RNA analysis of the modifications present at the 5′ end of mutant SL RNAs.
Contrasting results were also reported for the importance of SL intron sequences in cap 4 formation and trans-splicing. In L. tarentolae the sequence of stem–loop II, the Sm site, and the structure of stem–loop III were important for cap 4 modification and trans-splicing (75). On the other hand, in Leptomonas the following results were obtained: substitution of stem–loop II did not affect cap 4 formation, but the structure rather than the sequence of stem–loop II was required for trans-splicing; the Sm site and stem–loop III were dispensable for trans-splicing; stem–loop III was expendable for cap 4 formation, but the effect of the Sm mutant on cap 4 formation was not assayed (41). As pointed out above, it is difficult to find a basis for these contradictory results and further investigations are clearly needed to assign functional importance to SL RNA subregions.
An alternative approach toward an understanding of cap 4 biosynthesis has been to examine the temporal progression of cap synthesis in vivo (46). The experimental set-up was to capture prematurely terminated SL RNA chains, generated by inclusion of the transcription terminator 3′-O-methyl-GTP in permeable T. brucei cells, and to determine the modification state of the SL RNA fragments by direct RNA analysis. m7G capping of the 140-nt-long SL RNA occurred on SL RNA transcripts 30 nucleotides in length or longer, entirely consistent with data from Drosophila nuclear run-on and vaccinia virus systems (27,63). But more importantly, the T. brucei studies provided the first in-depth look at the progression of cap 4 formation in that subsequent modifications were added successively in a 5′ to 3′ direction. In particular, short transcripts (between 56 and 67 nucleotides) were partially modified and carried methyl groups exclusively on the first two adenosine residues, whereas a fully modified cap 4 structure was present on RNA chains 117 nucleotides in length and longer. Thus, both modifications leading to a cap 4 structure (i.e., addition of a m7G cap and subsequent methylation of base and sugar moieties) are cotranscriptional events. Naturally, this opens up an interesting avenue of investigations, namely the interaction of the cap 4 biosynthetic machinery with the SL RNA transcriptional complex.
Another noteworthy aspect of the transcriptional arrest studies in T. brucei was that m7G capping and modification of the first four nucleotides of the SL RNA was independent of core ribonucleoprotein formation (46). The SL RNA was only precipitable with anti-common protein antibodies, when the transcripts were longer than 112 nucleotides. Satisfactorily, this would place the structural determinant(s) for common protein binding in a region that was previously proposed to be analogous to the Sm binding site (81).
PERSPECTIVE
After some detours it turns out that trypanosomatids have mechanisms of pre-mRNA splicing quite similar to those of other eukaryotes. trans-Splicing has been joined by cis-splicing and a complete set of spliceosomal U-snRNAs has been characterized. Certainly, the addition of cis-splicing has taken away a major attraction, namely that trans-splicing was the only type of snRNP-mediated splicing present in these organisms. Although in vivo assays for both splicing events are available, the lack of a cell-free system for trans-splicing, or for that matter for cis-splicing, complicates an in-depth analysis of these mechanisms. Nevertheless, there are numerous areas that have emerged in the study of pre-mRNA processing worthy of further investigation. TMG cap formation on the RNA polymerase III-transcribed U-snRNAs is intriguing and raises numerous questions about the specificity and regulation of the capping mechanism. Another important aspect that needs to be addressed is whether there is a connection between Sm proteins and the cap modification machinery. In particular, as pointed out by others (21), there might be different combinations of Sm proteins that interact with methyltransferase(s) responsible for generating particular cap structures. Finally, the cap 4 structure on the SL RNA is unique among eukaryotes, and initial studies of its synthesis have already yielded the first insights into the mechanism of capping beyond the initial addition of a guanosine residue.
ACKNOWLEDGMENTS
We thank Shulamit Michaeli for critical comments on the manuscript. Work carried out in our laboratory received support from National Institutes of Health Grant AI28798 to E.U. and AI43594 to C.T. E.U. is the recipient of a Burroughs Wellcome Fund Scholar Award in Molecular Parasitology and C.T. is the recipient of a Burroughs Wellcome Fund New Investigator Award in Molecular Parasitology.
REFERENCES
- 1. Agabian N. Trans splicing of nuclear pre-mRNAs. Cell 61:1157–1160; 1990. [DOI] [PubMed] [Google Scholar]
- 2. Agami R.; Aly R.; Halman S.; Shapira M. Functional analysis of cis-acting DNA elements required for expression of the SL RNA gene in the parasitic protozoan Leishmania amazonensis . Nucleic Acids Res. 22:1959–1965; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alfonzo J. D.; Thiemann O.; Simpson L. The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 25:3751–3759; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ares M. Jr.; Weiser B. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog. Nucleic Acid Res. Mol. Biol. 50:131–159; 1995. [DOI] [PubMed] [Google Scholar]
- 5. Bangs J. D.; Crain P. F.; Hashizume T.; McCloskey J. A.; Boothroyd J. C. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805–9815; 1992. [PubMed] [Google Scholar]
- 6. Bell M.; Bindereif A. Cloning and mutational analysis of the Leptomonas seymouri U5 snRNA gene: Function of the Sm site in core RNP formation and nuclear localization. Nucleic Acids Res. 27:3986–3994; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bell M.; Wohner R.; Bindereif A. U4 small nuclear RNA genes of trypanosomes: Cloning of the Leptomonas seymouri gene and mutational analysis of core snRNP assembly. Gene 247:77–86; 2000. [DOI] [PubMed] [Google Scholar]
- 8. Ben-Shlomo H.; Levitan A.; Beja O.; Michaeli S. The trypanosomatid Leptomonas collosoma 7SL RNA gene. Analysis of elements controlling its expression. Nucleic Acids Res. 25:4977–4984; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boothroyd J. C.; Cross G. A. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene 20:281–289; 1982. [DOI] [PubMed] [Google Scholar]
- 10. Campbell D. A.; Sturm N. R.; Yu M. C. Transcription of the kinetoplastid spliced leader RNA gene. Parasitol. Today 16:78–82; 2000. [DOI] [PubMed] [Google Scholar]
- 11. Cho E. J.; Takagi T.; Moore C. R.; Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319–3326; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colgan D. F.; Manley J. L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755–2766; 1997. [DOI] [PubMed] [Google Scholar]
- 13. Coppola J. A.; Field A. S.; Luse D. S. Promoter-proximal pausing by RNA polymerase II in vitro: Transcripts shorter than 20 nucleotides are not capped. Proc. Natl. Acad. Sci. USA 80:1251–1255; 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crenshaw-Williams K.; Bellofatto V. In vivo transcriptional analysis of the spliced leader RNA gene in the trypanosomatid Leptomonas seymouri . Parasitol. Res. 85:700–706; 1999. [DOI] [PubMed] [Google Scholar]
- 15. Djikeng A.; Ferreira L.; D’Angelo M.; Dolezal P.; Lamb T.; Murta S.; Triggs V.; Ulbert S.; Villarino A.; Renzi S.; Ullu E.; Tschudi C. Characterization of a candidate Trypanosoma brucei U1 snRNA gene. Mol. Biochem. Parasitol. 113:109–115; 2001. [DOI] [PubMed] [Google Scholar]
- 16. Dungan J. M.; Watkins K. P.; Agabian N. Evidence for the presence of a small U5-like RNA in active trans-spliceosomes of Trypanosoma brucei . EMBO J. 15:4016–4029; 1996. [PMC free article] [PubMed] [Google Scholar]
- 17. Fantoni A.; Dare A. O.; Tschudi C. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol. Cell. Biol. 14:2021–2028; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garlapati S.; Aly R.; Shapira M. Genus-specific expression from the SL RNA promoter of Leishmania amazonensis . Exp. Parasitol. 89:266–270; 1998. [DOI] [PubMed] [Google Scholar]
- 19. Goldring A.; Michaeli S. The U6 snRNA-encoding gene of the monogenetic trypanosomatid Leptomonas collosoma . Gene 156:139–144; 1995. [DOI] [PubMed] [Google Scholar]
- 20. Goldring A.; Zimmer Y.; Ben-Yehuda E.; Goncharov I.; Michaeli S. Stable transfection in the monogenetic trypanosomatid Leptomonas collosoma—transcription barrier of heterologous trypanosomatid SL RNA genes and expression of a chimeric SL RNA molecule. Exp. Parasitol. 84:28–41; 1996. [DOI] [PubMed] [Google Scholar]
- 21. Goncharov I.; Palfi Z.; Bindereif A.; Michaeli S. Purification of the spliced leader ribonucleoprotein particle from Leptomonas collosoma revealed the existence of an Sm protein in trypanosomes. Cloning the SmE homologue. J. Biol. Chem. 274:12217–12221; 1999. [DOI] [PubMed] [Google Scholar]
- 22. Günzl A.; Bindereif A.; Ullu E.; Tschudi C. Determinants for cap trimethylation of the U2 small nuclear RNA are not conserved between Trypanosoma brucei and higher eukaryotic organisms. Nucleic Acids Res. 28:3702–3709; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Günzl A.; Cross M.; Bindereif A. Domain structure of U2 and U4/U6 small nuclear ribonucleoprotein particles from Trypanosoma brucei: Identification of trans-spliceosomal specific RNA–protein interactions. Mol. Cell. Biol. 12:468–479; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Günzl A.; Cross M.; Palfi Z.; Bindereif A. Assembly of the U2 small nuclear ribonucleoprotein from Trypanosoma brucei. A mutational analysis. J. Biol. Chem. 268:13336–13343; 1993. [PubMed] [Google Scholar]
- 25. Günzl A.; Tschudi C.; Nakaar V.; Ullu E. Accurate transcription of the Trypanosoma brucei U2 small nuclear RNA gene in a homologous extract. J. Biol. Chem. 270:17287–17291; 1995. [DOI] [PubMed] [Google Scholar]
- 26. Günzl A.; Ullu E.; Dörner M.; Fragoso S. P.; Hoffmann K. F.; Milner J. D.; Morita Y.; Nguu E. K.; Vanacova S.; Wünsch S.; Dare A. O.; Kwon H.; Tschudi C. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 85:67–76; 1997. [DOI] [PubMed] [Google Scholar]
- 27. Hagler J.; Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science 255:983–986; 1992. [DOI] [PubMed] [Google Scholar]
- 28. Hajduk S. L.; Adler B.; Madison-Antenucci S.; McManus M.; Sabatini R. Insertional and deletional RNA editing in trypanosome mitochondria. Nucleic Acids Symp. Ser. 15–18; 1997. [PubMed] [Google Scholar]
- 29. Hannon G. J.; Maroney P. A.; Ayers D. G.; Shambaugh J. D.; Nilsen T. W. Transcription of a nematode trans-spliced leader RNA requires internal elements for both initiation and 3′ end-formation. EMBO J. 9:1915–1921; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harris K. A. Jr.; Crothers D. M.; Ullu E. In vivo structural analysis of spliced leader RNAs in Trypanosoma brucei and Leptomonas collosoma: A flexible structure that is independent of cap4 methylations. RNA 1:351–362; 1995. [PMC free article] [PubMed] [Google Scholar]
- 31. Hartree D.; Bellofatto V. Essential components of the mini-exon gene promoter in the trypanosomatid Leptomonas seymouri . Mol. Biochem. Parasitol. 71:27–39; 1995. [DOI] [PubMed] [Google Scholar]
- 32. Henry R. W.; Ford E.; Mital R.; Mittal V.; Hernandez N. Crossing the line between RNA polymerases: Transcription of human snRNA genes by RNA polymerases II and III. Cold Spring Harb. Symp. Quant. Biol. 63:111–120; 1998. [DOI] [PubMed] [Google Scholar]
- 33. Ho C. K.; Sriskanda V.; McCracken S.; Bentley D.; Schwer B.; Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 273:9577–9585; 1998. [DOI] [PubMed] [Google Scholar]
- 34. Huie J. L.; He P.; Bellofatto V. In vitro transcription of the Leptomonas seymouri SL RNA and U2 snRNA genes using homologous cell extracts. Mol. Biochem. Parasitol. 90:183–192; 1997. [DOI] [PubMed] [Google Scholar]
- 35. Jove R.; Manley J. L. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J. Biol. Chem. 259:8513–8521; 1984. [PubMed] [Google Scholar]
- 36. Kambach C.; Walke S.; Young R.; Avis J. M.; de la Fortelle E.; Raker V. A.; Luhrmann R.; Li J.; Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96:375–387; 1999. [DOI] [PubMed] [Google Scholar]
- 37. Kastner B.; Bach M.; Lührmann R. Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: Evidence for a common structure-determining principle in the major U snRNP family. Proc. Natl. Acad. Sci. USA 87:1710–1714; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee M. G.; Van der Ploeg L. H. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu. Rev. Microbiol. 51:463–489; 1997. [DOI] [PubMed] [Google Scholar]
- 39. Li L.; Otake L. R.; Xu Y.; Michaeli S. The transspliceosomal U4 RNA from the monogenetic trypanosomatid Leptomonas collosoma. Cloning and identification of a transcribed tRNA-like element that controls its expression. J. Biol. Chem. 275:2259–2264; 2000. [DOI] [PubMed] [Google Scholar]
- 40. Lücke S.; Klockner T.; Palfi Z.; Boshart M.; Bindereif A. Trans mRNA splicing in trypanosomes: Cloning and analysis of a PRP8-homologous gene from Trypanosoma brucei provides evidence for a U5-analogous RNP. EMBO J. 16:4433–4440; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lücke S.; Xu G. L.; Palfi Z.; Cross M.; Bellofatto V.; Bindereif A. Spliced leader RNA of trypanosomes: In vivo mutational analysis reveals extensive and distinct requirements for trans splicing and cap 4 formation. EMBO J. 15:4380–4391; 1996. [PMC free article] [PubMed] [Google Scholar]
- 42. Luo H.; Bellofatto V. Characterization of two protein activities that interact at the promoter of the trypanosomatid spliced leader RNA. J. Biol. Chem. 272:33344–33352; 1997. [DOI] [PubMed] [Google Scholar]
- 43. Luo H.; Gilinger G.; Mukherjee D.; Bellofatto V. Transcription initiation at the TATA-less spliced leader RNA gene promoter requires at least two DNA-binding proteins and a tripartite architecture that includes an initiator element. J. Biol. Chem. 274:31947–31954; 1999. [DOI] [PubMed] [Google Scholar]
- 44. Lutz C. S.; Murthy K. G.; Schek N.; O’Connor J. P.; Manley J. L.; Alwine J. C. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 10:325–337; 1996. [DOI] [PubMed] [Google Scholar]
- 45. Mair G.; Shi H.; Li H.; Djikeng A.; Aviles H. O.; Bishop J. R.; Falcone F. H.; Gavrilescu C.; Montgomery J. L.; Santori M. I.; Stern L. S.; Wang Z.; Ullu E.; Tschudi C. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA 6:163–169; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mair G.; Ullu E.; Tschudi C. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275:28994–28999; 2000. [DOI] [PubMed] [Google Scholar]
- 47. Maroney P. A.; Yu Y. T.; Jankowska M.; Nilsen T. W. Direct analysis of nematode cis- and trans-spliceosomes: A functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs. RNA 2:735–745; 1996. [PMC free article] [PubMed] [Google Scholar]
- 48. McCracken S.; Fong N.; Yankulov K.; Ballantyne S.; Pan G.; Greenblatt J.; Patterson S. D.; Wickens M.; Bentley D. L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357–361; 1997. [DOI] [PubMed] [Google Scholar]
- 49. McNally K. P.; Agabian N. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol. Cell. Biol. 12:4844–4851; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mottram J.; Perry K. L.; Lizardi P. M.; Lührmann R.; Agabian N.; Nelson R. G. Isolation and sequence of four small nuclear U RNA genes of Trypanosoma brucei subsp. brucei: Identification of the U2, U4, and U6 RNA analogs. Mol. Cell. Biol. 9:1212–1223; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakaar V.; Dare A. O.; Hong D.; Ullu E.; Tschudi C. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol. 14:6736–6742; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakaar V.; Günzl A.; Ullu E.; Tschudi C. Structure of the Trypanosoma brucei U6 snRNA gene promoter. Mol. Biochem. Parasitol. 88:13–23; 1997. [DOI] [PubMed] [Google Scholar]
- 53. Nakaar V.; Tschudi C.; Ullu E. An unusual liaison: Small nuclear and cytoplasmic RNA genes team up with tRNA genes in trypanosomatid protozoa. Parasitol. Today 11:225–228; 1995. [Google Scholar]
- 54. Newman A. J.; Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68:743–754; 1992. [DOI] [PubMed] [Google Scholar]
- 55. Nilsen T. W. trans-splicing: An update. Mol. Biochem. Parasitol. 73:1–6; 1995. [DOI] [PubMed] [Google Scholar]
- 56. Nunes L. R.; Carvalho M. R.; Shakarian A. M.; Buck G. A. The transcription promoter of the spliced leader gene from Trypanosoma cruzi . Gene 188:157–168; 1997. [DOI] [PubMed] [Google Scholar]
- 57. Palfi Z.; Bindereif A. Immunological characterization and intracellular localization of trans-spliceosomal small nuclear ribonucleoproteins in Trypanosoma brucei . J. Biol. Chem. 267:20159–20163; 1992. [PubMed] [Google Scholar]
- 58. Palfi Z.; Günzl A.; Cross M.; Bindereif A. Affinity purification of Trypanosoma brucei small nuclear ribonucleoproteins reveals common and specific protein components. Proc. Natl. Acad. Sci. USA 88:9097–9101; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palfi Z.; Lücke S.; Lahm H. W.; Lane W. S.; Kruft V.; Bragado-Nilsson E.; Seraphin B.; Bindereif A. The spliceosomal snRNP core complex of Trypanosoma brucei: Cloning and functional analysis reveals seven Sm protein constituents. Proc. Natl. Acad. Sci. USA 97:8967–8972; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Palfi Z.; Xu G. L.; Bindereif A. Spliced leader-associated RNA of trypanosomes. Sequence conservation and association with protein components common to trans-spliceosomal ribonucleoproteins. J. Biol. Chem. 269:30620–30625; 1994. [PubMed] [Google Scholar]
- 61. Plessel G.; Fischer U.; Lührmann R. m3G cap hyper-methylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: Evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol. Cell. Biol. 14:4160–4172; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Plessel G.; Lührmann R.; Kastner B. Electron microscopy of assembly intermediates of the snRNP core: Morphological similarities between the RNA-free (E.F.G) protein heteromer and the intact snRNP core. J. Mol. Biol. 265:87–94; 1997. [DOI] [PubMed] [Google Scholar]
- 63. Rasmussen E. B.; Lis J. T. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. USA 90:7923–7927; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reddy R.; Singh R.; Shimba S. Methylated cap structures in eukaryotic RNAs: Structure, synthesis and functions. Pharmacol. Ther. 54:249–267; 1992. [DOI] [PubMed] [Google Scholar]
- 65. Saito R. M.; Elgort M. G.; Campbell D. A. A conserved upstream element is essential for transcription of the Leishmania tarentolae mini-exon gene. EMBO J. 13:5460–5469; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schnare M. N.; Gray M. W. A candidate U1 small nuclear RNA for trypanosomatid protozoa. J. Biol. Chem. 274:23691–23694; 1999. [DOI] [PubMed] [Google Scholar]
- 67. Schnare M. N.; Gray M. W. Spliced leader-associated RNA from Crithidia fasciculata contains a structure resembling stem/loop II of U1 snRNA. FEBS Lett. 459:215–217; 1999. [DOI] [PubMed] [Google Scholar]
- 68. Schnare M. N.; Gray M. W. Structural conservation and variation among U5 small nuclear RNAs from trypanosomatid protozoa. Biochim. Biophys. Acta 1490:362–366; 2000. [DOI] [PubMed] [Google Scholar]
- 69. Segault V.; Will C. L.; Sproat B. S.; Lührmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 14:4010–4021; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shi X.; Chen D. H.; Suyama Y. A nuclear tRNA gene cluster in the protozoan Leishmania tarentolae and differential distribution of nuclear-encoded tRNAs between the cytosol and mitochondria. Mol. Biochem. Parasitol. 65:23–37; 1994. [DOI] [PubMed] [Google Scholar]
- 71. Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66:1–40; 2000. [DOI] [PubMed] [Google Scholar]
- 72. Silva E.; Ullu E.; Kobayashi R.; Tschudi C. Trypanosome capping enzymes display a novel two-domain structure. Mol. Cell. Biol. 18:4612–4619; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stark H.; Dube P.; Lührmann R.; Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409:539–542.; 2001. [DOI] [PubMed] [Google Scholar]
- 74. Stuart K.; Allen T. E.; Heidmann S.; Seiwert S. D. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 61:105–120; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sturm N. R.; Campbell D. A. The role of intron structures in trans-splicing and cap 4 formation for the Leishmania spliced leader RNA. J. Biol. Chem. 274:19361–19367; 1999. [DOI] [PubMed] [Google Scholar]
- 76. Sturm N. R.; Fleischmann J.; Campbell D. A. Efficient trans-splicing of mutated spliced leader exons in Leishmania tarentolae . J. Biol. Chem. 273:18689–18692; 1998. [DOI] [PubMed] [Google Scholar]
- 77. Tschudi C.; Krainer A. R.; Ullu E. The U6 small nuclear RNA from Trypanosoma brucei . Nucleic Acids Res. 16:11375; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tschudi C.; Richards F. F.; Ullu E. The U2 RNA analogue of Trypanosoma brucei gambiense: Implications for a splicing mechanism in trypanosomes. Nucleic Acids Res. 14:8893–8903; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ullu E.; Tschudi C. Accurate modification of the trypanosome spliced leader cap structure in a homologous cell-free system. J. Biol. Chem. 270:20365–20369; 1995. [DOI] [PubMed] [Google Scholar]
- 80. Ullu E.; Tschudi C. Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc. Natl. Acad. Sci. USA 88:10074–10078; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ullu E.; Tschudi C.; Günzl A. Trans-splicing in trypanosomatid protozoa. In: Smith D. F.; Parsons M., eds. Molecular biology of parasitic protozoa. Oxford, UK: Oxford University Press; 1996:115–133. [Google Scholar]
- 82. Watkins K. P.; Dungan J. M.; Agabian N. Identification of a small RNA that interacts with the 5′ splice site of the Trypanosoma brucei spliced leader RNA in vivo. Cell 6:171–182; 1994. [DOI] [PubMed] [Google Scholar]
- 83. Wieland B.; Bindereif A. Unexpected diversity in U6 snRNA sequences from trypanosomatids. Gene 161:129–133; 1995. [DOI] [PubMed] [Google Scholar]
- 84. Xu G. L.; Wieland B.; Bindereif A. trans-spliceosomal U6 RNAs of Crithidia fasciculata and Leptomonas seymouri: Deviation from the conserved ACAGAG sequence and potential base pairing with spliced leader RNA. Mol. Cell. Biol. 14:4565–4570; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xu Y.; Ben-Shlomo H.; Michaeli S. The U5 RNA of trypanosomes deviates from the canonical U5 RNA: The Leptomonas collosoma U5 RNA and its coding gene. Proc. Natl. Acad. Sci. USA 94:8473–8478; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xu Y.; Liu L.; Michaeli S. Functional analyses of positions across the 5′ splice site of the trypanosomatid spliced leader RNA—implications for base-pair interaction with U5 and U6 snRNAs. J. Biol. Chem. 2000. [DOI] [PubMed] [Google Scholar]
- 87. Yu M. C.; Guy Roberts T.; Sturm N. R.; Campbell D. A. In vitro transcription of mutated Leishmania tarentolae spliced leader RNA genes approximates in vivo patterns. Mol. Biochem. Parasitol. 111:391–399; 2000. [DOI] [PubMed] [Google Scholar]
- 88. Yu M. C.; Sturm N. R.; Saito R. M.; Roberts T. G.; Campbell D. A. Single nucleotide resolution of promoter activity and protein binding for the Leishmania tarentolae spliced leader RNA gene. Mol. Biochem. Parasitol. 94:265–281; 1998. [DOI] [PubMed] [Google Scholar]
- 89. Yue Z.; Maldonado E.; Pillutla R.; Cho H.; Reinberg D.; Shatkin A. J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:12898–12903; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


