Abstract
Prostate cancer (PC) is the most common cancer and the third leading cause of cancer death in men worldwide. Despite its high incidence and mortality, the likelihood of a cure is low for late-stages of PC. There is an unmet need for more effective agents for treating PC. Here, we present a drug repositioning system, GenoPredict, for finding innovative drug candidates for treating PC. GenoPredict leverages upon a large amount of disease genomics data and a large-scale drug treatment knowledge base (TreatKB) that we recently constructed. We first constructed a genetic disease network (GDN) that comprised of 882 nodes and 200,758 edges and applied a network-based ranking algorithm to find diseases from GDN that are genetically related to PC. We developed a drug prioritization algorithm to reposition drugs from PC-related diseases to treat PC. When evaluated in a de-novo prediction setting using 27 FDA- approved PC drugs, GenoPredict found 25 of 27 FDA-approved PC drugs and ranked them highly (recall: 0.925, mean ranking: 27.3%, median ranking: 15.6%). When compared to PREDICT, a comprehensive drug repositioning system, in novel predictions, GenoPredict performed better than PREDICT across two evaluation datasets. GenoPredict achieved a mean average precision (MAP) of 0.447 when evaluated with 172 PC drugs extracted from 172,888 clinical trial reports, representing a 164.5% improvement as compared to a MAP of 0.169 for PREDICT. When evaluated with 72 PC drugs extracted from 43,811 ongoing clinical trial reports, GenoPredict achieved a MAP of 0.278, representing a 231.1% improvement as compared to a MAP of 0.084 for PREDICT.
The data is publicly available at: http://nlp.case.edu/public/data/PC_GenoPredict and http: //nlp.case.edu/public/data/treatKB.
Introduction
Prostate cancer (PC) is the most common cancer and the third leading cause of cancer death in men worldwide. In the United States alone, an estimated 238,590 men were diagnosed with PC in 2013 and 29,720 died from their disease [1]. Despite its high incidence and mortality, the likelihood of a cure is low for patients diagnosed with late-stage PC [2]. Docetaxel, the standard treatment for metastatic PC, provides a median overall survival improvement of roughly 3 months. While the PC market is expected to grow to $8.2 billion in 2019, there is an unmet need for more effective drug treatments for PC [2].
Computation-based drug repositioning approaches that assimilate vast amounts of genetic, genomic, chemical, and phenotypic data for thousands of drugs and diseases can greatly speed up the traditional drug discovery process [3]. Here we present a computational drug repositioning system, GenoPredict, that capitalizes on comprehensive disease genetic data generated by Genome-Wide Association Studies (GWAS) [4] and a unique large-scale drug treatment database that we recently constructed [5–7]. Our study is based on the premise that the genetic inter-releationships among diseases often reflect pathophysiological relevance. Though the majority of such shared pathophysiological features remain unknown, treatment insights from one disease may be used to inform our knowledge of others and potentiate their treatments. Existing approaches have directly inferred drug targets from disease genetics generated by GWAS studies [8–11]. Here, we present a complementary strategy to indirectly infer repositioned drug candidates from disease genomics data and demonstrated its utility in PC drug repositioning: diseases that share high genetics with PC were used as a starting point for both discovering repositioned drug candidates and gaining insights into the common mechanisms of action underlying identified candidates.
In order to systematically reposition drug treatments from one disease to another, it is critical to have a comprehensive drug treatment knowledge base. We have recently constructed a large-scale drug-disease treatment knowledge base (TreatKB) from multiple heterogeneous and complementary data resources [5–7]. We previously showed that TreatKB is critical for our computational drug repositioning tasks [12–13]. We evaluated GenoPredict in identifying and prioritizing FDA-approved PC drugs and compared GenoPredict to PREDICT, a comprehensive drug repositioning system [14], in novel predictions for PC. We demonstrated that GenoPredict is effective in prioritizing FDA-approved PC drugs and achieved better performance than PREDICT in novel predictions.
Materials and methods
Data
Drug-disease treatment relationship knowledge bases (TreatKBs) TreatKB contains 208,330 unique drug-disease treatment pairs, representing 2,484 drugs and 24,511 unique disease concepts [5–7]. TreatKB includes 9,216 drug- disease treatment pairs extracted from all FDA drug labels, 111,862 pairs extracted from the FDA Adverse Event Reporting System (FAERS), a database supporting the FDA’s post-marketing drug safety surveillance; 34,306 pairs extracted from 21 million published biomedical literature abstracts, and 69,724 pairs extracted from 172,888 clinical trials. The data is publicly available at http://nlp.case.edu/public/data/treatKB/.
Disease genetics data from the GWAS catalog Disease-gene association data from the Catalog of Published Genome- Wide Association Studies (the GWAS catalog) was used to construct disease networks and to identify PC-related diseases. The GWAS catalog is an exhaustive source containing the description of disease- and trait-associated single nucleotide polymorphisms (SNPs) from published GWAS data and currently contains 22,470 disease-gene pairs for 882 diseases and 8,689 genes [4]. Recent studies have shown that disease genomics data in the GWAS catalogy is a rich resource for drug discovery [8–11].
ICD10 for disease classification We identified diseases that are genetically related to PC (PC-related diseases) and analyzed PC-related diseases using the 10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD10) [15]. The 22 highest-level disease classes (e.g., “Neoplasms” and “Diseases of the nervous system”) were used in categorizing PC-related diseases.
The Anatomical Therapeutic Chemical (ATC) classification system for drug classificaiton The ATC system was used to classify and analyze repositioned drugs. The ATC classification system consists of 13 first-level codes, 94 second-level codes, 267 third-level codes, 882 fourth-level codes, and 4580 fifth-level codes, which are individual drugs [16].
Drug target genes and genetic pathways data Drugs’ gene targets were obtained from DrugBank [17], a comprehensive database containing information on drugs and drug targets. Gene-associated pathways were obtained from the Molecular Signatures Database (MSigDB), which contains 10,295 annotated genetic pathways [18].
Methods
The experiment framework of GenoPredict consists of the following steps: (1) we constructed a genetic disease network (GDN) and applied a network-based ranking algorithm to find diseases that are genetically related to PC; (2) we developed a drug prioritization algorithm to identify candidate drugs; GenoPredict was tested using 27 FDA-approved PC drugs and was compared to PREDICT in novel predictions; and (3) we performed drug classification and pathway enrichment analysis to analyze top ranked repositioning drug candidates.
Construct genetic disease network (GDN) and find PC-related diseases from GDN
Construct GDN We constructed GDN using disease-gene association data from the GWAS catalog. On GDN, two diseases were connected if their associated genes overlapped. The edge weights were determined by the cosine similarity coefficients of disease-associated genes [19]. GDN comprised of 882 disease nodes and 200,758 edges.
Apply network-based ranking algorithm to find diseases that share high genetic relevance with PC (PC-related diseases) We have recently applied a commonly used network-based ranking algorithm [20] for disease genetics discovery [21–23], human gut microbial metabolite prediction for diseases [24–25], and drug repositioning [12–13]. In this study, we directly used the network-based ranking algorithm to identify PC-related diseases from GDN. The iterative ranking algorithm is defined as: pt+1 = (1 — r)Mpt + rp0, wherein M is the column-normalized adjacency matrix of GDN, γ is a preset probability of restarting from the initial seed node (γ=0.1 in this study), and pt is a vector in which the ith element holds the normalized ranking score of disease i at tth iteration. The initial probability vector p0 contains normalized probability values for input. In our study, p0 contains PC, with a probability of 1.0. Diseases were ranked according to values in the steady-state probability vector, which was obtained by iterating the algorithm until the change between pt+1 and pt was less than 10-6.
We expect that top-ranked PC-related diseases should be enriched for cancers since cancers are known to share genetics with each other. To evaluate the network-based disease ranking approach and to systematically study PC-related diseases, we classified PC-related diseases based on the ICD10 classification scheme[15], and examined distributions of 22 disease classes at ten ranking cutoffs (top 10%, 20%,… 100%).
Drug prioritization algorithm
Drug repositioning algorithm We developed an approach to systematically reposition drugs from PC-related diseases to treat PC. We prioritized drugs based on the number of PC-related diseases that they could treat as well as the ranking scores of these diseases. For example, if drug X treats 25 top-ranked PC-related diseases, it will rank higher than drug Y, which treats only a few lower-ranked diseases. The drug prioritization algorithm is defined as: Rdrug = wherein n is the number of PC-related diseases that a drug can treat and R_disease_i is the disease ranking score (output from the network-based disease ranking algorithm).
De-novo validation using FDA-approved PC drugs We evaluated GenoPredict using 27 FDA-approved PC drugs. The drug prioritization algorithm used only PC-related diseases and their drug treatments. PC and its known drug treatments were not used, therefore the evaluation was a de-novo validation. We calculated recall, mean and median rankings of these 27 FDA-approved drugs among all drugs.
Compare GenoPredict to PREDICT in novel predictions We compared GenoPredict with PREDICT using two evaluation datasets constructed from TreatKB: (1) 172 PC drugs extracted from 172,888 clinical trials; and (2) 72 PC drugs extracted from 43,811 ongoing clinical trials. The 27 FDA-approved PC drugs were removed from the evaluation datasets. The output from GenoPredict is a ranked list of 2484 drugs with the 27 FDA-approved PC drugs removed. In PREDICT, 48 drugs were classified as positives for PC among a total of 593 drugs. These 48 drugs along with their corresponding probabilities (ranging from 0.543-0.994) are publicly available [14]. For the remaining 545 drugs that were predicted as negatives by PREDICT, we assigned to each drug a value that was randomly picked from 0.0 to 0.5 (the random process was repeated 10 times).
Precision-Recall (PR) curves were used to evaluate and compare GenoPredict to PREDICT. Studies have shown that in domains where the number of negatives greatly exceeds the number of positives such as in drug repositioning and many other biomedical classification problems, PR curves are better than the more commonly used receiver operating characteristic (ROC) curves in avoiding overly optimistic view of an algorithm’s performance [26]. Using each of the two evaluation datasets as gold standard, we calculated precisions at 11 different recall cutoffs (0.0, 0.1, 0.2, … 1.0) for both GenoPredict and PREDICT and plotted the PR curves. Mean Average Precision (MAP), which approximates the area under the precision-recall curve [27], was used to quantatively compare GenoPredict to PREDICT.
Analyze top-ranked repositioned drug candidates
Analyze drug classes for top-ranked drug candidates To understand the commonalities shared by top-ranked drug candidates, we examined class distributions of top 124 (top 5%) repositioned candidate drugs. Drugs were classified based on the ATC classification system [16]. For each ATC code, we assessed its probability of being associated with these 124 drugs (e.g., the code “immunosuppressants” is associated with 30 of the 124 drugs) as compared to its probability of being associated with the same number of randomly selected drugs (e.g., the same code is on average associated with 5 out of 149 randomly selected drugs). The random process is repeated 1000 times and a t-test was used to assess the statistical significance.
Analyze genetic pathways targeted by repositioned drug candidates We analyzed genetic pathways targeted by top- ranked 124 drug candidates. Enriched pathways can provide novel insights into coimnon molecular mechanisms underlying top-ranked drug candidates. We obtained drugs’ gene targets from DrugBank [17] and gene-associated pathways from MSigDB [18]. The signficance of each pathway being associated with top 124 drugs was calucated in the same manner as above for drug classe enrichment analysis.
Results
Cancers and metabolic diseases are enriched among top-ranked PC-related diseases
It is known that cancers often share genetics. We analyzed the distributions of cancers among diseases at different ranking cutoffs to evaluate the network construction and disease-ranking algorithms. As shown in Fig.1, cancers are enriched among top-ranked diseases. For example, at the 100% cut-off (all 822 retrieved diseases), 7.24% of the diseases are “Neoplasms”. At top 10% cutoff (top 82 diseases), 16.28% are “Neoplasms,” representing a 124.8% increase as compared to that for all diseases. The enrichment of cancers among top-ranked diseases demonstrated the validity of both network construction and ranking algorithms. Interestingly, the disease class “Endocrine, nutritional and metabolic diseases” was also enriched among top-ranked diseases. Recent studies demonstrated that certain metabolic disorders - including obesity, high blood pressure and high cholesterol - can impact the development and progression of prostate cancer [28]. Other disease classes were not enriched among top-ranked diseases.
GenoPredict found 25 of the 27 FDA-approved PC drugs and ranked them highly
We validated GenoPredict in predicting known PC drugs using 27 FDA-approved PC drugs. GenoPredict achieved a recall of 0.925, an average ranking of 27.3%, and a median ranking of 15.6%. There is a big difference between the mean and median rankings, demonstrating a skewed ranking distribution of these FDA-approved drugs (Fig. 2). While 16 of the 25 PC drugs were ranked within top 20%, some drugs were ranked very low, including degarelix at top 70.49%, abarelix at 86.76%, triptorelin at 89.21%, and histrelin at 91.22%.
Figure 2:
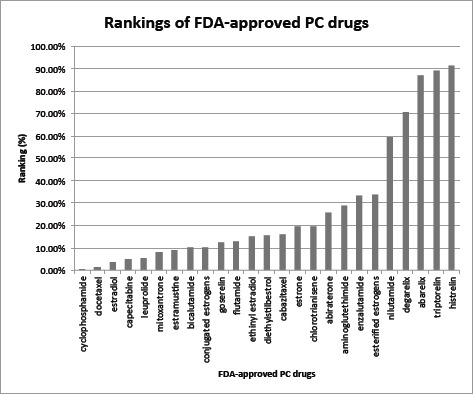
Rankings of FDA-approved PC drugs.
Compare GenoPredict to PREDICT in novel predictions
We plotted PR curves for both GenoPredict and PREDICT using 172 novel PC drugs extracted from 172,888 clinical trial reports as the evaluation dataset. As shown in Fig. 3, the PR curve for GenoPredict clearly dominates that for PREDICT. The mean average precision (MAP) is 0.447 for GenoPredict and 0.169 for PREDICT, representing a significant 164.5% improvement.
Figure 3:
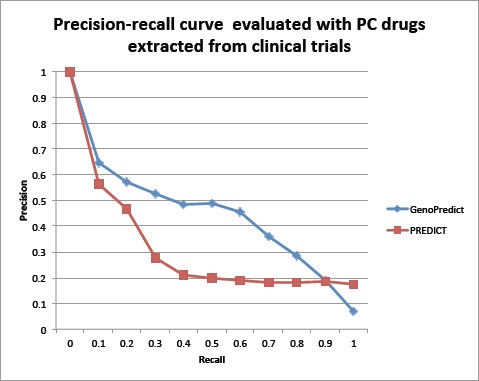
The Precision-Recall curves for GenoPredict and PREDICT. The evaluation data consists of 172 PC drugs extracted from 172,888 clinical trial reports.
Fig. 4 shows the PR curves when 72 novel PC drugs extracted from ongoing clinical trials were used as the evaluation set. While the MAPs for both algorithms were lower than previous ones, the curve for GenoPredict dominates that for PREDICT. The MAP is 0.278 for GenoPredict and 0.084 for PREDICT, representing a significant 231.1% improvement.
Figure 4:
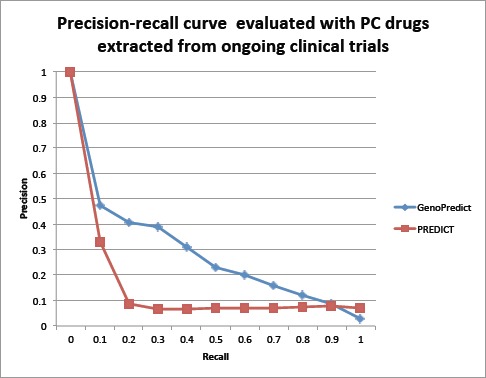
The Precision-Recall curves for GenoPredict and PREDICT. The evaluation data consists of 72 novel PC drugs extracted from 43,811 ongoing clinical trials.Top drug classes of repositioned candidate drugs
Analyze top-ranked drug candidates: drug classification and genetic pathway analysis
We examined drug classes enriched among top-ranked drugs. The highly enriched drug classes may offer insight into the mechanisms of action underlying repositioned drug candidates. Fig. 5 shows the top 15 drug classes. A total of 18.8% of top 124 drug candidates belong to the class “other antineoplastic agents” as compared to 3.9% based on random expectation. The other top-ranked drug classes include “immunosuppressants” and “corticosteroids for systemic use, plain.” We performed the same enrichment analysis for top 248 (top 10%) and top 372 (top 15%) drug candidates and obtained similar results (data not shown).
Figure 5:
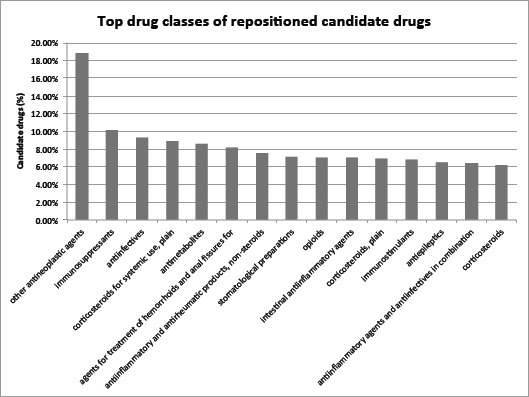
Top fifteen drug classes (ATC codes) representing top 124 drug candidates.
We then examined genetic pathways that are predominantly targeted by top-ranked (top 124) drug candidates. As shown in Fig. 6, the pathway “pathways in cancer” was targeted by 19.05% of the top 124 candidate drugs as compared to 7.92% based on random expectation. The pathway “prostate cancer” was also ranked at top, demonstrating the validity of our pathway analysis. In addition, four of the top fifteen pathways are related to immune systems. Currently, Immunotherapy holds great promise for cancer treatments. For example, the drug ipilimumab targets a checkpoint molecule called CTLA-4 on certain immune cells. This drug is now being tested in men with advanced PC, with early results showing treatment benefits [29–30]. Consistent with these published clinical studies, GenoPredict ranked ipilimumab at top 15% among 2484 drugs.
Figure 6:
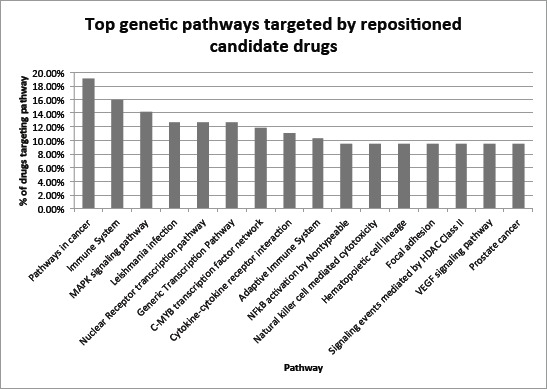
Top fifteen pathways targeted by top 124 (top 5%) repositioned candidate drugs.
Discussion
GenoPredict exploited genetic and drug treatment connections among tens of thousands of diseases. We applied GenoPredict to identify repositioning drug candidates for PC and performed in-deption analysis of top repositioned drug candidates. GenoPredict found 25 of 27 FDA-approved PC drugs and ranked them highly. GenoPredict performed significantly better than PREDICT, a comprehensive drug repositioning system, in novel predictions for PC. However, the performance of GenoPredict is not optimal as seen in its PR curves for drug repositioning for PC. We are further improving GenoPredict by incorporating more comprehensive disease genetics data and will evaluate it in broad types of diseases (e.g., all diseases in the GWAS catalog).
Our cunent study is restricted by the limited number of diseases (882 diseases) in the GWAS catalog, even though TreatKB includes 24,511diseases and their drug treatments. With new studies being continually added to the GWAS catalog, additional drug repositioning opportunities will arise through human genetic analysis. Additionally, disease genetics from rare Mendelian disorders represents another valuable source of novel drug targets [31]. Another rich resource of human disease genetics is computation-based candidate disease genetics discovery. We recently showed that computationally-predicted disease genetics can lead to novel drug discovery [21–22],
This study focused on integrating disease genetics with drug treatment data for drug repositioning. To build a more comprehensive prediction system, additional data resources such as disease phenotypes, gene expression, drug side effects, and chemical structures can be incorporated into GenoPredict to further improve the performance.
Author’s contributions
Xu and Wang have jointly conceived the idea, designed and implemented the algorithms and prepared the manuscript. All authors read and approved the final manuscript.
Figure 1:
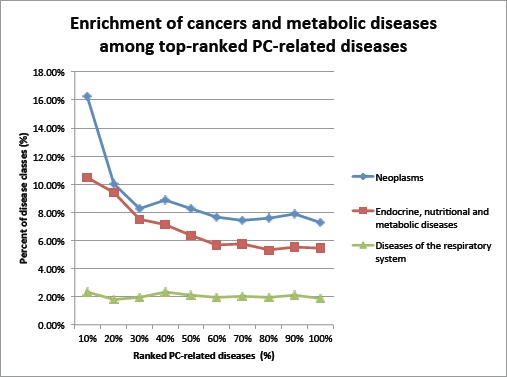
Enrichment of “Neoplams” and “Endocrine, nutritional and metabolic diseases” among top-ranked PC- related diseases. The disease class “Diseases of the respiratory system” (shown as a negative control) and other disease classes (not shown) are not enriched.
Acknowledgements
RX is funded by Case Western Reserve University/Cleveland Clinic CTSA Grant (UL1 RR024989), the NIH Director’s New Innovator Award under the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health (DP2HD084068, Xu), American Cancer Society Research Scholar Grant (RSG-16- 049-01 - MPC, Xu), the Landon Foundation-AACR INNOVATOR Award for Cancer Prevention Research (15-20-27- XU), Mary Kay Foundation Grant (057-15, Xu), and Pfizer 2015 ASPIRE Rheumatology and Dennatology Research Award (WI206753, Xu). QuanQiu’s effort on this project is funded by DP2HD084068.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Trewartha D., Carter K. Advances in prostate cancer treatment. Nature reviews Drug discovery. 2013;12(11):823–824. doi: 10.1038/nrd4068. [DOI] [PubMed] [Google Scholar]
- 3.Hurle M.R., Yang L., Xie Q., Rajpal D.K., Sanseau R, Agarwal R. Computational drug repositioning: from data to therapeutics. Clinical Pharmacology & Therapeutics. 2013;93(4) doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 4.Welter D., MacArthur J., Morales J., Burdett T., Hall R, Junkins H., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42(D1):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu R., Wang Q. Large-scale extraction of accurate drug-disease treatment pairs from biomedical literature for drug repurposing. BMC bioinformatics. 2013;14(1):181. doi: 10.1186/1471-2105-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu R., Li L., Wang Q. Towards building a disease-phenotype knowledge base: extracting disease-manifestation relationship from literature. Bioinformatics. 2013;29(17):2186–2194. doi: 10.1093/bioinformatics/btt359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu R., Wang Q. Automatic signal extraction, prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from the FDA Adverse Event Reporting System (FAERS>. Journal of biomedical informatics. 2014;47:171–177. doi: 10.1016/j.jbi.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanseau P., Agarwal P., Barnes M.R., Pastinen T., Richards J.B., Cardon L.R., Mooser V. Use of genome-wide association studies for drug repositioning. Nature biotechnology. 2012;30(4):317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 9.Okada Y, Wu D., Trynka G., Raj T., Terao C., Ikari K., Graham R.R. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plenge R.M., Scolnick E.M., Altshuler D. Validating therapeutic targets through human genetics. Nature reviews Drug discovery. 2013;12(8):581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 11.Nelson M.R., Tipney H., Painter J.L., Shen J., Nicoletti P., Shen Y., Cardon L.R. The support of human genetic evidence for approved drug indications. Nature genetics. 2015;47(8):856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 12.Xu R., Wang Q. PhenoPredict: A disease phenome-wide drug repositioning approach towards schizophrenia drug discovery. Journal of biomedical informatics. 2015;56:348–355. doi: 10.1016/j.jbi.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Xu R. In AMIA Annual Symposium Proceedings. Vol. 2015. American Medical Informatics Association; 2015. DenguePredict: An Integrated Drug Repositioning Approach towards Drug Discovery for Dengue; p. 1279. [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlieb A., Stein G.Y., Ruppin E., Sharan R. Predict: a method for inferring novel drug indications with application to personalized medicine. Molecular systems biology. 2011;7(1) doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. International statistical classification of diseases and related health problems. 2009 [Google Scholar]
- 16.World Health Organization. The anatomical therapeutic chemical classification system with defined daily doses (ATC/DDD) Norway: WHO. 2006 [Google Scholar]
- 17.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Tang A. DrugBank 4.0: shedding new light on drug metabolism. Nucleic acids research. 2014;42(D1):D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberzon A., Birger C., Thorvaldsdttir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database hallmark gene set collection. Cell systems. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J., Pei J., Kamber M. Elsevier; 2011. Data mining: concepts and techniques. [Google Scholar]
- 20.Page L., Brin S., Motwani R., Winograd T. The PageRank citation ranking: Bringing order to the web. Stanford InfoLab. 1999 [Google Scholar]
- 21.Chen Y., Xu R. Context-sensitive network based disease genetics prediction and its implications in drug discovery. Bioinformatics. 2017:btw737. doi: 10.1093/bioinformatics/btw737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Xu R. Phenome-driven Disease Genetics Prediction Towards Drug Discovery. Bioinformatics. 2015;31(12):i276–i283. doi: 10.1093/bioinformatics/btv245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Xu R. Network-based Gene Prediction for Plasmodium falciparum Malaria Towards Genetics-based Drug Discovery. [11 June 2015];BMC Genomics. 2015 16(Suppl 7):S9. doi: 10.1186/1471-2164-16-S7-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu R., Wang Q., Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC genomics. 2015;16(7):S4. doi: 10.1186/1471-2164-16-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu R., Wang Q. Towards understanding brain-gut-microbiome connections in Alzheimers disease. BMC Systems Biology. 2016;10(3):63. doi: 10.1186/s12918-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis J., Goadrich M. In Proceedings of the 23rd international conference on Machine learning. ACM; 2006. Jun, The relationship between Precision-Recall and ROC curves; pp. 233–240. [Google Scholar]
- 27.Manning C.D., Raghavan P., Schtze H. 1. Vol. 1. Cambridge: Cambridge university press; 2008. Introduction to information retrieval; p. 496. [Google Scholar]
- 28.Sourbeer K.N., Howard L.E., Andriole G.L., Moreira D.M., CastroSantamaria R., Freedland S.J., Vidal A.C. Metabolic syndromelike components and prostate cancer risk: results from the Reduction by Dutasteride of Prostate Cancer Events (REDUCE) study. BJU international. 2015;115(5):736–743. doi: 10.1111/bju.12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slovin S.F., Higano C.S., Hamid O., Tejwani S., Harzstark A., Alumkal J.J., Beer T.M. Ipilimumab alone or in combination with radiotherapy in metastatic castration-resistant prostate cancer: results from an open-label, multicenter phase I/II study. Annals of Oncology. 2013;24(7):1813–1821. doi: 10.1093/annonc/mdt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon E.D., Drake C.G., Scher H.I., Fizazi K., Bossi A., Van den Eertwegh A.J., Ng S. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. The lancet oncology. 2014;15(7):700–712. doi: 10.1016/S1470-2045(14)70189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z.Y., Zhang H.Y. Rational drug repositioning by medical genetics. Nature biotechnology. 2013;31(12):1080–1082. doi: 10.1038/nbt.2758. [DOI] [PubMed] [Google Scholar]


