Abstract
Successful diagnosis and management of neurological dysfunction relies on proper communication between the neurologist and the primary physician (or other specialists). Because this communication is documented within medical records, the ability to automatically infer the clinical correlations for a patient from his or her medical records would provide an important step towards enabling health care systems to automatically identify patients requiring additional follow-up as well as flagging any unexpected clinical correlations for review. In this paper, we present a Deep Section Recovery Model (DSRM) which applies deep neural learning on a large body of EEG reports in order to infer the expected clinical correlations for a patient from the information in a given EEG report by (1) automatically extracting word- and report- level features from the report and (2) inferring the most likely clinical correlations and expressing those clinical correlations in natural language. We evaluated the performance of the DSRM by removing the clinical correlation sections from EEG reports and measuring how well the model could recover that information from the remainder of the report. The DSRM obtained a 17% improvement over the top-performing baseline, highlighting not only the power of the DSRM but also the promise of automatically recognizing unexpected clinical correlations in the future.
Introduction
Diagnosing and managing neurological dysfunction often hinges on successful communication between the neurologist performing a diagnostic test (such an Electroencephalogram or EEG), and the primary physician or other specialists. In 2005, Glick et al.1 studied malpractice claims against neurologists and found that 71% of the claims arose from “an evident failure of communication by the neurologist” and that the majority of the claims resulted from deficient communication between the neurologist and the primary physician or other specialists. In addition, Glick et al. found that 62.5% of claims included diagnostic errors and that 41.7% involved errors in “ordering, interpreting, and reporting of diagnostic imaging, follow-through and reporting mechanisms.” It is expected that these types of errors could be reduced, and communication could be improved by developing tools capable of automatically analyzing medical reports2. Moreover, a recent Institute of Medicine Report3 advocated the need for decision-support tools operating on electronic health records for primary care and emergency room providers to manage referral steps for further evaluation and care of persons with epilepsy. Specifically, the ability to automatically extract and analyze the clinical correlations between any findings documented in a neurological report and the over-all clinical picture of the patient, could enable future automatic systems to identify patients requiring additional follow-up by the primary physician, neurologist, or specialist. Furthermore, systems capable of automatic analysis of the clinical correlations documented in a large number of reports could ultimately provide a foundation for automatically identifying reports with incorrect, unusual, or poorly-communicated clinical correlations mitigating misdiagnoses and improving patient care2. It should be noted, however, that automatically identifying incorrect, unusual, or poorly-communicated clinical correlations has two critical requirements: (1) inferring what the expected clinical correlations would be for the patient and (2) quantifying the degree of disagreement or contradiction between the clinical correlations documented in a report and the expected clinical correlations for the patient. In this initial study, we focus on the first requirement by considering the clinical correlation sections documented in EEG reports.
The role of the clinical correlation section is not only to describe the relationships between findings in the EEG report and the patient’s clinical picture, but to also explain and justify the relationships so as to convince any interested health care professionals. Consequently, the clinical correlation section of an EEG report is expressed through natural language, meaning that the clinical correlations documented in the clinical correlation section are qualified and contextualized through all the subtlety and nuance enabled by natural language expression4. For this reason, while it might appear sufficient to simply extract individual findings or medical concepts from the clinical correlation section, describing and justifying the clinical correlations requires producing coherent natural language2. This requirement makes inferring the expected clinical correlation section from an EEG report a challenging problem because it requires not only identifying the correct clinical correlations, but also expressing those correlations through natural language which is by the content of the EEG report as well as the neurologist’s medical knowledge and accumulated experience.
In this paper, we present a novel Deep Section Recovery Model (DSRM) which applies deep neural learning on a large body of EEG reports in order to infer the expected clinical correlations for a patient based solely on the natural language content in his or her EEG report. The DSRM was trained and evaluated using the Temple University Hospital (TUH) EEG Corpus5 by (a) identifying and removing the clinical correlation section written by the neurologist and (b) training the DSRM to infer the entire clinical correlation section from the remainder of the report. At a high level, the DSRM can be viewed as operating through two general steps:
- Step 1:
word- and report- level features are automatically extracted from each EEG report to capture contextual, semantic, and background knowledge; and
- Step 2:
the most likely clinical correlation section is jointly (a) inferred and (b) expressed through automatically generated natural language.
Our experimental results against a number of competitive baseline models indicate the generative power of the DSRM, as well as the promise of automatically recognizing unusual, incorrect, or incomplete clinical correlations in the future. It should be noted that although we evaluated the DSRM by recovering the clinical correlation sections from EEG reports, the model automatically extracts its own features based on the words in a given report and (clinical correlation) section. Consequently, we believe the DSRM could be easily adapted to not only process addition types of medical reports, but to also to infer and generate medical language for other purposes, e.g., generating explanations for CDS systems, providing automated second opinions, and assessing and tracking documentation quality.
Background
The Deep Section Recovery Model (DSRM) presented in this paper was originally envisioned as part of a larger project to design an automatic patient cohort retrieval system (operating on natural language) for EEG reports6. This system assigns different weights or importance to each section in an EEG report, with the clinical correlation section being the most important. Unfortunately, we found that as many as 1 in 10 EEG reports were missing a clinical correlation section. In previous work7, we designed a binary classification model for automatically inferring the over-all impression (normal or abnormal) for an EEG report. This model was extended and adapted to produce natural language, forming the basis for the DSRM presented in this paper. As a natural language generator, the DSRM incorporates advances from Natural Language Generation, Machine Translation, and Automatic Summarization. We briefly review each of these topics below.
Natural Language Generation. Natural language generation (NLG) is an area of study on how automatic systems can produce high-quality natural language text from an internal representation8. Traditionally, NLG systems rely on a pipeline of sub-modules including content selection – determining which information the model should generate – and surface realization – determining how the model should express the information in natural language. These systems typically require supervision at each individual stage and cannot scale to large domains9. In health care, NLG has traditionally focused on surface realization through a number of applications2, including generating explanations10, advice11 or critiques12 in expert systems, as well as generating explanatory material for patients13. These systems largely rely on templates and rule-based mechanisms for producing natural language content. By contrast, the DSRM jointly performs content selection (via latent feature extraction) and surface realization (using a deep neural language model) without requiring predefined rules or templates.
Machine Translation. Perhaps the most ubiquitous application of NLG, machine translation has been an active area of research for the last 50 years14. While the earliest systems were largely rule-based, statistical machine translation (SMT) systems have become the focus of the field. Statistical machine translation systems typically rely on gold-standard word or sentence alignments between parallel texts in a source and target language and use machine learning to train models which can automatically translate between them15. More recently, the advent of deep learning has enabled the design of systems which jointly learn to align and translate16. The canonical work by Bahdanau et al. (2015)16 introduces the notion of neural attention, which allows the model to learn how words in the target language should be aligned to words in the source language without supervision. The DSRM extends this idea by incorporating an attention layer to learn the association between words in the clinical correlation section and those in the rest of the report.
Automatic Summarization. Automatic summarization systems can be typically divided into two categories: extractive summarization systems, which aim to select individual words or sentence from a document and “stitch” them together to form a summary17, and abstractive summarization systems which consider structural and/or semantic information to produce a summary that can containing words not mentioned in the document. It has been shown18 that extractive summarization may not be sufficient for health care needs; rather, abstract summarization efforts should be preferred. Fortunately, as with SMT, advances in deep learning have allowed allowed summarization systems to learn an internal or embedded representation of a document which can be used as the basis for NLG17 using so-called Sequence-to-Sequence19 models. Consequently, the DSRM model adapts the notion of abstractive summarization and combines and extends Sequence-to-Sequence models with the attention mechanisms used by SMT systems.
Data
The experiments reported in this paper use the Temple University Hospital (TUH) EEG Corpus5 with a standard 3:1:1 split for training, validation, and testing sets. The TUH EEG Corpus is the largest publicly available collection of EEG reports and the first publicly released collection which includes both the raw EEG signal data as well as the EEG report associated with each EEG session. The EEG reports were authored according to The American Clinical Neurophysiology Society (ACNS) guidelines20 for writing an EEG report which stipulate that all EEG reports should contain (a) an introduction, (b) a description of the EEG recording, and (c) an interpretation regarding the normality or abnormality of findings as well as a correlation to the patient’s overall clinical picture. In the TUH EEG Corpus, the introduction was typically divided into three sections: (1) the CLINICAL HISTORY section indicating the age and gender of the patient as well as a brief history of any medical conditions which may affect the EEG recording; (2) the MEDICATIONS section which consists of a comma-separated list of any medications the patient is regularly taking that could influence the EEG recording; and (3) the INTRODUCTION section itself which describes the setting of the EEG, the configuration of electrodes, the patient’s state of consciousness, whether the patient had been fasting, and any other pertinent information about the EEG setting. The description of the EEG recording was represented by the DESCRIPTION section which provides a “complete and objective”20 list of any notable findings including details about all waveforms in the record as well as a description of the patient’s background electro-cerebral activity. The neurologist’s interpretation was documented in two sections in the TUH EEG Corpus: (1) the IMPRESSION section in which the neurologist documents whether the EEG recording indicates normal or abnormal brain activity as well as – particularly in the cast of abnormal brain activity – a list of the most important findings that lead to this conclusion; and (2) the CLINICAL CORRELATION section in which the neurologist ties the findings in the report to the over-all clinical picture of the patient. Table 1 provides an example of each section from the TUH EEG Corpus.
Table 1:
Examples of EEG Report sections from the TUH EEG Corpus (each section was taken from a different EEG report).
| CLINICAL HISTORY: An elderly woman with change in mental status, waxing and waning mental status, COPD, morbid obesity, and markedly abnormal EEG. Digital EEG was done on XXXX XX, XXXX. |
| INTRODUCTION: The EEG was performed using the standard 10/20 electrode placement system with an EKG electrode and anterior temporal electrodes. The EEG was recorded during wakefulness and photic stimulation, as well as hyperventilation, activation procedures were performed. |
| MEDICATIONS: Keppra, Aricept, Senna, Aricept, ASA, famotidine |
| DESCRIPTION: In wakefulness, the background EEG is very low voltage, relatively featureless with some 10 Hz activity in the background and a posterior dominant rhythm, which may be estimated at 7 Hz. The patient seems to have very brief lapses into sleep with diffuse 10 to 13 Hz activity and then spontaneous arousals. This pattern is a beta spindle and then an arousal can be identified throughout the record. Later portions of the record seem to demonstrate more sustained sleep, but with ongoing eye movements. HR: 66 BPM. |
| IMPRESSION: Abnormal EEG due to: |
| 1. Slow and disorganized background. |
| 2. Left occipital sharp waves, at times becoming somewhat periodic in sleep. |
| 3. Some additional epileptiform discharges with more of a mid to posterior temporal localization. |
| CLINICAL CORRELATION: This tracing raises the possibility of a mechanism for seizures outside of the area of the abscess described above. The photoparoxysmal response is unusual and may be accentuated by the previous surgery in the posterior brain regions. |
Inferring the Clinical Correlation Section
When writing the clinical correlation section of an EEG report, the neurologist considers the information described in the previous sections, such as relevant clinical history or notable epileptiform activities, as well as their accumulated medical knowledge and experience with interpreting EEGs. This type of background knowledge is difficult to capture with hand-crafted features because it is rarely explicitly stated; rather, it is implied through the subtlety, context, and nuance afforded the neurologist by natural language. Consequently, to approach this problem, we present a deep neural network architecture which we refer to as the Deep Section Recovery Model (DSRM). Illustrated in Figure 1, the DSRM consists of two major components:
the Extractor which learns how to automatically extract (a) feature vectors representing contextual and background knowledge associated with each word in a given EEG report as well as (b) a feature vector encoding semantic, background, and domain knowledge about the entire report; and
the Generator which learns how to use the feature vectors extracted by the Extractor to produce the most likely clinical correlation section for the given report while also considering the semantics of the natural language it is generating.
Figure 1:
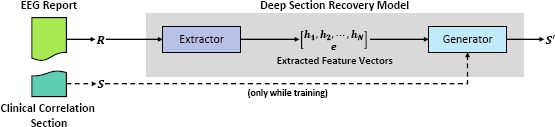
Simplified Architecture of the Deep Section Recovery Model (DSRM).
In order to train and evaluate the DSRM, we identified all EEG reports in the TUH EEG Corpus which contained a CLINICAL CORRELATION section and removed that section from the report. The model was trained to recover the missing clinical correlation section in the training set and evaluated based on the clinical correlation sections it inferred for reports in the test set. In the remainder of this section, we describe (1) the natural language pre-processing steps applied to the data, (2) the mathematical problem formulation, (3) the Extractor, (4) the Generator, (5) how the parameters of the model are learned from the training set, and (6) how the learned parameters are used to infer the most likely clinical correlation section for a (new) EEG report.
Natural Language Pre-processing
Before applying the Deep Section Recovery Model, we pre-processed each EEG report with three basic natural language processing steps: (1) sentence boundaries were identified using the OpenNLP* sentence splitter; (2) word boundaries were detected using the GENIA 21 tokenizer, and (3) section boundaries were identified using a simple regular expression search for capitalized characters ending in a colon. These three pre-processing steps allowed us to represent each section of an EEG report as a sequence of words in which the symbols ⟨s⟩ and ⟨/s⟩ were used to indicate the start and end of each sentence, ⟨p⟩ and ⟨/p⟩ were used to indicate the start and end of each section, and ⟨d⟩ and ⟨/d⟩ were used to indicate the start and end of each report.
Problem Formulation
In order to formally define the problem, it is necessary to first define the vocabulary as the set of all words observed at least once in any section (including the clinical correlation section) of any EEG report in the training set. Let V indicate the size or number of words in the vocabulary. This allows us to represent an EEG report as sequence of V - length one-hot vectors corresponding to each word in the report, i.e., where N is the length or number of words in the report. Likewise, we also represent a clinical correlation section as a sequence of V - length one-hot vectors; in this case, where M is the number of words in the clinical correlation section. The goal of the Deep Section Recovery Model is to infer the most likely clinical correlation section for a given EEG report. Let θ be the learn-able parameters of the model. Training the model equates to finding the values of θ which assign the highest probabilities to the gold-standard (neurologist-written) clinical correlation sections for each EEG report in the training set; formally:
| (1) |
We decompose the probability of a particular clinical correlation section being produced for a given EEG report (i.e., correctly identifying and describing the clinical correlations in the report) into two factors:
| (2) |
where the the first factor is implemented by the Extractor and the second factor is implemented by the Generator.
The Extractor
The language in the clinical correlation section is intended to relate findings and observations described in the previous sections of the record to the over-all clinical picture of the patient. Consequently, in order to automatically produce the clinical correlation section, the goal of the Extractor is to automatically (1) identify important neurological findings and observations (e.g., “background slowing”), (2) identify descriptions of the patient’s clinical picture (e.g., “previous seizure”), and (3) determine the inferred relationship(s) between each finding and the clinical picture as described by the EEG report or implied by medical knowledge and experience (e.g., “observed epileptiform activity is consistent with head trauma”). It should be noted that the length and content of each EEG report varies significantly throughout the collection, both in terms of the sections included in each report as well as the content in each section. Moreover, when producing an EEG report, each neurologist writes in a different style, ranging between terse 12-word sections to 600-word sections organized into multiple paragraphs. Consequently, the roleof the Extractor is to overcome these barriers and extract meaningful feature vectors which characterize semantic, contextual, and domain knowledge. To address these requirements, we implemented the Extractor using the deep neural architecture illustrated in Figure 2. The Encoder relies on five neural layers to produce feature vectors for each word in the report (h1,-,hN) as well as a feature vector characterizing the entire report (e):
Layer 1: Embedding. The role of the embedding layer is to embed each word in the EEG report Ri (represented as a V - length 1-hot vector) into a K-length continuous vector (where ). This is accomplished by using a fully connected linear projection layer, where correspond to the vocabulary projection matrix and bias vector learned by the Extractor.
Layer 2: Bidirectional Recurrent Neural Network. Layer 2 implements a bidirectional recurrent neural network (RNN) using two parallel RNNs trained on the same inputs: (1) a forward RNN which processes words in the EEG report in left-to-right order and (2) a backward RNN which processes words in the EEG report in right-to-left order. This allows the forward RNN to extract features capturing any short- or long-range contextual information about each word in R provided by any preceding words in the EEG report (e.g. that “slowing” is negated in “no background slowing”). Likewise, the backward RNN extracts features capturing any short- or long-range contextual information provided by successive words in the EEG report (e.g. that “hyperventilation” described in the introduction section may influence the inclusion of “spike and wave discharges” in the EEG impression or description sections). Formally, the forward RNN maps the series word embeddings to a series of “forward” word-level feature vectors , while the backward RNN maps to a series of “backward” word-level feature vectors . In our model, the forward and backward RNNs were implemented as a series of shared Gated Recurrence Units22 (GRUs)*.
Layer 3: Concatenation. The concatenation layer combines the forward and backward word-level feature vectors to produce a single feature vector for each word, namely, where [x.y] indicates the concatenation of vectors x and y
Layer 4: 2nd Bidirectional Recurrent Neural Network. In order to allow the model to extract more expressive features, we use a second bidirectional RNN layer. This layer operates identically to the bidirectional RNN in Layer 2, except that the word-level feature vectors produced in Layer 3, i.e., , are used as the input to the bidirectional RNN (instead of used in Layer 2). Likewise, the memory states produced in Layer 4 are denoted as f2 and b2, corresponding to the forward RNN and the backward RNN, respectively. Unlike the bidirectional RNN used in Layer 2, we use the final memory of the forward RNN (i.e. f2) as the report-level feature vector e which will be used by the Generator.
Layer 5: 2nd Concatenation. As in Layer 3, the second concatenation layer combines the forward and backward word-level features vectors produced in the previous layer. In the case of Layer 5, however, we used the resulting feature vectors h 1,…, hN as the word-level feature vectors which will be provided to the Generator.
Figure 2:
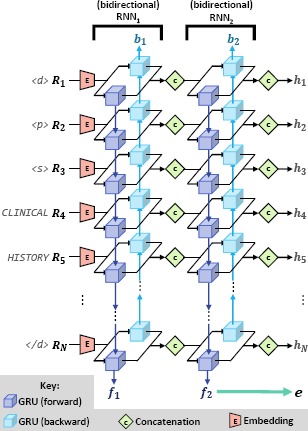
Detailed Architecture of the Extractor.
The Generator
The role of the Generator is to generate the most likely clinical correlation section for a given EEG report using the feature vectors extracted by the Extractor. It is important to note that because the clinical correlation sections vary both in terms of their length and content, the number of possible clinical correlations sections that could be produced is intractably high (VMmax where Mmax is the maximum length of a clinical correlation section). Consequently, we substantially reduce the complexity of the problem by modeling the assumption that each word in the clinical correlation section can be determined based solely on (1) the word-level feature vectors h1,…,hN extracted by the Extractor, (2) the report-level feature vector e extracted by the Extractor, and (3) any preceding words produced by the Generator. This assumption allows us to define the probability of any clinical correlation section, S', having been produced by a neurologist for a given EEG report (i.e., the second factor in Equation 2) as:
To compute Equation 3, we designed the Generator to act as a type of Recurrent Neural Language Model23 (RNLM) which incorporates a Recurrent Neural Network (RNN) to produce one word in the clinical correlation section at-a-time while maintaining and updating an internal memory of which words have already been produced.
To improve training efficiency, the Generator has two similar but distinct configurations: one for training, and one for inference (e.g., testing). Figure 3 illustrates the architecture of the Generator under both configurations. The primary difference between each configuration is the input to the RNN: when training, the model embeds the previous word from the gold-standard clinical correlation section (e.g. Sj_j) to predict Sj' while during inference the RNN operates on the embedding of the previously generated word (e.g. Sj-1') to predict S'j. The Generator produces the natural language content of a clinical correlation section for a given EEG report using four layers (with the preliminary embedding layer in the training configuration acting as an extra “zero”-th layer):
Layer 0: Embedding. The embedding layer, which is only used when the Generator is in training configuration, embeds each word in the gold-standard clinical correlation section Sj (represented by V - length 1-hot vectors) into an L-length continuous vector (pace, , where . This is accomplished by using a fully connected linear projection layer, where correspond to the vocabulary projection matrix and vocabulary bias vector learned by the Generator.
Layer 1: Concatenation. The first layer used in both configurations of the Generator is a concatenation layer which combines the embedded representation of the previous word with e, the report-level feature vector extracted by the Extractor, where [x,y] indicates the concatenation of vectors x and y and s0(0) is defined as a zero vector.
Layer 2: Gated Recurrent Unit. The second layer used by both configurations is a Gated Recurrent Unit (GRU). The GRU allows the model to accumulate memories encoding long-distance relationships between each produced word of the clinical correlation section, S', and any words previously produced by the model. This is performed by updating and maintaining an internal memory within the GRU which is shared across all words in the clinical correlation section. We denote the output of the GRU as .
-
Layer 3: Attention. In order to improve the quality and coherence of natural language produced by the Generator, an attention mechanism was introduced. The attention mechanism allows the Generator to consider all of the world-level feature vectors h1,…,hN produced by the Extractor for the given report, and learns the degree that each word in the EEG report influences the selection of (or aligns with) S'j; formally:
such that αi,j is an alignment vector used in the alignment model βij which determines the degree that the ith word in the EEG report R (represented by hi) influences the jth word of the clinical correlation section S'j (represented by sj(2)).
Layer 4: Addition. The role of the fourth layer is to combine the result of the previous attention layer with the result of the GRU in Layer 2, i.e.,
- Layer 5: Softmax Projection. In order to measure the probability of each word S'j being produced for the given EEG report, we use a final softmax projection layer to produce a vocabulary-length vector sj(5) in which the vth element indicates the probability that S'j should be generated as the vth word in the vocabulary, where , and . This allows us to complete the definition of Equation 3:
4
Figure 3:
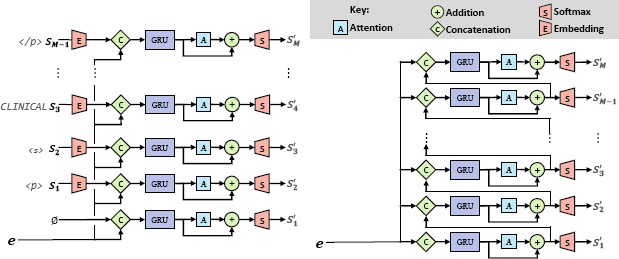
Detailed Architecture of the Generator under (a) Training and (b) Inference Configurations.
Training the Deep Section Recovery Model
Training the Deep Section Recovery Model (DSRM) is achieved by finding the parameters θ which are most likely to produce the gold-standard clinical correlation sections for each EEG report in the training set T. Formally, we model this by minimizing the cross-entropy loss between the vocabulary-length probability vectors produced by the model (sj(5)) and the one-hot vectors corresponding to each word in the gold-standard clinical correlation section (Sj).
| 5 |
The model was trained using Adaptive Moment Estimation (ADAM)24 (with an initial learning rate η=0.001).
Inferring Clinical Correlations
Given θ learned from the training set, the clinical correlation section S can be generated for a new EEG report R using the inference configuration illustrated in Figure 3b. In contrast to the training configuration in which S'j is selected using the previous word from the gold-standard clinical correlation section (Sj_1), during inference, the model predicts S'j using the word previously produced by the model (Sj-1). It is important to note that, unlike training, we do not know the length of the clinical correlation section we will generate. Consequently, the model continually generates output until it produces the END-OF-SECTION symbol ⟨/p⟩. Thus, the length of the inferred clinical correlation section M is determine dynamically by the model. When inferring the most likely clinical correlation section, it is necessary to the convert the vocabulary probability vectors to one-hot vocabulary vectors S'j that can be directly mapped to natural language.*
Experiments
We evaluated the performance of the Deep Section Recovery Model (DSRM) using the Temple University Hospital EEG Corpus5 (described in the Data section) using a standard 3:1:1 split for training, validation, and testing sets. The performance of our model was compared against four baseline systems:
NN:Cosine. In this nearest-neighbor baseline, we represented each EEG report as a bag-of-words vector. This baseline infers the clinical correlation for a given EEG report by copying the clinical correlation associated with the EEG report in the training set whose bag-of-words vector had the least cosine distance to the bag-of-words vector representation of the given EEG report.
NN:LDA. In the second nearest-neighbor baseline, we represented each EEG report as a latent topic vector which was computed by applying Latent Dirichlet Allocation25 to the EEG reports in the training set. This allowed us to infers the clinical correlation for a given EEG report by copying the clinical correlation associated with the EEG report in the training whose topic-vector representation has the least Euclidean distance to the topic-vector representation of the given EEG report.
DL:Attn-RNLM. The first deep-learning baseline considers a recurrent neural language model23 (RNLM) using the standard attention mechanism operating on the embedded word-representations of a given EEG report. This baseline closely resembles the DSRM if the Extractor component were removed.
DL:Basic-S2S. The second deep-learning baseline uses a standard Sequence-to-Sequence19 model without attention. This baseline closely resembles the DSRM if word-level feature vectors (i.e., h1…,hN) were not extracted and only the report-level feature vector is considered by the Generator.
Implementation Details
Our model and the two deep learning baselines were implemented in Tensorflow* version 1.0. For all deep learning models, we used a mini-batch size of 10 EEG reports, a maximum EEG report length of 800 words, a maximum clinical correlation section length of 60 words, 200-dimensional vectors for word embeddings, and 256 hidden units in all RNNs based on a grid search over the validation set.
Experimental Setup and Results
Evaluating the quality of automatically produced natural language (such as the inferred clinical correlation sections) is an open problem in the natural language processing community. Consequently, to quantify the quality of the clinical correlation sections inferred by all four baseline systems as well as the DSRM, we considered standard metrics used to evaluate machine translation, automatic summarization, and speech recognition.
We measured the surface-level accuracy of an automatically inferred clinical correlation section in two ways: (1) the Word Error Rate26 (WER) which measures how many “steps” it takes to transform the inferred clinical correlation section into the gold-standard clinical correlation section produced by the neurologist, where steps include (a) insertion, (b) deletion, or (c) replacement of individual words in the inferred clinical correlation; (2) the Bilingual Evaluation Understudy27 (BLEU) metric which is a commonly used analogue for Precision in language generation tasks. The surface-level completeness of each inferred clinical correlation section was measured using the Recall-Oriented Understudy for Gisting Evaluation28 (ROUGE), a commonly used analogue for Recall (i.e. Sensitivity) in language generation tasks. Finally, we measured the surface-level coherence by additionally computing the bi-gram and tri-gram variants of BLEU and ROUGE, which have been shown to correspond to human notions of coherence. It is important to note that the WER, BLEU, and ROUGE metrics do not take into account the similarity between individual words nor the semantics of multi-word expressions. For example, if the gold-standard clinical correlation contains “absence of epileptiform features”, then the excerpt “no epileptiform activity” would have BLEU-2 and ROUGE-2 scores of zero and a WER of 2 despite the fact that both excerpts express the same information. Consequently, these surface-level metrics should be interpreted as strict lower-bounds on the performance of each evaluated system. Table 2 presents these results.
Table 2:
Evaluation of automatically inferred clinical correlation sections.
| System/Model | BLEU-1 | BLEU-2 | BLEU-3 | ROUGE-1 | ROUGE-2 | ROUGE-3 | WER |
|---|---|---|---|---|---|---|---|
| NN:Cosine | .55334*** | .40274*** | .32137*** | .54284*** | .38516*** | .31508*** | 2.521*** |
| NN:LDA | .51730*. | .36316*** | .28199*** | .52389*** | .36863*** | .28686*** | 2.891*** |
| DL:Attn-RNLM | .57907** | .41619* | .32433* | .58196*** | .41960* | .32575*** | 2.315*** |
| DL:Basic-S2S | .58992** | .36829*** | .26806*** | .47487*** | .31170*** | .23445*** | 2:658*** |
| DSRM | .68792 | .54686 | .46323 | .63523 | .50459 | .42894 | 1.631 |
*p <0.05, ** p<0:01, *** p<0:001; statistical significance against DSRMusing theWilcoxon signed-rank test.
It can be seen that the DSRM achieved the best over-all performance. Moreover, it can be observed that the Attention Decoder (DL:Attn-Decoder) achieved the second-best performance. The Basic Sequence-to-Sequence model (Basic-S2S) as well as the Cosine and LDA nearest neighbor approaches achieved comparable, but only moderate performance. The high performance of the DSRM compared to the Basic S2S model indicates the importance of incorporating attention, allowing the model to discover latent relationships between words in the EEG report and each word in the clinical correlation section. Moreover, the improvement in performance shown by the DSRM compared to the Attention Decoder indicates that the clinical correlation cannot be generated solely from word-level features: report-level information should be considered as well.
Discussion
In order to analyze the automatically inferred clinical correlation sections produced by the DSRM, we manually reviewed 100 randomly selected EEG reports from the test set by comparing the inferred clinical correlation sections to the gold-standard clinical correlation sections written by the neurologists. The over-all quality of the inferred clinical correlation sections was assessed using the Likert scale illustrated in Table 4, with the DSRM obtaining an average score 3.491, indicating that the inferred clinical correlation sections are generally accurate, but may contain minor additional erroneous information or have minor omissions.
Table 4:
Likert scale used to assess over-all quality of inferred clinical correlation sections.
| 1: | (strongly disagree) clinical correlation section is incomprehensible |
| 2: | (disagree) clinical correlation section is not correct |
| 3: | (weakly agree) clinical correlation section is generally correct, but omits important information or contains additional false or inconsistent information |
| 4: | (agree) clinical correlation section is correct but omits minor details |
| 5: | (strongly agree) clinical correlation section is effectively equivalent to the gold-standard |
Table 3 illustrates the inferred clinical correlation as well as the gold-standard clinical correlation section for three EEG reports in the test set. Example 1 illustrates an example of a correct, but incomplete inferred clinical correlation section. Both the inferred and gold-standard clinical correlation sections agree that (1) no epileptiform discharges were observed, and (2) thata repeat EEG focusing on extensive sleep is needed. However, the gold-standard clinical standard includes additional details about asymmetry and asymmetry of wicket activity which the DSRM omitted.
Table 3:
Comparisons of inferred and gold-standard clinical correlation sections for three EEG reports.
| Example 1 | Report: 00005044_s03 |
| Inferred: No epileptiform features are identified. If epilepsy is an important consideration, a repeat EEG capturing deeper stages or sleep deprivation prior to the EEG may be helpful to identify epileptiform activity. | |
| Gold: There are no definitive epileptiform discharges, but there is an amplitude asymmetry and there is an asymmetry of wicket activity. Additional recording capturing more extensive sleep may be helpful to identify epileptiform activity. | |
| Example 2 | Record: 00010462_s01 |
| Inferred: This EEG supports a severe underlying encephalopathy and diffuse disturbance of cerebral dysfunction involving both gray and white matter. Contributing factors can include some of the renal failure, acute or metabolic processes. The focal features described above should be correlated with imaging. | |
| Gold: This abnormal EEG demonstrates a severe, diffuse disturbance of cerebral function involving both gray and subcortical white matter. This EEG pattern was communicated to the primary care team. | |
| Example 3 | Report: 000004928_s02 |
| Inferred: This EEG is not suggestive of a metabolic or intermittent encephalopathy. The rare left with focal feature suggests conforms with underlying metabolic pattern. | |
| Gold: As discussed with the team on the date of this recording, this EEG is most compatible with a metabolic encephalopathy. |
Example 2 illustrates an inferred clinical correlation section which accurately expresses the diffuse disturbance of cerebral function. However, the inferred clinical correlation section additionally indicates a “severe underlying encephalopathy” which was not expressed in the gold-standard clinical correlation section. Moreover, the inferred clinical correlation section attempts to correlate the findings with the patients “renal failure, and acute, and/or metabolic processes” and indicates that these findings should be correlated with imaging. While these inclusions highlight the model’s ability to accumulate knowledge across the large corpus of EEGs in the training set in order to simulate experience, they also demonstrate that the model occasionally struggles to determine which information is (or is not) relevant.
The inferred clinical correlation illustrated in Example 3 illustrates a relatively rare (15% of reviewed EEG reports) but significant error: contradiction within the inferred clinical correlation sections. While the first sentence (incorrectly) states that the EEG does not suggest metabolic encephalopathy, the second sentence indicates that it does. This error strongly suggests that the performance of the model could be improved by developing and incorporating a more sophisticated loss function: the average cross-entropy loss (shown in Equation 5) considers each individual word in the inferred clinical correlation equally; thus, the incorrect inclusion of “not” in the first sentence has a very small impact on the loss despite it inverting the meaning of the entire sentence.
Conclusion
In this paper, we have presented a deep learning approach for automatically inferring the clinical correlation section for a given EEG report, which we call the Deep Section Recovery Model (DSRM). While traditional approaches for inferring clinical correlations would require hand-crafting a large number of sophisticated features, the DSRM learns to automatically extract word- and report- level features from each EEG report. Our evaluation on over 3,000 EEG reports revealed the promise of the DSRM: achieving an average of 17% improvement over the top-performing baseline. These promising results provide a foundation towards automatically identifying unusual, incorrect, or inconsistent clinical correlations from EEG reports in the future. Immediate avenues for future work include (1) considering more sophisticated loss functions which incorporate contextual and semantic information and (2) an in-depth study and evaluation of metrics for qualifying the degree of disagreement between a given clinical correlation section and the inferred or expected clinical correlation section.
Acknowledgements
Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under award number 1U01HG008468. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
https://opennlp.apache.org/
A GRU is a block of coordinated sub-layers in a neural network which learn to transform an input vector (e.g. r(0)i) into an output vector (e.g. r(2f)i or r(2b)i) by maintaining and updating an internal memory state. The memory state used in the forward RNN is denoted by f1 while the memory state used in the backward RNN is denoted by b1.
Let is defined as the one-hot vector in which the value is 1 and all other values are zero.
https://www.tensorflow.org/
References
- 1.Glick TH, Cranberg LD, Hanscom RB, Sato L. Neurologic patient safety: an in-depth study of malpractice claims. Neurology. 2005;65(8):1284–1286. doi: 10.1212/01.wnl.0000180961.82198.0d. [DOI] [PubMed] [Google Scholar]
- 2.Cawsey AJ, Webber BL, Jones RB. The Oxford University Press; 1997. Natural language generation in health care. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.England MJ, Liverman CT, Schultz AM, Strawbridge LM. Epilepsy across the spectrumml: Promoting health and understanding. A summary of the Institute of Medicine report. Epilepsy & Behavior. 2012;25(2):266–276. doi: 10.1016/j.yebeh.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albright D, Lanfranchi A, Fredriksen A, Styler WF, Warner C, Hwang JD, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. Journal of the American Medical Informatics Association. 2013;20(5):922–930. doi: 10.1136/amiajnl-2012-001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harati A, Choi SM, Tabrizi M, Obeid I, Picone J, Jacobson M. In: Global Conference on Signal and Information Processing (GlobalSIP), 2013 IEEE. IEEE; 2013. The Temple University Hospital EEG Corpus; pp. 29–32. [Google Scholar]
- 6.Goodwin T, S H. Multi-modal Patient Cohort Identification from EEG Report and Signal Data. AMIA Annual Symposium. 2016:1694–1803. [PMC free article] [PubMed] [Google Scholar]
- 7.Goodwin T, S H. Deep Learning from EEG Reports for Inferring Underspecified Information. AMIA CRI. 2017;2017 [PMC free article] [PubMed] [Google Scholar]
- 8.Varile GB, Zampolli A. Vol. 13. Cambridge University Press; 1997. Survey of the state of the art in human language technology. [Google Scholar]
- 9.Iyer S, Konstas I, Cheung A, Zettlemoyer L. Summarizing source code using a neural attention model. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics. 2016;1:2073–2083. [Google Scholar]
- 10.Swartout WR. In: Computer-assisted medical decision making. Springer; 1985. Explaining and justifying expert consulting programs; pp. 254–271. [Google Scholar]
- 11.Carberry S, Harvey T. Generating coherent messages in real-time decision support: Exploiting discourse theory for discourse practice. In: Nineteenth Annual Conference of the Cognitive Science Society. 1997:79–84. [Google Scholar]
- 12.Gertner AS, Webber BL, Clarke JR, Hayward CZ, Santora TA, Wagner DK. On-line assurance in the initial definitive management of multiple trauma: evaluating system potential. Artificial Intelligence in Medicine. 1997;9(3):261–282. doi: 10.1016/s0933-3657(96)00377-6. [DOI] [PubMed] [Google Scholar]
- 13.Buchanan BG, Moore JD, Forsythe DE, Carenini G, Ohlsson S, Banks G. An intelligent interactive system for delivering individualized information to patients. Artificial intelligence in medicine. 1995;7(2):117–154. doi: 10.1016/0933-3657(94)00029-r. [DOI] [PubMed] [Google Scholar]
- 14.Slocum J. A survey of machine translation: its history, current status, and future prospects. Computational linguistics. 1985;11(1):1–17. [Google Scholar]
- 15.Brown PF, Pietra VJD, Pietra SAD, Mercer RL. The mathematics of statistical machine translation: Parameter estimation. Computational linguistics. 1993;19(2):263–311. [Google Scholar]
- 16.Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. In: ICLR. 2015 [Google Scholar]
- 17.Rush AM, Chopra S, Weston J. A neural attention model for sentence summarization. EMNLP. 2015 Sep;:379–389. [Google Scholar]
- 18.Pivovarov R, Coppleson YJ, Gorman SL, Vawdrey DK, Elhadad N. In: AMIA Annual Symposium Proceedings. Vol. 2016. American Medical Informatics Association; 2016. Can Patient Record Summarization Support Quality Metric Abstraction? p. 1020. [PMC free article] [PubMed] [Google Scholar]
- 19.Cho K, Van Merriënboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, et al. Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation. EMNLP. 2014 Oct;:1724–1734. [Google Scholar]
- 20.Tatum WO, Selioutski O, Ochoa J, Clary HM, Cheek J, Drislane F, et al. American Clinical Neurophysiology Society Guideline 7. Guidelines for EEG Reporting. 2016 doi: 10.1080/21646821.2016.1245576. [DOI] [PubMed] [Google Scholar]
- 21.Tsuruoka Y, Tateishi Y, Kim JD, Ohta T, McNaught J, Ananiadou S, et al. In: Panhellenic Conference on Informatics. Springer; 2005. Developing a robust part-of-speech tagger for biomedical text; pp. 382–392. [Google Scholar]
- 22.Glorot X, Bordes A, Bengio Y. Deep sparse rectifier neural networks. In: International Conference on Artificial Intelligence and Statistics. 2011:315–323. [Google Scholar]
- 23.Mikolov T, Karafiát M, Burget L, Cernockỳ J, Khudanpur S. Recurrent neural network based language model. In: INTERSPEECH. 2010;2:3. [Google Scholar]
- 24.Kingma D, Ba J. Adamml: A method for stochastic optimization. ICLR. 2015 [Google Scholar]
- 25.Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. Journal of machine Learning research. 2003;3(Jan):993–1022. [Google Scholar]
- 26.Jurafsky D, James H. Speech and Language Processing: an Introduction to Natural Language Processing, Computational Linguistics, and Speech. Pearson Education. 2000 [Google Scholar]
- 27.Papineni K, Roukos S, Ward T, Zhu WJ. In: Proceedings of the 40th annual meeting on association for computational linguistics. Association for Computational Linguistics; 2002. BLEU: a method for automatic evaluation of machine translation; pp. 311–318. [Google Scholar]
- 28.Marie-Francine Moens SS., editor. Text Summarization Branches Out: Proceedings of the ACL-04 Workshop. Barcelona, Spain: Association for Computational Linguistics; 2004. ROUGE: A Package for Automatic Evaluation of Summaries; pp. 74–81. [Google Scholar]


