Abstract
After transcription by RNA polymerase (pol) III, nascent Pol III transcripts pass through RNA processing, modification, and transport machineries as part of their posttranscriptional maturation process. The first factor to interact with Pol III transcripts is La protein, which binds principally via its conserved N-terminal domain (NTD), to the UUU-OH motif that results from transcription termination. This review includes a sequence Logo of the most conserved region of La and its refined modeling as an RNA recognition motif (RRM). La protects RNAs from 3′ exonucleolytic digestion and also contributes to their nuclear retention. The variety of modifications found on La-associated RNAs is reviewed in detail and considered in the contexts of how La may bind the termini of structured RNAs without interfering with recognition by modification enzymes, and its ability to chaperone RNAs through multiple parts of their maturation pathways. The CTD of human La recognizes the 5′ end region of nascent RNA in a manner that is sensitive to serine 366 phosphorylation. Although the CTD can control pre-tRNA cleavage by RNase P, a rate-limiting step in tRNASer UGA maturation, the extent to which it acts in the maturation pathway(s) of other transcripts is unknown but considered here. Evidence that a fraction of La resides in the nucleolus together with recent findings that several Pol III transcripts pass through the nucleolus is also reviewed. An imminent goal is to understand how the bipartite RNA binding, intracellular trafficking, and signal transduction activities of La are integrated with the maturation pathways of the various RNAs with which it associates.
Keywords: tRNA processing, RNA modification, RNase P, RNA recognition motif (RRM), Protein structure modeling, Transcription, Nucleolus, Autoantigen, 5′-ppp, Walker A motif, Lhp1, Sla1
RNA polymerase (pol) III synthesizes many small RNAs including 5S ribosomal (r)RNA and transfer (t)RNAs, as well as a variety of less abundant, small nuclear (sn), small nucleolar (sno), and small cytoplasmic (sc) RNAs. Newly synthesized Pol III transcripts undergo a maturation process that involves cleavage and/or modification and, in some cases, addition of nucleotides. Some Pol III transcripts undergo little processing while others must endure an elaborate maturation process (e.g., tRNAs).
Unlike the case for pre-mRNAs, which are transcribed by Pol II, neither RNA processing nor modification has been demonstrated to occur during Pol III transcription. Therefore, maturation of a Pol III transcript would appear to begin after synthesis is completed and the nascent RNA is released from the transcription complex. The final stage of transcription occurs in response to an oligo(dA) tract found at the 3′ ends of all pol III-dependent genes, which causes Pol III to terminate synthesis (16). Because the proximal part of the oligo(dA) signal is transcribed, all nascent Pol III transcripts share the common motif, oligo(rU)-OH, at their 3′ termini. Termination within the oligo(dA) tract generates heterogeneous ends, usually U(1-4)U-OH (hereafter referred to as UUU-OH), the distribution of which may vary depending on oligo(A) length and context (21,41,89,90,103,105,106,145). Because terminal uridylates comprise a high-affinity binding site for the La protein, all nascent Pol III transcripts are bound, at least transiently, by La (122). Indeed, the first protein that can be detected in association with newly synthesized Pol III transcripts is La.
La was first identified as an autoantigen in patients suffering from rheumatic disorders such as systemic lupus erythematosus and Sjogren’s syndrome, although homologues have been identified in every eukaryote examined, including protozoa (3), trypanosomes (87,137), yeasts (79,131,143), insects (10,99,143), amphibians (113), and mammals (24,127). Nascent Pol III transcripts that are known to associate with La are the precursors to tRNAs, 5S rRNA, U6 snRNA, 7SL (SRP) scRNA, 7SK snRNA, hY scRNAs, 7-2 (MRP) snoRNA, RNase P RNA, 4.5S I RNA, B1-Alu, Alu RNAs, and others, including Euplotes telomerase RNA (3,25,35,51,77,80,82,89,103,105,106,117) (see below).
Presumably by binding to UUU-OH, La can facilitate clearance of the newly terminated RNA from the transcription complex (40,41,86). Human La has also been shown to act as an ancillary factor that stimulates Pol III transcription in some in vitro systems, promoting efficient recycling of Pol III (27,34,38-41,84,86), although this is controversial (35,78,136,144). Another proposed activity of La, involvement in the translation of viral and cellular mRNAs that contain internal ribosome entry sites, has been reviewed previously (11,56,85), and will not be discussed here.
La has been considered as a molecular chaperone in multiple contexts: (i) that it may stabilize RNAs in the correct conformation (i.e., an activity related to RNA folding or RNA structure), (ii) that it can stabilize associated transcripts against exonuclease digestion or other degradative processes, and (iii) that it can serve as a platform on which associated transcripts may be recognized by modification and processing factors that contribute to the maturation of the RNA (i.e., that it accompanies transcripts during modification) (33,57,83,85,97,98,144). Examples of each of these will be considered below.
La also associates with certain RNAs that are synthesized by pol II. The first of these to be described were the leader transcripts generated from vesicular stomatitis virus and a U1 snRNA precursor in HeLa cells (70,81), followed by reports of others including the 5′ regions of viral and cellular mRNAs, several of which were represented by probes that contained terminal uridylates [reviewed in (85)]. La associates with precursor intermediates of U1-U5 snRNAs as well as U3 snoRNA, in large part via short tracts of terminal uridylates that result from incomplete trimming of their 3′ ends (69,142). La was also found to associate with an intermediate in a histone mRNA decay pathway whose 3′ processing occurred at a position that is predicted to expose terminal uridylates (91). Thus, for many of the above transcripts, binding appears to be driven primarily by recognition of 3′ terminal uridylates. Although recently published evidence suggests that La can also recognize RNAs that contain a CACAA sequence, via an RNA binding activity that will be considered in the context of pre-tRNAe Met stability in a later section, the elements in La responsible for this binding also reside in the NTD and overlap and/or cooperate with UUU-OH recognition (4,64). Thus, substantial evidence indicates that the major mode of RNA binding, recognition of UUU-OH, is mediated by the most highly conserved region of La, the N-terminal domain (NTD) (85).
As diagrammed in Figure 1 and discussed in more detail in a later section, the human La protein has been shown to recognize the 5′-ppp end region of RNA, a mode of binding that is clearly distinct from the NTD-mediated interaction with UUU-OH, as this 5′ end binding activity is mediated by the C-terminal domain (CTD) of La [(33,57), reviewed in (85)].
Figure 1.
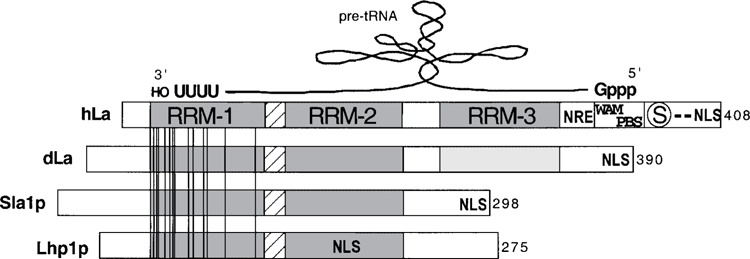
Schematic representation of human La, its apparent mode of bipartite binding to a nascent precursor tRNA, and comparison to other La proteins. Cartoon alignment of the La proteins of human (hLa), D. melanogaster (dLa), S. pombe (Sla1p), and S. cerevisiae (Lhplp). Their lengths, in aa, are indicated at the right. RRM-1, -2 and -3 domains are shaded and labeled for hLa only. Invariant residues in 13 sequences available in the database are shown as vertical lines; note that these are limited to RRM-1. The invariant residues are numbered according to hLa: Q20, E22, Y24, F25, N29, D33, F35, L36, G45, V47, F55, R57, A71, and R91. A putative Walker A motif (WAM) and phosphate binding site (PBS), and serine 366 (encircled S), are shown (see text). Positions of the NLS are indicated in bold font according to Rosenblum et al. (109). An element that functions as a nuclear retention element (NRE, see text) (119) is also shown. A schematized pre-tRNA has been modeled above to illustrate its proposed orientation and bipartite mode of binding, according to prior work (33,57) as described in the text. Reproduced from Maraia and Intine (85).
THE FUNCTIONAL REPERTOIRE OF La EXTENDS TO 5′ END METABOLISM OF PRE-tRNA
As detailed previously, a Walker A motif (WAM)-like sequence in the CTD of human La contributes to pre-tRNA 5′-ppp end recognition and control of processing by RNase P (85). This mode of recognition could potentially explain how La can protect a variety of pre-tRNA species from 5′ processing despite little if any conservation in their leader sequences, because the pppG/A moiety represents a unique determinant on each transcript (33). The BLOCKS program identifies aa 348–368 of hLa as a potential phosphate binding site (PBS), just downstream of the WAM, that is rich in conserved basic residues (53) (Fig. 1). Further support for this mode of interaction came from in vivo studies, which revealed that only when the WAM and the PBS sequences were intact was there protection of pre-tRNA from 5′ processing (57). The bipartite mode of 3′ and 5′ end binding by La also explains data obtained in vivo. Pre-tRNA 3′ end stabilization is mediated by La proteins that lack RRM-3 or downstream sequences, provided that they have an intact NTD, while 5′ end stabilization occurs only if the WAM-PBS and RRM-3 are also intact (33,57).
SIGNAL TRANSDUCTION THROUGH SERINE 366 PHOSPHORYLATION OF THE ACCESSORY CTS OF HUMAN La
The 5′-ppp end recognition activity of La and its modulation by serine 366 phosphorylation has been shown to play a functional role of tRNA expression in the intact cell [reviewed in (85)]. Indeed, human La is faithfully phosphorylated, specifically on S366 in fission yeast cells, and this modification promotes tRNA maturation, indicating the potential for regulation of tRNA processing in widely divergent species (57). Note that this mode of binding need not be limited to pre-tRNAs because all nascent Pol III transcripts initiate with 5′-ppp and terminate with UUU-OH (33,57). The CTD could afford La the ability to retain a nascent transcript (or its 5′ end region) in a stable form until it receives a signal (i.e., in the form of S366 phosphorylation) to release it (33). This feature, could potentially be used to control the handing off of a nascent RNA from La to another protein as occurs during RNP assembly, a period during which unchaperoned RNAs can succumb to exonucleolytic digestion as discussed above (69,97,142).
A CLASS OF La MOTIF-CONTAINING (LMC) POLYPEPTIDES THAT ARE STRUCTURALLY AND FUNCTIONALLY DISTINCT FROM GENUINE La PROTEINS
The most highly conserved region of the La proteins of many species resides in the N-terminal part of the protein, in a ∼80- amino acid region referred to as the La motif or RRM-1 (85). The N-terminal position of the La motif is a characteristic that appears to distinguish genuine (i.e., Pol III transcript-associated) La proteins from other polypeptides that contain homology to the La motif, in which cases the homology region is located at a more central or C-terminal position (121). As reviewed previously and summarized below, this region of La proteins appears to comprise an RNA recognition motif (RRM) that mediates UUU-OH recognition (85), suggesting a similar mode of RNA binding by the La motif-containing (LMC) proteins. Two of the LMC proteins have been identified in yeast, Sro9p and Slflp, and homologs have been identified in other species (121). Unlike genuine La proteins, the LMC proteins appear not to be involved with nascent Pol III transcripts but instead were associated with translating ribosomes (121). Indeed, genetic analyses have shown that the functions of these LMC proteins do not appear to overlap with the function of the genuine yeast La protein, Lhp1p (121). Sro9p has been identified as a suppressor of a variety of mutations, suggesting involvement in a wide range of genetic interactions [reviewed in (121)]. The ability of high copy Sro9p to suppress a mutation in a subunit of RNA polymerase II led Woychik and colleagues to the conclusion that this LMC protein increased general mRNA levels by increasing transcription as well as transcript stability (124). Although the characterization of the LMC class of proteins is still in its early stages, it seems clear that these proteins function independently of La and in a distinct functional pathway. For the remainder of this review, the focus will be on the genuine La proteins, all of which contain a La motif (a.k.a. RRM-1) near their N-terminus and many of which (Drosophila, human, rodent, S. cerevisiae, S. pombe, X. laevis) have been characterized to be UUU-OH binding proteins that associate with nascent Pol III transcripts.
IN THE ABSENCE OF DETERMINED STRUCTURE, CONSIDER A MODEL OF THE CONSERVED NTD OF La
Although the capacity of the La motif to form a RRM-like structure has been detailed before (85), the present review includes a new analysis of the predicted secondary structure of this region and a new perspective provided by the sequence Logo (115). We believe that this analysis and perspective will be useful both for sequence comparisons and targeted mutagenesis of yet undiscovered La proteins as well as LMC proteins, as new sequences become available. In addition, this analysis has led us to propose here that the lack of solvent-exposed aromatic residues in the β3 region of the La RRM-1 motif (see below) may be a specific solution to the problem of having to recognize a short tract of pyrimidines [i.e., oligo(U)], because exposed aromatic side chains might select for purines as a result of greater stacking potential (below).
La belongs to a large family of proteins that contain an ∼80 amino acid domain known as the RNA recognition motif (RRM) (13,19,65,132). The canonical RRM in human La that was initially noted (65) is now referred to as RRM-2, because two additional RRMs, designated RRM-1 and RRM-3, have been identified in the human La protein (13,64). The most highly conserved feature of all known La proteins is an ∼80 aa region located near the N-terminus, referred to as the La motif or RRM-1, which appears to be critical for UUU-OH binding (64,85). Although there is not yet a crystal structure for La, the structures of other RRMs have revealed a globular domain with a β1-α1-β2-β3-α2-β4 topology (28,31,49,55,96,116). In the RRM structure, the four β strands form a platform that contacts RNA on one surface while the other surface is packed against by the α helices, contributing to a hydrophobic core that is important for structural integrity of the RRM. A hallmark of the RRM is the presence of two short conserved sequence motifs, RNP-1 and RNP-2, which reside on β3 and β1, respectively (1,65,111), although some RRM proteins do not contain consensus RNP motifs (13,28,65). Some of the amino acids in the RNP motifs are oriented inward, toward and contributing to the hydrophobic core, while others face outward, exposed to the solvent, to contact RNA. In general, the buried hydrophobic residues in the RNP motifs alternate with the solvent-exposed residues that contact RNA (13,65).
In Figure 2, the RRM-1 regions of 13 La sequences were aligned and displayed as a sequence Logo (115). In this display, the single letter aa designations are stacked on top of each other for each position in the aligned sequence, the height of each letter being proportional to its frequency, and the letters are sorted with the most common one on top. The height of each stack is then adjusted by the Logo program, according to the information content (i.e., apparent conservation), and plotted on a logarithmic scale (base 2) on the Y axis, in bits (114). A sequence Logo reveals the consensus sequence as the top letter at each position, as well as the frequency of residues and the information content at each position (114). In the sequence Logo in Figure 2, single letters comprise the 14 tallest stacks, reflecting their complete invariance among the RRM-1 sequences (85).
Figure 2.
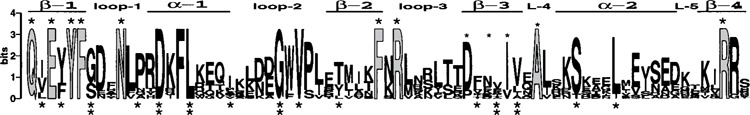
A sequence Logo and predicted structural features provide a model of the conserved RRM-1 of La protein. Sequences representing the RRM-1 regions of 13 La proteins were aligned and a sequence Logo was generated (114,115). The sequence Logo shown corresponds to residues 20–92 of hLa. The RRM topology, β1-loop1-α1-loop2-β2-loop3-β3-loop4-α2-loop5-β4 as proposed for La RRM-1, was placed above the sequence Logo (see text). Candidate residues potentially involved in RNA recognition are light shaded and topped with a bold asterisk. The putative RNP-2 and RNP-1 reside on β1 and β3, respectively (see text). Asterisks were placed below the positions corresponding to the conserved RRM core structure; two asterisks indicate a match to the consensus at that position and one asterisk indicates similarity (see text) (13). Apparent insertions in some of the La sequences (e.g., A.t. loop-3) were deleted prior to input into the Logo program. The sequences and accession numbers are: Homo sapiens, P05455; Bos taurus, P10881; Rattus norvegicus, P38656; Mus musculus, P32067; Xenopus laevis, P28049; Xenopus laevis, P28048; Aedes albopictus, Q26457; Caenorhabditis elegans, T30953; Drosophila melanogaster, P40796; Trypanosoma brucei, AAF34598.1; Schizosaccharomyces pombe, AAB 82145.1; Saccharomyces cerevisiae, P33399; A. thaliana, AAF09063.
The Logo output generated for La RRM-1 was annotated for Figure 2 in the following way: (i) differential shading of letters, (ii) placement of asterisks above or below certain positions, and (iii) addition of secondary structure elements that corresponded to the β1-loop1-α1-loop2-β2-loop3-β3-loop4-α2-loop5-β4 RRM topology as previously described (85). With regard to the latter, all of the α helices and connecting loops were previously predicted in the corresponding regions of the La sequences, while the regions of only three of the four putative β strands were predicted to contain an extended β strand (85). Examination of the consensus produced by Logo revealed an extended β strand in the β3 region that could not be predicted by previous methods (32) (data not shown), providing novel evidence to indicate that all of the secondary structure elements conforming to the RRM topology, β1-loop1-α1-loop2-β2-loop3-β3-loop4-α2-loop5-β4, have now been predicted for La RRM-1 as reflected in Figure 2.
Asterisks were placed below positions that correspond to all of the conserved residues that contribute to the hydrophobic core of the generic RRM structure, as discerned from examination of 70 RRMs (13,85). Although several of the hydrophobic core residues were described simply as hydrophobic, a hierarchy of sequence conservation at these positions had been noted. Some positions that contribute to the hydrophobic core were noted to be heavily represented by a single residue (e.g., the G found in loop 2 of 75–90% of the 70 RRMs analyzed), while identity at other core positions were moderately conserved (e.g., D or E in α1 occurs in 44% of the 70 RRM sequences) (13). Some of the conserved core RRM residues are thought to make defined contributions to the RRM structure; for example, the highly conserved G found in the loop 2 regions of 83% of 70 RRMS (invariant in La), may be required for the turn into β2 (13). Therefore, to assimilate this information into Figure 2, the number of asterisks under the hydrophobic core positions was varied; two asterisks were placed where a match between the top letter in the sequence Logo and the conserved residue in the core consensus was observed. This revealed that a significant fraction of the invariant and conserved residues in La RRM-1 correspond to conserved identity structural core positions (two asterisks below the Logo). This analysis suggests that the significance of a substantial portion of the sequence information in the most conserved region of La is that it specifies structural integrity of the RRM-1 domain. In addition, there is also a very good match between La RRM-1 and the RRM core residues that are distinguished simply by the shared hydrophobicity of their side chains (one asterisk below the Logo). Overall, this further indicates that the conserved regions of La proteins have conserved the capacity to form RRM-like structures.
Several highly conserved and invariant positions in the RRM-1 sequence do not correspond to core residues (and therefore do not have asterisks under them). Because these may not correspond to conserved core residues, it was reasoned that their conservation may reflect a role in RNA recognition, and these are indicated by a light shaded letter topped by an asterisk in Figure 2. This display suggests that residues topped by asterisks might project from one surface of RRM-1, toward the RNA ligand, while the residues with asterisks below would project toward the hydrophobic core. This perspective is generally consistent with the putative RNP motifs described in the next paragraph.
A reasonably good RNP-2 motif can be fit to β1, with invariant residues projecting putatively toward the RNA interaction surface, in general alternating with buried core residues. Arguing for the assignment of a RNP-1 motif appears weaker than for RNP-2 because solvent-exposed aromatic residues, a hallmark feature of prototypic RNP-1 motifs, are lacking in the β3 region, and those aromatic residues that are apparent correspond to buried positions. However, this should not necessarily detract from the RRM-1 model because some RRMs do not have prototypic RNP-1 motifs (13,28,65). Moreover, avoidance of aromatic residues in RNP-1 may be beneficial to the type of RNA binding exhibited by La proteins. Recall that aromatic residues on RNP-1 motifs of other RRM proteins contribute to RNA recognition via stacking, an interaction that is more favorable for purines than for pyrimidines (31,49,96,132). Thus, it should not be surprising that the putative RNP-1 would lack solvent-exposed aromatic residues because their inclusion might bias toward purine recognition. Therefore, the absence of solvent-exposed aromatic residues on RNP-1 might be a useful discriminatory factor when specificity must be limited to three or four uridylates.
Assignment of the hydrophobic core residues in β3 (13) leads to the putative designation of the conserved D and I residues in β3 along with the other position topped by a small asterisk in this region, as the solvent-exposed residues, because these alternate with the core positions. We suspect that these residues might contribute to RNA binding via protein backbone interactions with RNA as seen for other RNP-1 motifs (28,96).
In summary, the sequence Logo in Figure 2 reveals the most conserved residues in all La proteins, and should therefore be useful for comparisons of candidate La proteins including mutants. Additionally, the sequence and predicted secondary structure were evaluated in terms of the potential fit as a RRM. Our conclusion is that the fit is moderately good with the expectation that substantial differences between the predicted and real structures of the La RRM-1 region will be revealed upon knowing the atomic coordinates of a crystal structure. With these limitations, the proposed model would be most useful if tested by structure probing methods, mutagenesis, and functional studies.
Pol III TERMINATION-MEDIATED RECRUITMENT OF La PROTECTS NASCENT TRANSCRIPTS FROM 3′ EXONUCLEASE DIGESTION
Although mechanisms exist that allow nascent Pol III transcripts to endure in the absence of La, when present, it increases the efficiency of expression (i.e., increases the fraction of newly synthesized RNA that is converted to a functional product) of a variety of RNAs, especially under certain conditions (20,57,79,97,144). The number of Us required for optimal La binding matches the template requirement for termination by Pol III (122) and, accordingly, the number of terminal Us on nascent Pol III transcripts (41,90,106,145). Alteration of the terminal 3′-OH group to a phosphate decreases RNA binding by yeast, insect, amphibian, and human La proteins (122,125,143). This remarkably high degree of conservation of this otherwise subtle feature suggests that (i) recognition of the very end of a nascent transcript is critical to La function and (ii) some of the conserved residues of La might contribute specific recognition of the terminal ribose 3′-OH moiety. The high degree of conservation of 3′-OH discrimination by La discussed above, in conjunction with other experimental data, indicates that La’s conserved function is to cap the 3′ ends of nascent RNAS, protecting them from 3′-5′ exonucleolytic digestion (33,38,78,86,144).
Because the principal recognition site for La is produced by transcription termination by pol III, it seems reasonable to suspect that these transcripts are directed to La as a result of a link between La and Pol III termination or because they would otherwise be vulnerable, as newly terminated transcripts, to 3′ exonuclease digestion. In either case, Pol III termination leads to recruitment of La to the transcripts. Therefore, it seems worthwhile to consider a hypothetical thread that can putatively link La, Pol III termination, and RNA 3′ nucleolytic cleavage, which is suggested by recent but otherwise disparate observations. Pol III cleaves its associated nascent RNA at the 3′-OH end under certain circumstances, similar to bacterial RNA polymerase and pols I and II upon encountering pauses or other blocks to elongation (14,15,17,58,104,129,138). A small subunit of pol III, RPC11, facilitates termination by stimulating this 3′-OH cleavage activity (26). Thus, it is a remarkable coincidence that La protein, a factor shown to function in Pol III termination, exhibits an activity that can protect nascent RNAs from a polymerase-intrinsic 3′ cleavage reaction that is important for termination. Indeed, La has been found to copurify with a human Pol III holo-enzyme, suggesting intimate association with the synthetic machinery (135). It will be intriguing in the future to consider La in the context of the RPCII-associated termination activity.
RNA 3′ cleavage is certainly not limited to the hydrolytic activity of RNA polymerases, as exonucleases abound in nuclei, and it seems likely that protection of nascent transcripts after release from the transcription complex is a most important function of La (33,38,69,78). Indeed, Pol III termination is not a prerequisite for La binding, because, as mentioned above, La can protect snRNAs synthesized by pol II (69,142). Nonetheless, because Pol III produces 3′ oligo-U, there is an inextricable link between termination and La binding.
Normal cells rely on an orderly passage of nascent RNA from La to the next protein in a pathway, as the RNA matures. While the pathway of pre-tRNA maturation is quite elaborate (141), 5S rRNA maturation may appear less complex but nonetheless involves La. Evidence from mammalian and Xenopus cells indicated that 5S rRNA is first bound by La and then by the ribosomal protein L5 (or TFIIIA in Xenopus oocytes) prior to further transport (47,123). Similar involvement of La was suggested by studies of 7-2 (MRP) RNA, U6 snRNA, and B1-Alu RNA (51,82,125). Genetic evidence from yeast mutants indicates that La stabilizes newly synthesized U6 sn-RNA, increasing its chances for transfer or assembly into a Lsm8p-containing U6 snRNP (97). Indeed, multiple genetic interactions have been demonstrated between yeast La and the Lsm2–Lsm8 complex, to stabilize nascent U6 snRNA (98). Additional mutants indicate a similar chaperone activity of La for precursors of U3 snoRNA and U4 snRNA, even though they are synthesized by pol II (69,142). It is noteworthy that the proteins that replace La on U3 and U4 precursors may also stabilize the 3′ ends of the RNAs from digestion (69,98,142).
PRE-tRNA MODIFICATIONS OCCUR ON La-RNPs
It is becoming clear that facilitating RNA assembly into stable RNPs is not the extent of La’s function as a chaperone (97), because some evidence suggests that La may facilitate RNA folding and other evidence suggests that RNAs appear to remain associated with La during transient recognition by other proteins. Steitz and colleagues showed that La-associated transcripts contain a variety of modifications, including ribose and base methylation, dihydrouridylation, and pseudouridylation (52,75,105). As it was later shown that most La-associated RNAs were transient precursors to 5S rRNA and tRNAs (106), it would appear that the pre-tRNAs were modified before or while they were associated with La. Because, as discussed above, La appears to bind the RNAs before or immediately after transcription termination by Pol III, the cumulative evidence suggests that modifications occur while the nascent transcripts are associated with La [recently reviewed in (83)].
According to a bipartite binding model (Fig. 1), interactions with La would be limited principally to the terminal regions of a pre-tRNA, leaving the remainder of the tRNA accessible to modification enzymes and processing factors (33). As briefly reviewed previously (83) and described in greater detail below, the extent to which physical association with La may support or interfere with the activities of pre-tRNA modification and processing enzymes can be considered from perspectives afforded by five data sets: (i) recent results linking La and pre-tRNAi Met m1A58 modification (6,20), (ii) differences in the modifications found on a human pre-tRNAe Met that is stabilized by La, and its mature counterpart (50,64), (iii) results of Nishikura and DeRobertis who documented the modifications found on the nascent precursor and various intermediate species of tRNATyr (94), (iv) the finding that La specifically modulates access of the pre-tRNA substrate to RNase P in a manner that is sensitive to serine 366 phosphorylation in the intact cell (57), and (v) evidence that association of pre-tRNA with La protects against 3′ exonucleolytic digestion and thereby facilitates 3′ endonucleolytic cleavage (144).
With regard to (i) above, data indicate that La cooperates with the m1A58 modifying enzyme and that m1A58 is one of the earliest modifications of pre-tRNAs (6,20). Because La associates with many if not all pre-tRNAs, we expect similar cooperation with other modifying enzymes. Scrutiny of preexisting data also suggests that La may interfere with certain modifications (below).
With regard to (ii) above, human pre-tRNAe Met was initially identified as a tRNA precursor of unusually high stability (50). This pre-tRNA has been independently isolated as the most abundant La-associated transcript in HeLa cells (64) [see (4)]; this characteristic, in conjunction with its length of 86 nt and modification profile, suggests that it may be the “hb” RNA previously reported by Hendrick and colleagues (52). This stable pre-tRNAe Met contains the modified bases, D, T, m1A, m5C, M2 2G, and Cm, which are found mostly on the outer aspect of the elbow (D and T loops) and the lower half of the tRNA structure (Fig. 3), while the mature tRNAe Met species contains these plus two M2G residues (M2Ci6 & M2G1O, indicated as M2G in the oval in Fig. 3) in the acceptor stem and D stem, respectively, and i6A37 adjacent to the anticodon (50). With regard to (iii), this pattern of modifications is similar to that observed during the expression of pre-tRNATyr (94). Specifically, the pre-tRNATyr species containing a 5′ leader, 3′ trailer, and intron bears several of the modifications found on pre-tRNAe Met, while additional modifications were found on the intron-containing species only after end processing occurred (94).
Figure 3.
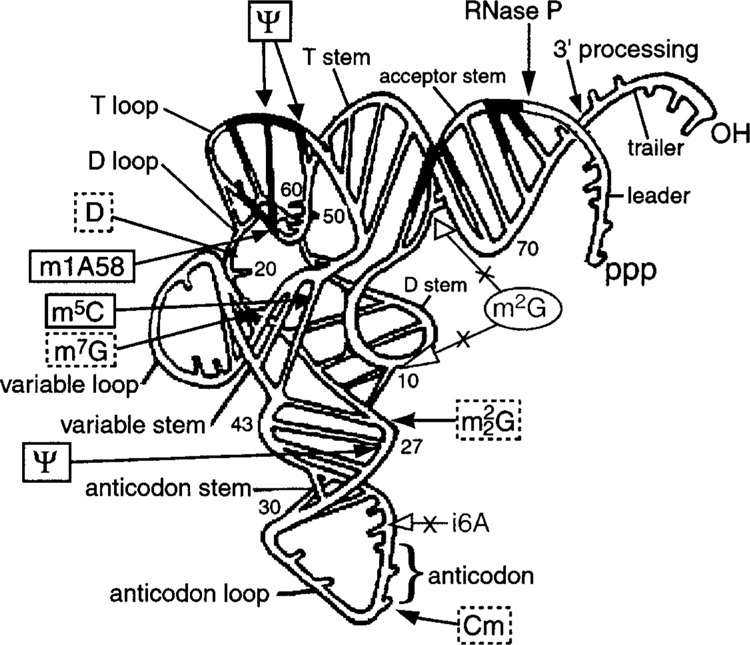
Model of a pre-tRNA with a summary of modifications found on precursors that have not yet had their 5′ leader or 3′ trailer removed, as previously described (50,94). Modifications in solid boxes were found on both pre-tRNAe Met and pre-tRNATyr, modifications in dashed boxes were found on pre-tRNAe Met (see text), and the m2G6 and m2G10 modifications (in the oval) were not present on pre-tRNAe Met or pre-tRNATyr, but were detected after the 5′ leaders and 3′ trailers were removed from the precursors. The data are consistent with the possibility that stable association with La specifically precludes access of the #6 and #10 position to the m2G modification machinery (see text). The blackened areas on the pre-tRNA backbone depict possible contact points for RNase P as previously reported (68). The model was modified from Krupp (68) and reproduced with permission.
We have superimposed these data onto a model of pre-tRNA structure. For this, we added non-base paired 5′ leader and 3′ trailer regions onto a preexisting model for tRNASer (Fig. 3) (68). The following observations suggest that it is not inappropriate to propose that pre-tRNAs that resemble the overall structure depicted in the Figure 3 model are bound by La. A La-pre-tRNA is known to be a substrate for RNase P, the latter of which recognizes folded pre-tRNA (5,33,63,68,74,139-141,144). La has been proposed to stabilize the correctly folded form of a pre-tRNA (144). While the sequences of the 5′ leaders and 3′ trailers are not strictly conserved among eukaryotic pre-tRNAs, their potential ability to base pair most often involves the terminal uridylates in naked RNA (73), which would otherwise be bound by La in vivo. Finally, eukaryotic tRNA genes are usually flanked by A+T-rich DNA and extensions tend not to interfere with formation of tRNA structure [(2,36), R.M., unpublished observation]. When considering this model, recall that while the specificity elements recognized by La are localized at the 5′ and 3′ ends of a pre-tRNA, these are thought to include the entire leader and trailer regions (Fig. 1).
For both pre-tRNAe Met and pre-tRNATyr, the m2G modifications (depicted in oval on right side of Fig. 3) appeared only after removal of the La binding sites, the 5′ and 3′ extensions, from the pre-tRNAs. Because these modifications were found on the intron-containing pre-tRNATyr species, they almost certainly occurred in the nucleus (94). One interpretation is that La sterically hinders access to these modification sites, preventing their addition until after the leader and trailer, and La, are removed. This would not be too unlike what has been observed for La and RNase P (33). This is consistent with the model depicted in Figure 3, which suggests that the m2G modification enzyme must access the confined inner aspect of the tRNA structure, which may be hindered by La.
While the stable pre-tRNAe Met was found to contain m2 2G26 as well as two dihydrouridines in the D loop, these modifications were not acquired by pre-tRNATyr until after end removal, although they did occur on the intron-containing species (50,94). These findings are not necessarily contradictory if we consider the differences in the two pre-tRNA substrates in the context of their relationship to La. As recently described, pre-tRNAe Met contains a CACAA motif, near its 3′ terminus, which is closely related to the sequence that constitutes a high-affinity binding site that was identified by RNA SELEX experiments (4,64). Because this CACAA sequence includes the pre-tRNAe Met 3′ processing site (CA↓CAA), association with La may preclude efficient processing and contribute to the remarkable stability of this precursor (50,64). Thus, we speculate that the m2 2G26 and dihydrouridines found on pre-tRNAe Met but not pre-tRNATyr may be somewhat aberrant due in part to the longer half-life of the pre-tRNAe Met. The presence of m2 2G26 on the pre-tRNAe Met would further suggest that the stable precursor La-RNP circulates through the nucleus because the m2 2G enzyme appears to reside at the nuclear periphery (107,108). Yet, neither of the two m2G modifications on mature tRNAe Met was detected on the stable precursor (50).
In summary, 12 modifications were found on the stable pre-tRNAe Met while three additional modifications were found on the mature tRNAe Met: m2G6, m2G1O, and i6A37 (50). In the time course study by Nashimoto and DeRobertis, the i6A37 modification did not appear to be impeded by the 5′ leader or 3′ trailer because the end-processed, intron-containing species also remained unmodified, while the mature tRNATyr species contained i6A37 (94). Consistent with this, pre-tRNAe Met does not contain an intron nor the i6A37 modification that is found on the mature species (50). These observations suggest that failure to acquire the i6A37 modification is probably not due to association with La, and may require another type of downstream signal or compartmentalization.
Although other interpretations exist, the data are consistent with the possibility that stable association with La specifically precludes access of the pre-tRNA G6 and G1O modification sites to the m2G methyltransferase (Fig. 3). Modification studies that employ transcripts corresponding to unprocessed species versus end processed may shed light on the possible influences of La on pre-tRNA modifications (45).
La CHAPERONES OTHERWISE VULNERABLE TRANSCRIPTS
It is becoming clear that La can stabilize certain imperfect pre-tRNAs that might not otherwise endure the maturation process. This is suggested by La’s effects on a mutant pre-tRNASer CGA that carries a destabilizing mismatch in the anticodon stem (144), a pre-tRNAi Met that is hypomethylated at A58 (6,20), and a pre-tRNASer UGAM species with suboptimal base pairing between the anticodon and intron (57). Thus, it seems that La stabilizes Pol III transcripts predominantly at the precursor stage, supporting them through a critical maturation process, while they might otherwise be vulnerable to degradation by exonucleases or other degradative processes (6,20,57,97,144). The absence of La has also been associated with decreased steady-state levels of several other Pol III transcripts, namely tRNASer CGA, NME1 RNA, and RPR1 RNA (the latter are components of RNases P and MRP, respectively) when combined with mutants in the nuclear protein, Gcdl0p-Gcdl4p, the 1-methyl-adenosine (m1A) methyltransferase (7,20). The lack of m1A methyltransferase activity in a gcd10Δ strain can be overcome by increasing expression of pre-tRNAi Met. This suggests that the tRNAi Met m1A58 modification is most important for correct precursor processing because the defective unmethylated but otherwise matured tRNAi Met can function at the ribosome (6,20). Because La can rescue certain gcd10-gcd14 mutants but not a gcd10Δ null strain, it may function by supporting increased levels of pre-tRNAi Met (6,20). It is not yet clear for the class of gcd10-gcd14 mutants that La can rescue, whether La protects pre-tRNAi Met and this allows methylation by a low activity (i.e., mutant) methylase, or if La promotes maturation of the unmethylated precursor, because neither the level of methylase activity nor the level of the tRNAi Met m1A58 modification in the mutant strains has been reported. In any case, it does seem that La protects against degradation (6,20).
Cumulative results indicate that the conserved function of La is to protect transcripts during the time between synthesis and incorporation into more stable RNPs, in some cases facilitating their assembly into mature RNPs and in other cases supporting them during modification; both processes would be expected to increase the stability of the RNA. While the period of transcript vulnerability would appear to begin upon termination of synthesis, when this period ends probably varies for different transcripts. La’s participation in the maturation pathways of the U RNAs occurs early in their lifetimes (69,142). While processing by RNase P of a La-associated pre-tRNA is thought to be among the earliest RNA cleavage events in the tRNA maturation pathway (57,141,144), pre-tRNA m1A58 methylation (and other modifications) appears to occur even earlier (50,94) (see Fig. 3). With regard to (iv) from the previous section, data that indicate La also affects pre-tRNA 3′ processing provide further evidence that La chaperones its pre-tRNA ligands extensively, through many parts of their maturation pathways (33,57,144) (Fig. 4).
Figure 4.
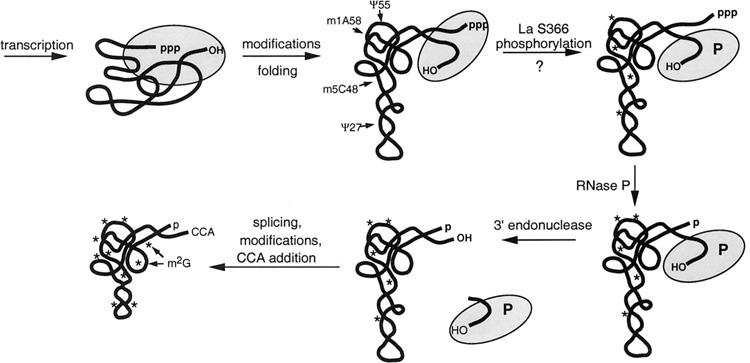
Cartoon view of the maturation pathway of a pre-tRNA. Asterisks represent modifications, some of which occur before end processing and some of which occur later (see text). La is represented by the yellow oval. Although it is unclear whether phosphorylation of serine 366 of human La occurs while associated with RNA or whether, in some cases, it may be phosphorylated prior to RNA binding, it is shown here. In any case, the data indicate that only the phosphorylated form of human La can support efficient cleavage by RNase P (see text). Although casein kinase 2 (CK2) is thought to be the kinase that phosphorylates serine 366 (31), the arrow indicates uncertainty as to which kinase mediates this activity.
Because modifications are thought to contribute to the stability of higher order structure, it would be consistent that these occur early in the lifetime of a pre-tRNA. This suggests that the ability of La to support tRNA modifications may explain, at least in part, La’s ability to facilitate pre-tRNA folding (144) and is consistent with La’s ability to cooperate with tRNA modifying enzymes.
Nascent Pol III Transcripts Are Synthesized in the Nucleoplasm
Newly made Pol III transcripts have been visualized directly and calculated to reside at ∼2000 discernible sites (distinct from pol II transcription loci) containing ∼5 nascent Pol III transcripts per site in HeLa nucleoplasm (102). Considering the quantity of newly synthesized Pol III transcripts in nuclei, there is a very large discrepancy between this number, 104 (102), and the much larger number of La molecules present in HeLa cells, 2 × 107. One consideration that could reconcile some of the discrepancy is that La may remain associated with a pool of transcripts, such that the number of RNAs associated with La is greater than the number of instantaneously labeled transcripts. It was noted that newly synthesized Pol III transcripts appeared unable to move away from their transcription sites, suggesting that a soluble factor that may normally mediate transcript clearance and/or transport was lost from the permeabilized cells (102). Thus, it is plausible that in living cells La would be involved in the clearance of nascent transcripts from their sites of synthesis and that La remains associated with a large reservoir of premature transcripts that would result from multiple rounds of synthesis per gene. This is consistent with the estimate that approximately 1–2 × 105 molecules of pre-5S RRNA are associated with La in HeLa cells (representing 1–2% of the amount of mature 5S rRNA) (106,123), transcribed from far fewer 5S rRNA genes, the latter estimated to be 2 × 103 per human haploid genome (120). Considering the possibility that an equally large number of precursor tRNAs and other premature transcripts remain bound to La, this would still lead to the conservative estimate that only about one tenth of La molecules would be associated with nascent RNA. Although this estimate is consistent with that of Habets et al., who noted that a substantial fraction of nuclear La may not be associated with Pol III transcripts (48), it is possible that the remaining La is either associated with other RNAs or free.
A point that has not been considered with regard to this issue is that approximately 15% of La in HeLa nuclear extract is unphosphorylated, a form that exhibits RNA binding characteristics subtly distinguishable from the phosphoprotein (33,57). Therefore, determination of the molar amounts of RNA associated with the phosphorylated and unphosphorylated forms of La is an issue that remains to be clarified.
Intracellular Trafficking of La
Although the majority of La is nuclear, a significant amount also resides in the cytoplasm and nucleolus as revealed by immunostaining (29,30,42,52). Most nuclear La appears to be freely soluble and readily distributes between cytoplasmic and nuclear fractions upon cellular disruption and biochemical preparation (48,52,105). Although mammalian La has been shown to reside in part in the nucleolus (22,30,42,52), identifying the determinants responsible for this localization has confirmed and extended our studies of the biology of La (manuscript in preparation). Species-specific differences in intracellular trafficking and the ability of La to serve as a nuclear retention factor for its RNA ligands have been reviewed recently (83).
Accumulating evidence indicates that the nuclear cytoplasmic partitioning of La can be disturbed under certain conditions, suggesting that subcellular localization may be used to regulate La’s activities. Infection with poliovirus and other viruses leads to an increase in the cytoplasmic staining of La [(9,92), also see (18)]. Poliovirus protease 3C cleaves La to leave a 358 amino acid (aa) protein (full length human La is 408 aa) that lacks its C-terminal nuclear localization signal (NLS), and this is associated with decrease of La in the nucleus and increase in the cytoplasm (118). However, because the amount of cleaved La appeared disproportionately lower than the amount of cytoplasmic accumulation (110), the data suggest that protease 3C cleavage alone may not be sufficient to explain the redistribution of La to the cytoplasm. This is consistent with the finding that poliovirus affects nuclear cytoplasmic redistribution in a global manner (134). Induced apoptosis also leads to cleavage of La, at or near residue 374, and is associated with cytoplasmic accumulation (8,110). In this case again, the amount of cytoplasmic accumulation is disproportionately higher than the amount of La that is cleaved, suggesting that the relationship between proteolytic cleavage and cytoplasmic accumulation is complex. Pruijn and colleagues showed that proteolysis of La, although limited to a fraction of molecules, is associated with dephosphorylation of phosphoserine 366 early in apoptosis, although phosphorylation alone does not appear to affect the nuclear cytoplasmic distribution of La (18,110).
An isoform of La mRNA, distinct from the form that encodes nuclear La, has been identified in lymphocytes (23,128). Some isoforms are tissue specific and when expressed in recombinant form give rise to proteins that distribute differentially to the nucleus or cytoplasm (43,44,54). Intriguingly, these La mRNA isoforms may be differentially expressed during inflammation, apoptosis, or certain viral infections (23).
A Fraction of Nascent Pol III Transcripts Localize to the Nucleolus
Several Pol III transcripts, including pre-tRNAs (12,46,66), U6 snRNA (71), and the RNA components of RNases P and MRP as well as SRP RNA and 5S rRNA have been observed in the nucleolus (12,59–62). Specific proteins known to be intimately associated with most of these RNAs have also been localized in the nucleolus (62,93,101). SRP RNA, RNase P RNA, and U6 snRNA pass transiently through the nucleolus (59,61,71). At least for one of these transcripts, U6, it is almost certain that some of its 2′-O-methylation and pseudouridylation modifications occur in the nucleolus [(37,71,130), reviewed in (83)].
Because components of RNase P, the endonuclease that matures the 5′ ends of pre-tRNAs, as well as pre-tRNAs themselves, have been visualized in the nucleolus (12,59,62), it has been proposed that 5′-pre-tRNA processing activity actually occurs there (67,76,141). Although most of the RNase P RNA and intron-containing pre-tRNAs detectable in yeast appeared nucleolar (12), other studies reveal yeast pre-tRNAs in the nucleoplasm as well as the nucleolus (46,66,112). In mammalian cells, a substantial amount of RNase P RNA is found in the nucleoplasm (59,72,88). Because transient nucleolar localization is common to several Pol III transcripts, including RNase P RNA, and associated proteins, pre-tRNAs and RNase P components may each be required to pass through this compartment without necessarily interacting there. In archaea, homologs of snoRNAs are associated with Archaeal fibrillarin and some of these exhibit complementarity to the regions of tRNAs known to be 2′-O-methylated, suggesting that eukaryotic pre-tRNAs may be similarly modified in the nucleolus (95). Some evidence suggests that other tRNA modifications may occur in the nucleolus (126). Indeed, our preliminary results in fission yeast, using a La-dependent opal suppressor tRNA system, suggest that passage through the nucleolus has no effect on efficient 5′ processing by RNase P (manuscript in preparation). In summary, while it is possible that RNAse P functions in pre-tRNA processing in the nucleolus, the evidence for this is circumstantial and alternative explanations may account for the localization of pre-tRNAs and RNAs P components there.
What Is La Doing in the Nucleolus?
As alluded to above, earlier reports that La resides in part in the nucleolus have been confirmed by multiple approaches including direct visualization of La-GFP fusion proteins and colocalization with known nucleolar proteins (manuscript in preparation). Because pol III transcripts pass through the nucleolus during what appears to be an early phase of their life cycle, it might seem that La would be involved in their nucleolar transit perhaps to chaperone and/or protect them in this compartment. However as recently reasoned (83), this need not be so for several of the RNAs (5S, MRP, RNase P, SRP) as other nucleolar-localizing proteins have been identified that interact with these RNAs and may be better candidates for their nucleolar localization. However, for other Pol III nascent transcripts (e.g., pre-U6 RNA and pre-tRNA) there are no known proteins other than La that associates with them that are also found in the nucleolus.
Indeed, as reviewed recently, there is indirect evidence suggesting that La associates with pre-U6 snRNA in the nucleolus (83). The small nucleolar (sno)RNAs and other factors that direct 2′-O-methylation and pseudouridylation of U6 snRNA almost certainly do so in the nucleolus (37,130). Precursors to tRNAs and U6 snRNA that are associated with La contain methylated bases, pseudouridine, dihidrouridine, and some 2′-O-methylated ribose. For the subset of U6 snRNA precursors found associated with La, fingerprint analysis suggested the presence of several residues modified in U6 (105), which recent data strongly imply are modified in the nucleolus (37,130). Specifically, several 2′-O-methylated and pseudouridylated U6 nucleotides that were recently shown to be directed by specific snoRNAs (37,130) appear to be present in La-associated pre-U6 RNA. This was evidenced by co-migration of the corresponding oligonucleotides isolated from La-associated pre-U6 snRNA with the known modified oligonucleotides isolated from mature U6 snRNA (although some of the La-associated oligonucleotides did appear to be undermodified, providing additional evidence of their precursor status) (105). U6 snRNA has recently been shown to transiently pass through the nucleolus (71). The cumulative data provide compelling yet indirect evidence that La travels with pre-U6 snRNA to the nucleolus.
As noted earlier, yeast La also associates with a U3 snoRNA intermediate and it is possible that La hands off this transcript to the next proteins in the U3 RNP assembly pathway in the nucleolus (69). Because biochemical evidence suggests that a fraction of La may be associated with a subset of small ribosomal subunits (100), we should also consider the possibility that La may join those subunits in the nucleolus. A fraction of La was observed in the nucleoli during the G1-early S phase of synchronized cells (29). It is noteworthy in this regard that Cfi1/Net1 sequesters Cdcl4 in the nucleolus during this time, contributing to the fidelity of the timing of mitotic exit (133). Although cumulative findings suggest several roles for La in the nucleolus, more definitive data are required before the function of nucleolar La is understood.
CONCLUSIONS AND PERSPECTIVES
La is a ubiquitous nuclear phosphoprotein that is found associated with the large variety of RNA ligands that end with 3′ terminal uridylates, the latter of which comprises the principal binding site for the conserved NTD of La proteins. The most clear and ubiquitous function of La is that it facilitates the expression of Pol III transcripts by stabilizing the nascent precursors that would otherwise be vulnerable to 3′ exonucleases and/or other degradative enzymes. Accumulating evidence suggests that La maintains close contact with its transcripts during several individual processing and/or modification events that involve transient interactions of the La-associated RNA with other factors. This proposal appears most reasonable for pre-tRNA modifying enzymes and RNase P. The yeast tRNA modifying enzyme, m1A58 methyltransferase, which acts early in tRNA maturation pathways, has been shown to cooperate with La in the expression of tRNAi Met, and La also appears to cooperate with RNAse P. La has also been described as a chaperone that may help with RNA folding; because some modifications appear to limit the conformational flexibility of tRNA (i.e., stabilize certain isoforms over others), it is possible that La may affect tRNA folding in part by its ability to cooperate with tRNA modifying enzymes and support pre-tRNAs during modification. It is also becoming clear that La can serve as a nuclear retention factor for its RNA ligands.
Structure–function and molecular modeling analyses of a growing number of La protein sequences have strengthened the idea that a tandem pair of RRMs comprise the conserved NTD of La and work together to bind RNA. While the NTD recognizes the UUU-OH 3′ terminal motifs of nascent Pol III transcripts, the CTD recognizes the 5′-ppp end region that results from transcription initiation. The 5′-ppp end recognition activity of human La can be modulated by phosphorylation of S366. Although it remains unknown whether the yeast La phosphoproteins also exhibit 5′-ppp RNA recognition, the signal transduction function of La would appear to be conserved because human La is faithfully phosphorylated in fission yeast and this modification is transduced in the yeast to control tRNA processing and maturation (57). The challenge for the future is clear: to understand how the bipartite mode of nascent RNA binding, intracellular trafficking, and the signal transduction functions of La are integrated with the pathways of expression of the variety of small nuclear and nucleolar RNA precursors with which it associates. An additional challenge is to understand how the signal transduction through La may be integrated with other aspects of intermediary metabolism and cellular proliferation.
ACKNOWLEDGMENTS
The authors thank Ram Reddy for providing the opportunity to prepare this review, D. Kenan and J. Keene for a foundation of RRM-1 structure analysis, A. Hinnebusch for discussion, and B. Peculis for comments on the manuscript. R.I. is a NICHD Fellow.
REFERENCES
- 1. Adam S. A.; Nakagawa T.; Swanson M. S.; Woodruff T. K.; Dreyfuss G. mRNA polyadenylate-binding protein: Gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol. Cell. Biol. 6:2932–2943; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adeniyi-Jones S.; Romeo P. H.; Zasloff M. Generation of long read-through transcripts in vivo and in vitro by deletion of 3′ termination and processing sequences in the human tRNAi met gene. Nucleic Acids Res. 12:1101–1115; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aigner S.; Lingner J.; Goodrich K. J.; Grosshans C. A.; Shevchenko A.; Mann M.; Cech T. R. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 19:6230–6239; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ali N.; Pruijn G. J.; Kenan D. J.; Keene J. D.; Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site (IRES)-mediated translation. J. Biol. Chem. 275:27531–27540; 2000. [DOI] [PubMed] [Google Scholar]
- 5. Altman S.; Kirsebom L. Ribonuclease P. In: Gesteland R. F.; Chech T. R.; Atkins J. F., eds. The RNA world (2nd ed.). Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1999:351–380. [Google Scholar]
- 6. Anderson J.; Phan L.; Cuesta R.; Carlson B. A.; Pak M.; Asano K.; Bjork G. R.; Tamame M.; Hin-nebusch A. G. The essential Gcdl0p-Gcdl4p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 12:3650–3662; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson J.; Phan L.; Hinnebusch A. G. The Gcdl0p/Gcdl4p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA 97:5173–5178; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ayukawa K.; Taniguchi S.; Masumoto J.; Hashimoto S.; Sarvotham H.; Hara A.; Aoyama T.; Sagara J. La autoantigen is cleaved in the COOH terminus and loses the nuclear localization signal during apoptosis. J. Biol. Chem. 275:34465–34470; 2000. [DOI] [PubMed] [Google Scholar]
- 9. Bachmann M.; Falke D.; Schroder H.-C.; Muller W. E. G. Intracellular distribution of the La antigen in CV-1 cells after herpes simplex virus type I infection compared with the localization of U small nuclear ribonucleoprotein particles. J. Gen. Virol. 70:881–891; 1989. [DOI] [PubMed] [Google Scholar]
- 10. Bai C.; Li Z.; Tolias P. P. Developmental characterization of a Drosophila RNA-binding protein homologous to the human systemic lupus erythematosus-associated La/SS-B autoantigen. Mol. Cell. Biol. 14:5123–5129; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beisham G. J.; Sonenberg N.; Svitkin Y. V. The role of the La autoantigen in internal initiation. Curr. Top. Microbiol. Immunol. 203:85–98; 1995. [DOI] [PubMed] [Google Scholar]
- 12. Bertrand E.; Houser-Scott F.; Kendall A.; Singer R. H.; Engelke D. R. Nucleolar localization of early tRNA processing. Genes Dev. 12:2463–2468; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Birney E.; Kumar S.; Krainer A. R. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21:5803–5816; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bobkova E. V.; Habib N.; Alexander G.; Hall B. D. Mutational analysis of the hydrolytic activity of yeast RNA polymerase Ill. J. Biol. Chem. 274:21342–21348; 1999. [DOI] [PubMed] [Google Scholar]
- 15. Bobkova E. V.; Hall B. D. Substrate specificity of the RNase activity of yeast RNA polymerase III. J. Biol. Chem. 272:22832–22839; 1997. [DOI] [PubMed] [Google Scholar]
- 16. Bogenhagen D. F.; Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell 24:261–270; 1981. [DOI] [PubMed] [Google Scholar]
- 17. Borukhov S.; Polyakov A.; Nikiforov V.; Goldfarb A. GreA protein: A transcription elongation factor from Escherichia coli . Proc. Natl. Acad. Sci. USA 89:8899–8902; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Broekhuis C. H.; Neubauer G.; van der Heijden A.; Mann M.; Proud C. G.; van Venrooij W. J.; Pruijn G. J. Detailed analysis of the phosphorylation of the human La (SS-B) autoantigen. (De)phosphorylation does not affect its subcellular distribution. Biochemistry 39:3023–3033; 2000. [DOI] [PubMed] [Google Scholar]
- 19. Burd C. G.; Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615–621; 1994. [DOI] [PubMed] [Google Scholar]
- 20. Calvo O.; Cuesta R.; Anderson J.; Gutierrez N.; Garcia-Barrio M. T.; Hinnebusch A. G.; Tamame M. GCDl4p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhplp in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae . Mol. Cell. Biol. 19:4167–4181; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campbell F. E.; Setzer D. R. Transcription termination by RNA polymerase III: Uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 12:2260–2272; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carmo-Fonseca M.; Pfeifer K.; Schroder H. C.; Vaz M. F.; Fonseca J. E.; Muller W. E.; Bachmann M. Identification of La ribonucleoproteins as a component of interchromatin granules. Exp. Cell Res. 185:73–85; 1989. [DOI] [PubMed] [Google Scholar]
- 23. Carter M. S.; Sarnow P. Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J. Biol. Chem. 275:28301–28307; 2000. [DOI] [PubMed] [Google Scholar]
- 24. Chambers J. C.; Keene J. D. Isolation and analysis of cDNA clones expressing human lupus La antigen. Proc. Natl. Acad. Sci. USA 82:2115–2119; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chambers J. C.; Kurilia M. G.; Keene J. D. Association between the 7 S RNA and the lupus La protein varies among cell types. J. Biol. Chem. 258:11438–11441; 1983. [PubMed] [Google Scholar]
- 26. Chedin S.; Riva M.; Schultz P.; Sentenae A.; Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like sub-unit and is important for transcription termination. Genes Dev. 12:3857–3871; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chu W. M.; Ballard R. E.; Schmid C. W. Palindromic sequences preceding the terminator increase polymerase III template activity. Nucleic Acids Res. 25:2077–2082; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Conte M. R.; Grune T.; Ghuman J.; Kelly G.; La-das A.; Matthews S.; Curry S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 19:3132–3141; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng J. S.; Takasaki Y.; Tan E. M. Nonhistone nuclear antigens reactive with autoantibodies. Immuno-fluorescence studies on distribution in synchronized cells. J. Cell Biol. 91:654–660; 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Deng J. S.; Tan E. M. Effect of actinomycin D on the expression of nuclear antigen SS-B/La. J. Invest. Dermatol. 84:225–228; 1985. [DOI] [PubMed] [Google Scholar]
- 31. Deo R. C.; Bonanno J. B.; Sonenberg N.; Burley S. K. Recognition of polyadenylate RNA by the poly (A)-binding protein. Cell 98:835–845; 1999. [DOI] [PubMed] [Google Scholar]
- 32. Di Francesco V.; Garnier J.; Munson P. J. Improving protein secondary structure prediction with aligned homologous sequences. Protein Sci. 5:106–113; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan H.; Goodier J. L.; Chamberlain J.; Engelke D. R.; Maraia R. J. 5′ processing of tRNA precursors can be modulated by the human La antigen phospho-protein. Mol. Cell. Biol. 18:3201–3211; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fan H.; Sakulich A. L.; Goodier J. L.; Zhang X.; Qin J.; Maraia R. J. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell 88:707–715; 1997. [DOI] [PubMed] [Google Scholar]
- 35. Francoeur A. M.; Mathews M. B. Interaction between VA RNA and the lupus antigen La: Formation of a ribonucleoprotein particle in vitro . Proc. Natl. Acad. Sci. USA 79:6772–6776; 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Furter R.; Snaith M.; Gillespie D. E.; Hall B. D. Endonucleolytic cleavage of a long 3′-trailer sequence in a nuclear yeast suppressor tRNA. Biochemistry 31:10817–10824; 1992. [DOI] [PubMed] [Google Scholar]
- 37. Ganot P.; Jady B. E.; Bortolin M. L.; Darzacq X.; Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 19:6906-6917; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goodier J. L.; Fan H.; Maraia R. J. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol. Cell. Biol. 17:5823–5832; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goodier J. L.; Maraia R. J. Terminator-specific recycling of a B1-Alu transcription complex by RNA polymerase III is mediated by the RNA terminus-binding protein La. J. Biol. Chem. 273:26110–26116; 1998. [DOI] [PubMed] [Google Scholar]
- 40. Gottlieb E.; Steitz J. A. Function of the mammalian La protein: Evidence for its action in transcription termination by RNA polymerase III. EMBO J. 8:851–861; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gottlieb E.; Steitz J. A. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 8:841–850; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Graus F.; Cordon-Cardo C.; Bonfa E.; Elkon K. B. Immunohistochemical localization of La nuclear antigen in brain. Selective concentration of the La protein in neuronal nucleoli. J. Neuroimmunol. 9:307–319; 1985. [DOI] [PubMed] [Google Scholar]
- 43. Grolz D.; Bachmann M. An altered intracellular distribution of the autoantigen La/SS-B when translated from a La mRNA isoform. Exp. Cell Res. 234:329–335; 1997. [DOI] [PubMed] [Google Scholar]
- 44. Grolz D.; Laubinger J.; Wilmer F.; Troster H.; Bachmann M. Transfection analysis of expression of mRNA isoforms encoding the nuclear autoantigen La/ SS-B. J. Biol. Chem. 272:12076–12082; 1997. [DOI] [PubMed] [Google Scholar]
- 45. Grosjean H.; Edqvist J.; Straby K. B.; Giege R. Enzymatic formation of modified nucleosides in tRNA: Dependence on tRNA architecture. J. Mol. Biol. 255:67–85; 1996. [DOI] [PubMed] [Google Scholar]
- 46. Grosshans H.; Hurt E.; Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 14:830–840; 2000. [PMC free article] [PubMed] [Google Scholar]
- 47. Guddat U.; Bakken A. H.; Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell 60:619–628; 1990. [DOI] [PubMed] [Google Scholar]
- 48. Habets W. J.; denBrok J. H.; Boerbooms A. M.; van de Putte L. B.; van Venrooij W. J. Characterization of the SS-B (La) antigen in adenovirus-infected and uninfected HeLa cells. EMBO J. 2:1625–1631; 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Handa N.; Nureki O.; Kurimoto K.; Kim I.; Sakamoto H.; Shimura Y.; Muto Y.; Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398:579–585; 1999. [DOI] [PubMed] [Google Scholar]
- 50. Harada F.; Matsubara M.; Kato N. Stable tRNA precursors in HeLa cells. Nucleic Acids Res. 12:9263–9269; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hashimoto C.; Steitz J. A. Sequential association of nucleolar 7-2 RNA with two different autoantigens. J. Biol. Chem. 258:1379–1382; 1983. [PubMed] [Google Scholar]
- 52. Hendrick J. P.; Wolin S. L.; Rinke J.; Lerner M. R.; Steitz J. A. Ro small cytoplasmic ribonucleo-proteins are a subclass of La ribonucleoproteins: Further characterization of the Ro and La small ribo nucleoproteins from uninfected mammalian cells. Mol. Cell. Biol. 1:1138–1149; 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Henikoff S.; Henikoff J. G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 19:6565–6572; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hilker M.; Troster H.; Grolz D.; Hake U.; Bachmann M. The autoantigen La/SS-B: Analysis of the expression of alternatively spliced La mRNA isoforms. Cell. Tissue Res. 284:383–389; 1996. [DOI] [PubMed] [Google Scholar]
- 55. Hoffman D. W.; Query C. C.; Golden B. L.; White S. W.; Keene J. D. RNA-binding domain of the A protein component of the U1 small nuclear ribo-nucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc. Natl. Acad. Sci. USA 88:2495–2499; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Holcik M.; Komeluk R. G. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: Role of La autoantigen in XIAP translation. Mol. Cell. Biol. 20:4648–4657; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Intine R. V. A.; Sakulich A. L.; Koduru S. B.; Huang Y.; Pierstorrf E.; Goodier J. L.; Phan L.; Maraia R. J. Transfer RNA maturation is controlled by phosphorylation of the human La antigen on serine 366. Mol. Cell 6:339–348; 2000. [DOI] [PubMed] [Google Scholar]
- 58. Izban M. G.; Luse D. S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′ → 5′ direction in the presence of elongation factor SII. Genes Dev. 6:1342–1356; 1992. [DOI] [PubMed] [Google Scholar]
- 59. Jacobson M. R.; Cao L. G.; Taneja K.; Singer R. H.; Wang Y. L.; Pederson T. Nuclear domains of the RNA subunit of Rnase P. J. Cell Sci. 110:829–837; 1997. [DOI] [PubMed] [Google Scholar]
- 60. Jacobson M. R.; Cao L. G.; Wang Y. L.; Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J. Cell Biol. 131:1649–1658; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jacobson M. R.; Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc. Natl. Acad. Sci. USA 95:7981–7986; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jarrous N.; Wolenski J. S.; Wesolowski D.; Lee C.; Altman S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J. Cell Biol. 146:559–572; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kahle D.; Wehmeyer U.; Krupp G. Substrate recognition by RNase P and by the catalytic MI RNA: Identification of possible contact points in pre-tRNAs. ENMO J. 9:1929–1937; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kenan D. J. RNA recognition by the human La protein and its relevance to transcription, translation and viral infectivity. Ph.D. thesis, Duke University, l995. [Google Scholar]
- 65. Kenan D. J.; Query C. C.; Keene J. D. RNA recognition: Towards identifying determinants of specificity. Trends Biochem. Sci. 16:214–220; 1991. [DOI] [PubMed] [Google Scholar]
- 66. Kendall A.; Hull M. W.; Bertrand E.; Good P. D.; Singer R. H.; Engelke D. R. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc. Natl. Acad. Sci. USA 97:13108–13113; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kressler D.; Linder P.; de La Cruz J. Protein transacting factors involved in ribosome biogenesis in Sac-charomyces cerevisiae . Mol. Cell. Biol. 19:7897–7912; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Krupp G. Enzymatic RNA synthesis and RNase P. Evolutionary aspects. Mol. Biol. Rep. 22:177–180; 1995. [DOI] [PubMed] [Google Scholar]
- 69. Kufel J.; Allmang C.; Chanfreau G.; Petfalski E.; Lafontaine D. L.; Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20:5415–5424; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kurilia M. G.; Keene J. D. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell 34:837–845; 1983. [DOI] [PubMed] [Google Scholar]
- 71. Lange T. S.; Gerbi S. A. Transient nucleolar localization of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell 11:2419–2428; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lee B.; Matera A. G.; Ward D. C.; Craft J. Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: A possible coordinate role in ribosome biogenesis. Proc. Natl. Acad. Sci. USA 93:11471–11476; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lee Y.; Kindelberger D. W.; Lee J. Y.; McCiennen S.; Chamberlain J.; Engelke D. R. Nuclear pre-tRNA terminal structure and RNase P recognition. RNA 3:175–185; 1997. [PMC free article] [PubMed] [Google Scholar]
- 74. Leontis N.; DaLio A.; Strobel M.; Engelke D. Effects of tRNA-intron structure on cleavage of precursor tRNAs by RNase P from Saccharomyces cerevisiae . Nucleic Acids Res. 16:2537–2552; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lerner M. R.; Steitz J. A. Snuips and seyrps. Cell 25:298–300; 1981. [DOI] [PubMed] [Google Scholar]
- 76. Lewis J. D.; Tollervey D. Like attracts like: Getting RNA processing together in the nucleus. Science 288:1385–1389; 2000. [DOI] [PubMed] [Google Scholar]
- 77. Li W. Y.; Reddy R.; Henning D.; Epstein P.; Busch H. Nucleotide sequence of 7 S RNA. Homology to Alu DNA and La 4.5 S RNA. J. Biol. Chem. 257:5136–5142; 1982. [PubMed] [Google Scholar]
- 78. Lin-Marq N.; Clarkson S. G. Efficient synthesis, termination and release of RNA polymerase III transcripts in Xenopus extracts depleted of La protein. EMBO J. 17:2033–2041; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin-Marq N.; Clarkson S. G. A yeast RNA binding protein that resembles the human autoantigen La. J. Mol. Biol. 245:81–85; 1995. [DOI] [PubMed] [Google Scholar]
- 80. Liu M. H.; Yuan Y.; Reddy R. Human RNase P RNA and nucleolar 7-2 RNA share conserved ‘To’ antigen-binding domains. Mol. Cell. Biochem. 130:75–82; 1994. [DOI] [PubMed] [Google Scholar]
- 81. Madore S. J.; Wieben E. D.; Pederson T. Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA-protein complexes. J. Biol. Chem. 259:1929–1933; 1984. [PubMed] [Google Scholar]
- 82. Maraia R.; Zasloff M.; Plotz P.; Adeniyi-Jones S. Pathway of B1-Alu expression in microinjected oocytes: Xenopus laevis proteins associated with nuclear precursor and processed cytoplasmic RNAs. Mol. Cell. Biol. 8:4433–4440; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Maraia R. J. La Protein and the trafficking of nascent RNA polymerase III transcripts. J. Cell. Biol. 153:F13–FI7; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Maraia R. J. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc. Natl. Acad. Sci. USA 93:3383–3387; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Maraia R. J.; Intine R. V. Recognition of nascent RNA by the human La antigen: Conserved and diverged features of structure and function. Mol. Cell. Biol. 21:367–379; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Maraia R. J.; Kenan D. J.; Keene J. D. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol. Cell. Biol. 14:2147–2158; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Marchetti M. A.; Tschudi C.; Kwon H.; Wolin S. L.; Ullu E. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J. Cell Sci. 113:899–906; 2000. [DOI] [PubMed] [Google Scholar]
- 88. Matera A. G.; Frey M. R.; Margelot K.; Wolin S. L. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the poly-pyrimidine tract-binding protein, hnRNP I. J. Cell Biol. 129:1181–1193; 1995. [Published erratum appears in J. Cell Biol. 130(2):497–500; 1995] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mathews M. B.; Francoeur A. M. La antigen recognizes and binds to the 3′-oligouridylate tail of a small RNA. Mol. Cell. Biol. 4:1134–1140; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mazabraud A.; Scherly D.; Muller F.; Rungger D.; Clarkson S. G. Structure and transcription termination of a lysine tRNA gene from Xenopus laevis . J. Mol. Biol. 195:835–845; 1987. [DOI] [PubMed] [Google Scholar]
- 91. McLaren R. S.; Caruccio N.; Ross J. Human La protein: A stabilizer of histone mRNA. Mol. Cell. Biol. 17:3028–3036; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Meerovitch K.; Svitkin Y. V.; Lee H. S.; Lejbkow-icz F.; Kenan D. J.; Chan E. K.; Agol V. I.; Keene J. D.; Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798–3807; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Michael W. M.; Dreyfuss G. Distinct domains in ribosomal protein L5 mediate 5 S RRNA binding and nucleolar localization. J. Biol. Chem. 271:11571–11574; 1996. [DOI] [PubMed] [Google Scholar]
- 94. Nishikura K.; De Robertis E. M. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J. Mol. Biol. 145:405–420; 1981. [DOI] [PubMed] [Google Scholar]
- 95. Omer A. D.; Lowe T. M.; Russell A. G.; Ebhardt H.; Eddy S. R.; Dennis P. P. Homologs of small nucleolar RNAs in Archaea. Science 288:517–522; 2000. [DOI] [PubMed] [Google Scholar]
- 96. Oubridge C.; Ito N.; Evans P. R.; Teo C. H.; Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the UIA spliceosomal protein completed with an RNA hairpin. Nature 372:432–438; 1994. [DOI] [PubMed] [Google Scholar]
- 97. Pannone B.; Xue D.; Wolin S. L. A role for the yeast La protein in U6 SNRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 17:7442–7453; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pannone B. K.; Kim S. D.; Noe D. A.; Wolin S. L. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA and the yeast La protein. Genetics (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pardigon N.; Strauss J. H. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J. Virol. 70:1173–1181; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Peek R.; Pruijn G. J.; Van Venrooij W. J. Interaction of the La (SS-B) autoantigen with small ribosomal subunits. Eur. J. Biochem. 236:649–655; 1996. [DOI] [PubMed] [Google Scholar]
- 101. Politz J. C.; Yarovoi S.; Kilroy S. M.; Gowda K.; Zwieb C.; Pederson T. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA 97:55–60; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Pombo A.; Jackson D. A.; Hollinshead M.; Wang Z.; Roeder R. G.; Cook P. R. Regional specialization in human nuclei: Visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 18:2241–2253; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Reddy R.; Henning D.; Tan E.; Busch H. Identification of a La protein binding site in a RNA polymerase III transcript (4.5 1 RNA). J. Biol. Chem. 258:8352–8356; 1983. [PubMed] [Google Scholar]
- 104. Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J. Biol. Chem. 267:3795–3800; 1992. [PMC free article] [PubMed] [Google Scholar]
- 105. Rinke J.; Steitz J. A. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 13:2617–2629; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rinke J.; Steitz J. A. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell 29:149–159; 1982. [DOI] [PubMed] [Google Scholar]
- 107. Rose A. M.; Belford H. G.; Shen W. C.; Greer C. L.; Hopper A. K.; Martin N. C. Location of N2,N2-dimethylguanosine-specific tRNA methyl-transferase. Biochimie 77:45–53; 1995. [DOI] [PubMed] [Google Scholar]
- 108. Rose A. M.; Joyce P. B.; Hopper A. K.; Martin N. C. Separate information required for nuclear and subnuclear localization: Additional complexity in localizing an enzyme shared by mitochondria and nuclei. Mol. Cell. Biol. 12:5652–5658; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Rosenblum J. S.; Pemberton L. F.; Bonifaci N.; Blobel G. Nuclear import and the evolution of a multifunctional RNA-binding protein. J. Cell. Biol. 143:887–899; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rutjes S. A.; Utz P. J.; der Heijden A.; Broekhuis C.; van Venrooij W. J.; Pruijn G. J. The La (SS-B) autoantigen, a key protein in RNA biogenesis, is dephosphorylated and cleaved early during apoptosis. Cell Death Differ. 6:976–986; 1999. [DOI] [PubMed] [Google Scholar]
- 111. Sachs A. B.; Bond M. W.; Komberg R. D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: Domain structure and expression. Cell 45:827–835; 1986. [DOI] [PubMed] [Google Scholar]
- 112. Sarkar S.; Hopper A. K. tRNA nuclear export in Saccharomyces cerevisiae: In situ hybridization analysis. Mol. Biol. Cell 9:3041–3055; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Scherly D.; Stutz F.; Lin-Marq N.; Clarkson S. G. La proteins from Xenopus laevis cDNA cloning and developmental expression. J. Mol. Biol. 231:196–204; 1993. [DOI] [PubMed] [Google Scholar]
- 114. Schneider T. D. Reading of DNA sequence logos: Prediction of major groove binding by information theory. Methods Enzymol. 274:445–455; 1996. [DOI] [PubMed] [Google Scholar]
- 115. Schneider T. D.; Stephens R. M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shamoo Y.; Abdul-Manan N.; Patten A. M.; Crawford J. K.; Pellegrini M. C.; Williams K. R. Both RNA-binding domains in heterogenous nuclear ribo-nucleoprotein A1 contribute toward single-stranded-RNA binding. Biochemistry 33:8272–8281; 1994. [DOI] [PubMed] [Google Scholar]
- 117. Shen C.-K. J.; Maniatis T. The organization, structure, and in vitro transcription of Alu family RNA polymerase III transcription units in the human α-like globin gene cluster: Precipitation of in vitro transcripts by lupus anti-La antibodies. J. Mol. Appl. Genet. 1:343–360; 1982. [PubMed] [Google Scholar]
- 118. Shiroki K.; Isoyama T.; Kuge S.; Ishii T.; Ohmi S.; Hata S.; Suzuki K.; Takasaki Y.; Nomoto A. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol. 73:2193–2200; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Simons F. H.; Broers F. J.; Van Venrooij W. J.; Pruijn G. J. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp. Cell Res. 224:224–236; 1996. [DOI] [PubMed] [Google Scholar]
- 120. Singer M.; Berg P. Genes and genomes. Mill Valley, CA: University Science Books; 1991. [Google Scholar]
- 121. Sobel S. G.; Wolin S. L. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol. Biol. Cell 10:3849–3862; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Stefano J. E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36:145–154; 1984. [DOI] [PubMed] [Google Scholar]
- 123. Steitz J. A.; Berg C.; Hendrick J. P.; La Branche-Chabot H.; Metspalu A.; Rinke J.; Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 106:545–556; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tan Q.; Li X.; Sadhale P. P.; Miyao T.; Woychik N. A. Multiple mechanisms of suppression circumvent transcription defects in an RNA polymerase mutant. Mol. Cell. Biol. 20:8124–8133; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tems M. P.; Lund E.; Dahlberg J. E. 3′-end-depen-dent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol. Cell. Biol. 12:3032–3040; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tolerico L. H.; Benko A. L.; Aris J. P.; Stanford D. R.; Martin N. C.; Hopper A. K. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics 151:57–75; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Topfer F.; Gordon T.; McCluskey J. Characterization of the mouse autoantigen La (SS-B). J. Immunol. 150:3091–3100; 1993. [PubMed] [Google Scholar]
- 128. Troster H.; Metzger T. E.; Semsei I.; Schwemmle M.; Winterpacht A.; Zabel B.; Bachmann M. One gene, two transcripts: Isolation of an alternative transcript encoding for the autoantigen La/SS-B from a cDNA library of a patient with primary Sjogrens’ syndrome. J. Exp. Med. 180:2059–2067; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tschochner H. A novel RNA polymerase independent RNase activity that shortens nascent transcripts from the 3′ end. Proc. Natl. Acad. Sci. USA 93:12914–12919; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tycowski K. T.; You Z. H.; Graham P. J.; Steitz J. A. Modification of U6 spliceosomal RNA is guided by other small RNAS. Mol. Cell 2:629–638; 1998. [DOI] [PubMed] [Google Scholar]
- 131. Van Horn D. J.; Yoo C. J.; Xue D.; Shi H.; Wolin S. L. The La protein in Schizosaccharomyces pombe: A conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA 3:1434–1443; 1997. [PMC free article] [PubMed] [Google Scholar]
- 132. Varani G.; Nagai K. RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct. 27:407–445; 1998. [DOI] [PubMed] [Google Scholar]
- 133. Visintin R.; Hwang E. S.; Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdcl4 phosphatase in the nucleolus. Nature 398:818-823; 1999. [DOI] [PubMed] [Google Scholar]
- 134. Waggoner S.; Sarnow P. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699–6709; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wang Z.; Luo T.; Roeder R. G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 11:2371–2382; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Weser S.; Bachmann M.; Seifart K. H.; Meißner W. Transcription efficiency of human polymerase III genes in vitro does not depend on the RNP-forming autoantigen La. Nucleic Acids Res. 28:3935–3942; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Westermann S.; Weber K. Cloning and recombinant expression of the La RNA-binding protein from Trypanosoma brucei . Biochim. Biophys. Acta 1492:483–487; 2000. [DOI] [PubMed] [Google Scholar]
- 138. Whitehall S. K.; Bardeleben C.; Kassavetis G. A. Hydrolytic cleavage of nascent RNA in RNA poly-merase III ternary transcription complexes. J. Biol. Chem. 269:2299–2306; 1994. [PubMed] [Google Scholar]
- 139. Willis I.; Frendewey D.; Nichols M.; Hottinger-Werien A.; Schaack J.; Soll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae . J. Biol. Chem. 261:5878–5885; 1986. [PubMed] [Google Scholar]
- 140. Willis I.; Nichols M.; Chisholm V.; Soll D.; Heyer W. D.; Szankasi P.; Amstutz H.; Munz P.; Kohli J. Functional complementation between mutations in a yeast suppressor tRNA gene reveals potential for evolution of tRNA sequences. Proc. Natl. Acad. Sci. USA 83:7860–7864; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wolin S. L.; Matera A. G. The trials and travels of tRNA. Genes Dev. 13:1–10; 1999. [DOI] [PubMed] [Google Scholar]
- 142. Xue D.; Rubinson D. A.; Pannone B. K.; Yoo C. J.; Wolin S. L. U snRNP assembly in yeast involves the La protein. EMBO J. 19:1650–1660; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Yoo C. J.; Wolin S. L. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: A yeast homolog of the La autoantigen is dispensable for growth. Mol. Cell. Biol. 14:5412–5424; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yoo C. J.; Wolin S. L. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89:393–402; 1997. [DOI] [PubMed] [Google Scholar]
- 145. Zasloff M.; Santos T.; Romeo P.; Rosenberg M. Transcription and precursor processing of normal and mutant human tRNAi met genes in a homologous cell-free system. J. Biol. Chem. 257:7857–7863; 1982. [PubMed] [Google Scholar]


