Abstract
Social networks, such as Twitter, have become important sources for active monitoring of user-reported adverse drug reactions (ADRs). Automatic extraction of ADR information can be crucial for healthcare providers, drug manufacturers, and consumers. However, because of the non-standard nature of social media language, automatically extracted ADR mentions need to be mapped to standard forms before they can be used by operational pharmacovigilance systems. We propose a modular natural language processing pipeline for mapping (normalizing) colloquial mentions of ADRs to their corresponding standardized identifiers. We seek to accomplish this task and enable customization of the pipeline so that distinct unlabeled free text resources can be incorporated to use the system for other normalization tasks. Our approach, which we call Hybrid Semantic Analysis (HSA), sequentially employs rule-based and semantic matching algorithms for mapping user-generated mentions to concept IDs in the Unified Medical Language System vocabulary. The semantic matching component of HSA is adaptive in nature and uses a regression model to combine various measures of semantic relatedness and resources to optimize normalization performance on the selected data source. On a publicly available corpus, our normalization method achieves 0.502 recall and 0.823 precision (F-measure: 0.624). Our proposed method outperforms a baseline based on latent semantic analysis and another that uses MetaMap.
Introduction
Pharmacovigilance is defined as “the science and activities relating to the detection, assessment, understanding and prevention of adverse effects or any other possible drug-related problems’“) The primary focus of pharmacovigilance is the monitoring of adverse drug reactions (ADRs). Due to the various limitations of pre-approval clinical trials, it is not possible to assess all the consequences of the use of a particular drug before it is released.2 Therefore, ADRs caused by prescription drugs is currently considered to be a major public health problem and various ADR monitoring mechanisms are currently in place, such as voluntary reporting systems, electronic health records, and, relatively recently, social media.3
Social media has emerged as an important source of information for various public health monitoring tasks. The increasing interest in social media is largely because of the abundance of data in the multitude of social networks— data that is directly generated by a vast number of consumers. It is estimated that about 75% of all U.S. adults have a social network account and about 50% worldwide. Data from social networks have been used in the past for a variety of tasks such as studying smoking cessation patterns on Facebook,4 identifying user social circles with common medical experiences (like drug abuse),5 and monitoring malpractice.6 In addition, recent research has utilized social media for the monitoring of ADRs from prescribed medications. From the perspective of pharmacovigilance, social media could be a platform of paramount importance, since it has been shown in past research that users discuss their health-related experiences, including use of prescription drugs, side effects and treatments on a regular basis. Users are known to share their personal experiences over social media sources, and as such, a large amount of health-related knowledge is generated within the realm of social media.3 However, while significant progress have been made in ADR text classification, and ADR mention extraction,7,8 the normalizing of user posted ADR mentions into a predefined set of concepts is still a largely unaddressed problem.
In this paper, we address the task of normalizing distinct ADR mentions to standardized concepts. This is an essential task given that the same ADR concept may have multiple lexical variants (e.g., “high blood pressure”, and “hypertension”). Therefore, following automatic ADR extraction approaches, automatic normalization techniques must be applied to obtain realistic estimates for the occurrences of ADRs. For social media data, this is particularly important because users often tend to express their problems using non-standard terms. For example, consider the following user posts:
“<DRUG NAME> makes me having the sleeping schedule of a vampire.”, “<DRUG NAME> evidently doesn’t care about my bed time”,
“… wired! Not sleeping tonight. # <DRUG NAME>”.
In the above examples, all the posts are referring to sleeplessness caused by the intake of a specific medication. Each expression is unique and non-standard, although referring to the same ADR concept. In the corpus that we use, each ADR concept is encoded using the Unified Medical Language System (UMLS) concept IDs. All standard and nonstandard lexical variants of an ADR are mapped to the most appropriate UMLS entry. The target of our approach, formally, is to predict the UMLS concept ID of a text-based ADR mention.
Concept normalization
The task of normalization of ADRs involves assigning unique identifiers to distinct ADR mentions, with different lexical variants of the same concept. The IDs are derived from any lexicon or knowledge base with sufficient coverage. In the case of our research, we use the UMLS concept identifiers to uniquely specify each ADR concept. The UMLS provides a vast vocabulary of medical concepts and the semantic groups into which the concepts can be classified. Each UMLS concept is assigned a unique ID, which represents all the lexical variants of the concept. From the previously mentioned example, all synonyms of the concept hypertension (e.g., hypertensive disorder, high blood pressure, high bp and so on) are assigned the ID c0020538. The UMLS Metathesaurus, due to its comprehensive coverage of medical terminologies, has been used to build corpora specialized for normalization in the past.9–11
The task of medical concept normalization can be regarded as a sub-field of biomedical named entity recognition (NER). Due to the abundance of text based medical data available, NER and concept normalization have seen growing research in the medical domain primarily through challenges such as BioCreative,12 BioNLP,13 TREC,14 and i2b2.15 Building on from these initiatives, the problem of concept normalization has seen substantial work for genes and proteins. Majority of the research on concept normalization relies on some variants of dictionary lookup techniques and string matching algorithms. Machine learning techniques have recently been employed, but mostly in the form of filtering techniques to choose the right candidates for normalization.16 A number of approaches17 rely on the use of tools/lexicons such as MetaMap18 as a first step for the detection of concepts. Due to the advances in machine learning techniques and also the increasing availability of annotated data, recent approaches tend to apply learning based algorithms to improve on banal dictionary lookup techniques. Very recently, Leaman et al.10 applied pairwise learning from a specialized disease corpus for disease name normalization. Prior works have involved list- wise learning, which learn the best list of objects associated with a concept and return the list rather than a single object, for tasks such as gene name normalization,19,20 graph-based normalization,21 conditional random fields,22 regression based methods,23 and semantic similarity based techniques.24 Semantic similarity or relatedness is a measure that shows how similar two concepts are. Such measures are often used for word sense disambiguation,25 where the term and its context information are utilized to assign a meaning to it. A number of techniques for computing semantic relatedness among medical entities have been proposed and compared in the past,26 some of which are mentioned in the next section. However, to the best of our knowledge, measures of semantic relatedness have not been previously used for normalizing ADR mentions.
Social media text normalization
While the task of normalization of medical concepts is itself quite challenging, in our case, the problem is exacerbated by the fact that our data originate from social media. Social media data is notoriously noisy.27 And while this hampers the performance of natural language processing (NLP) techniques, it is also the primary motivation behind the implementation of techniques for automatic correction and normalization of medical concepts in this type of text. Typos, ad hoc abbreviations, phonetic substitutions, use of colloquial language, ungrammatical structures and even the use of emoticons make social media text significantly different from texts from other sources.27
Past work on normalization of social media text focused at the lexical level, and has similarities to spell checking techniques with the primary difference that out of vocabulary terms in social media text are often intentionally generated. Text messages have been used as input data for normalization models, and various error models have been proposed, such as Hidden Markov Models28 and noisy channel models.29 Similar approaches targeted purely towards lexical normalization have been attempted on social media texts as well.30,31 For the research task we describe in this paper, although the primary goal is to perform concept level normalization, we apply several preprocessing techniques to perform lexical normalization before the application of our concept normalization pipeline.
Methods
The goal of this normalization task is to find the UMLS concept ID related to a text segment in a tweet that is pretagged as an ADR. For example, in the tweet: “had 2 quit job: tendons in lots of pain,” the phrase “tendons in lots of pain” is tagged as an ADR. The goal of our system is to normalize the annotated text to a concept in UMLS, which in this example is “c0231529-tenalgia”. Figure 1 shows the overall pipeline of the proposed normalization system. The system consists of syntactic and semantic matchers, synonym normalization and evaluation components. The pipeline is sequential, and so, as soon as a matching module finds a match between a lexical component and a UMLS concept, the remaining matchers in the pipeline are skipped and the flow goes to the synonym normalization and evaluation components.
Figure 1.
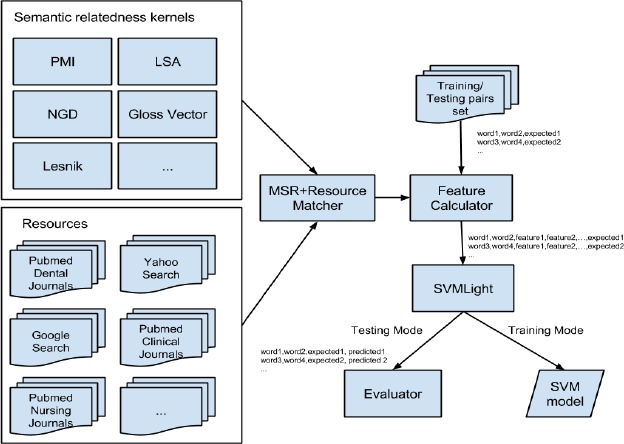
Overall architecture of the Hybrid Semantic Analysis technique.
For evaluation, we use a publicly available, annotated corpus of 2008 tweets mentioning drugs and adverse reactions.8 The corpus was generated by using Twitter API to search for tweets that contain the names of selected drugs. The dataset includes 1544 annotations using 345 unique concepts, of which 1272 are ADRs, 239 are indications/symptoms and 32 are medications. The annotations were performed by two trained biomedical informatics annotators, and all disagreements were resolved and the final corpus was validated by a pharmacology expert. In this work, we did not differentiate between annotation types (e.g., ADR vs. indication) and attempted to normalize all types using the same pipeline. More information about the corpus and annotations can be found in the publication associated with this dataset.
The first step in our normalization pipeline involves syntactic or lexical matching with concept names in UMLS. This part of the pipeline involves two steps: exact match and definition match. An exact match happens when an ADR mention in a user post exactly matches a UMLS concept name (i.e., when the user uses a standard lexical expression for a concept). This simple matching technique can detect many easy matches as standard terminologies are often used by the users. However, in many cases in informal text, ADR mentions are misspelled, and exact matches are not possible. Some of these misspellings can be caught by simple pre-processing techniques. For example, unnecessary character repetition can be removed, as in the tweet “I feel siiiiiiiiiiiiiiick”, “siiiiiiiiiiiiiiick” is matched with UMLS concept “c0231218-sick”.
The next step in syntactic matching utilizes the formal definitions of UMLS concepts. The UMLS metathesaurus provides one or more definitions for each concept. The definition is a passage that describes the concept in plain English. We use this information, in the semantic similarity component later in the pipeline, to create semantic vectors and calculate the similarity values. In the syntactic matching module, we check if the mention appears in the definition of a single concept only in UMLS. If it does, the mention is normalized to the concept. In most of the cases, a phrase appears in the definition of many concepts and no conclusion can be made.
Semantic match
ADR concepts that are not normalized by the syntactic matching components are passed on for semantic matching. The primary task of this component is to compute the similarities of potential ADR concepts with the UMLS concepts. We experiment with two Measures of Semantic Relatedness (MSR) methods. MSR methods or kernels are functions that accept a pair of phrases/words as input, and return a numeric value representing the relatedness score of the inputs. In this module, an MSR method is used to find semantic similarity of a mention and a subset of concepts in UMLS. The most similar concept, with a similarity above a specified threshold, will be chosen as the concept of the mention. We evaluated Latent Semantic Analysis (LSA),32 and our proposed hybrid method. In the semantic matching modules, only the UMLS concepts that are used in the annotations are considered for the prediction.
Latent semantic analysis
Latent Semantic Analysis (LSA) uses a term-document frequency matrix to estimate semantic similarity of two segments of text. LSA then harvests the matrix using Singular Value Decomposition (SVD), by selecting the k best SVD values. More details about LSA technique and various weighting techniques can be found in past publications.33–35,32 In our system, for the first step, the term vector space is generated from a corpus of plain text documents. Then this vector space is used to find a representative vector for each UMLS concept. The UMLS concept names are used to search for term vectors in the vector space. We evaluate some of the corpora, which are listed in Table 1 for creating the representative vectors for the UMLS concepts. After finding a representative vector for each UMLS concept, we search for a representative vector for each annotated text in the same vector space. The cosine similarity of each concept’s vector and the annotated text’s vector is computed. The concept with the highest similarity to the ADR is chosen as the normalized concept if the cosine similarity is above a certain threshold (⟹0.8).
Table 1.
Resources used by HSA for the experiments described in this paper.
| Term Count | Document Count | Topic | |
|---|---|---|---|
| PubMed Dental Journals | 182641 | 236767 | Dental |
| PubMed Nursing Journals | 74000 | 72494 | Nursing |
| PubMed Systematic Reviews | 219656 | 214252 | Clinical |
| BioNLP Corpus | 9483 | 908 | Biology |
| Reuters Corpus | 105675 | 694335 | News |
| ADR-Tweets Corpus | 6205 | 2008 | Drug |
| UMLS Definitions | 103933 | 188647 | Clinical |
Hybrid semantic analysis (HSA)
HSA uses machine learning to find the optimal combination of semantic relatedness function scores for each context, based on a set of calculated features. Using different free text sources, semantic representations for the concepts are learned, and then the different MSR scores are computed for the social media based lexical representations and the standard lexical representations of the concepts. Since different resources can be used, each MSR can return different values when applied to different resources. For example, we can apply PMI, and distinct resources like PMI-GENIA and PMI-I2B2ClinicalNotes. Each MSR method returns different scores when trained on different resources, and these scores are combined in a regression function as features.
For each pair of words/phrases, the Feature Calculator component of the system computes feature scores, which are the returned values from each MSR consisting of different corpora combinations. For example, one feature can be semantic relatedness returned for a pair by LSA-I2B2ClinicalNotes. After feature calculation, the regression model (SVM) is trained, and the model is evaluated against the test set.
Since the MSR and information resources are dynamic, and can be added or removed from the system, the feature set for the regression function is also dynamic and can be varied depending on the task. The output of regression function is a semantic relatedness score of two concepts in the given context, and the regression function is optimized using the labeled training data. The method is designed to easily adapt to new knowledge sources or corpora and adjust parameters accordingly. Also, it can be trained for a new text type or entity type.
For training the regression model, we prepared the training set from a subset of annotation (50% of the annotation). For each annotated text we created training examples for the annotated text and the UMLS concept names of the assigned concept with expected similarity of 100. For each annotation we generated 10 negative examples from the annotated text to random concepts in UMLS with expected similarity of 0. Figure 2 shows an example how HSA training examples are generated.
Figure 2.
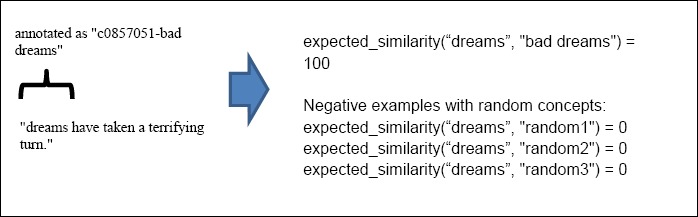
Example of HSA training from ADR normalization annotations.
The ratio of negative to positive examples can affect how the HSA regression model is trained. We used SVM with a linear kernel as the regression model, and trained HSA with the resources listed in Table 1 and LSA as the only MSR. SVM (SVMLight36) was used to create the regression model but other models such as neural network can be used and explored. We refrained from adding additional MSR methods as the intent of this experiment to study the effect of using the regression model with a single MSR and various additional resources. These resources are described in the next section.
After HSA is trained, the regression model is used to calculate the similarity of annotated phrases as ADR (which are the input to our system) to UMLS concept names. First, testing instances between the phrase and a set of selected UMLS concepts names are created. To limit the search space, UMLS concepts appeared in the training set annotations with frequencies of three or more are used for creating the test instances for HSA. Following that, for each example, the features are calculated. The features are all possible MSR and resource combinations defined in the system setup (e.g., LSA with PubMed). Next, the regression model is run on the test instances to calculate the similarity of the annotated text and each UMLS concept. The concept with the highest similarity and above a certain threshold ([>=90], note that the maximum and minimum similarities in the training set are 0 and 100) is chosen as the normalized concept. Since the method has to calculate several semantic similarities for normalizing each annotated text, the process is slower than using a single MSR. The output of the trained regression is not normalized to any boundary and can be any real value.
Corpora
The two semantic matching techniques discussed above require data from suitable corpora to generate their models. We used three textual corpora generated from three different queries on PubMed (provided as special queries: http://www.nlm.nih.gov/bsd/special_queries.html): Dental Journals (PubMed query: “(jsub-setd[text])”), Nursing Journals (PubMed query: “(jsubsetn[text])”) and Systematic Reviews. We filtered out articles that do not have publicly available abstracts. Table 1 shows the number of documents in each corpus. We are also interested in evaluating additional corpora instead of only those generated from PubMed. HSA uses all of the corpora matched with LSA as features to train the hybrid model. When using LSA independently for evaluation, without HSA, only one corpus is used for each run.
For the semantic similarity match step, we evaluate the following different settings:
Most similar concept returned by LSA using each of the corpora listed in Table 1.
Most similar concept returned by HSA
Evaluation criteria
For strict evaluation, we consider a prediction correct when the predicted concept is exactly the same as the expected concept. In contrast, in the relaxed evaluation mode, before calculating the evaluation metrics, we change the predicted class to the expected class if the predicted class has any of these relationships in the UMLS: “synonym”, “is-a”, “mapped-to” relations with the expected class. This means that if the system predicts a concept which is, for example, the synonym, child or parent of the expected concept, we consider it as a true positive. Considering the size of UMLS graph, we only do this normalization by distance of 2—meaning that if a concept “A” has an ‘is-a’ relation with a concept “B”, and the concept “B” has a “mapped-to” relation with a concept “C”, the concept “A” and “C” would be considered the same for the evaluation purpose. The following list shows some other examples of match in the relaxed evaluation:
A -(is-a)→ B -(is-a) → C: A will match with C
A -(is-a)→ B -(mapped-to)→ C: A will match with C
A -(mapped-to)→ B - (mapped-to)→ C: A will match with C
A -(mapped-to)→ B - (synonym)→ C: A will match with C
Results
From the perspective of evaluation, each UMLS concept is considered to be a class. We compute the final precision, recall, and F-measure as the micro-average of all the classes. For each class true positive (TP), false positive (FP), true negative (TN) and false negative (FN) are defined as below: TP is when the expected class is equal to the predicted class and the evaluated class. FP is when the predicted class is equal to the evaluated class but not equal to the expected class. FN is when the expected class is equal to the evaluated class but the predicted class is not equal to the expectedclass. TN is when both predicted and expected classes are not equal to the evaluated class. The following table illustrates an example for the evaluation strategy. The micro-averaged precision and recall are calculated using the following formula:
Precision = (∑c∊Classes TPc)/(∑c∊Classes (TPc + FPc))
Recall = (∑c∊Classes TPc)/(∑c∊Classes (TPc + FNc))
F—measure is the harmonic average of the micro-averaged precision and recall:
Table 3 shows the results for syntactic matcher, LSA using different corpora and the proposed hybrid model. HSA yielded the best F-measure of 62.37 and the best recall of 50.20. The next best precision after syntactic match is achieved by LSA with UMLS definitions corpus. Among LSA with various corpus, ADR-Tweets resulted in the best F- measure. In the investigated normalization problem, the ADR-Tweets corpus yielded the best performance for LSA method. Syntactic matcher has the highest precision, which was expected. Adding LSA-ADR-Tweets matcher on top of syntactical matcher decreases the precision but increases the recall resulting in a higher F-measure. Using HSA instead of LSA decreases the precision slightly more than LSA but the gain on recall is higher and results in a higher F- measure. MetaMap, which is designed for public medical literature data, suffers from very poor recall and therefore overall F-measure. We used the relaxed evaluation method in all of the reported results.
Table 3.
Results obtained by our system using the proposed pipeline and the relaxed evaluation technique.
| Precision | Recall | F-Measure | |
|---|---|---|---|
| Syntactic | 88.0 | 35.7 | 50.8 |
| LSA-PubM-Nursing | 83.1 | 38.6 | 52.7 |
| LSA-UMLS-Defs | 86.5 | 40.3 | 55.0 |
| LSA-Reuters | 81.5 | 44.9 | 57.9 |
| LSA-PubM-Systematic | 83.6 | 44.4 | 58.0 |
| LSA-ADR-Tweets | 84.6 | 47.7 | 61.0 |
| MetaMap | 82.6 | 18.7 | 30.5 |
| HSA | 82.3 | 50.2 | 62.4 |
Discussion
Figure 3 shows the sources of false positives and true positives. Semantic match generates most of false positives followed by exact match. Exact match returns majority of true positives followed by semantic match. As expected, when only the syntactic matching module is employed, we obtain high precision but very low recall. Searching for exact match in definition helps to find alternative representation of the concept. For example, “urge to vomit” is normalized correctly to “c0027497-Nausea” when we search the definition of “c0027497”: “unpleasant sensation in the stomach usually accompanied by the urge to vomit”. Exact match fails when the words in a phrase are expressed in complex orders and another concept matches exactly with the annotated phrase. For example, in the following tweet: “dreams have taken a terrifying turn.”, “dreams” is annotated as “c0857051-bad dreams” but exact match matches the phrase with “c0028084-dreams”.
Figure 3.
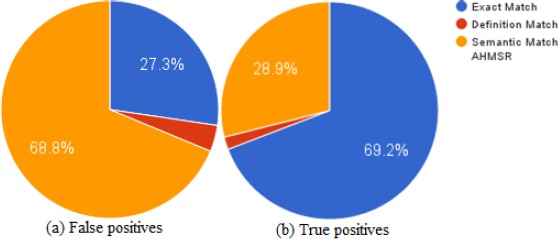
Sources of correct and incorrect predictions. Left chart shows percentage of false positives from each component (a) and the right chart shows true positive percentages (b).
In contrast to syntactic matching, semantic methods are designed to compute estimates of similarity, and match concepts that are not necessarily the same, but are similar. As such, they are expected to have high recall. In our experiments, the se-mantic matchers LSA and HSA have the higher numbers of false positives but yield higher recalls than syntactic match. This was expected since most of the hard to normalize concepts reach the semantic matchers modules. Most of the errors are caused by concepts with very similar meanings. For example, “antidepressant” in a tweet is tagged as “c0011570-mental depression,” but LSA returns “c0005586-manic depression” as the most similar concept.
Table 4 shows examples of correct and incorrect predictions by HSA. The hybrid model is very good at normalizing when the same word is represented in a different variation (“antidepressant” vs. “depression”) or match similar words which appear frequently in corpora (“fewer” vs. “loss”, “increase” vs. “gain”). In contrast, HSA performance is limited to the information in the provided resources and MSR technique. Since in this experiment we only used LSA, HSA would perform solely based on co-occurrences of terms in the resources. If there are not enough numbers of co-occurrences of two terms in the provided resources, we expect to have a very low similarity of the terms. In addition to using larger corpora, adding more diverse techniques that can leverage other resource types (such as graph-based techniques) can significantly boost this limit.
Table 4.
Examples of correct and incorrect predictions by HSA. The first four rows are correct predictions followed by three rows of incorrect predictions. The last row is correct based on the relaxed evaluation criteria.
| Annotated Phrase | Expected | Predicted |
|---|---|---|
| Antidepressant | c0011570-Depression | c0011570 |
| increase my weight | c0043094-Weight gain | c0043094 |
| gain so much weight | c0043094-Weight gain | c0043094 |
| fewer hours sleep | c0235161-Sleep loss | c0235161 |
| feel like need to throw up | c0027497-Nausea | c0917799-Hypersomnia |
| just eat, and eat | c0232461-Apetite increase | c0015672-Fatigue |
| falling asleep every day | c0541854-Daytime sleepiness | c0917801-Insomnia |
| it’s 4:30am. at this point ima just throw out a big “f*** you” | c0917801-Insomnia | c0917799-Hypersomnia |
Conclusion
In this work, we proposed a natural language processing pipeline for the problem of normalizing extracted mentions of ADRs from colloquial texts to UMLS concepts. We compared two semantic similarity techniques: LSA and a proposed hybrid approach (HSA). The hybrid approach shows improvement over a single similarity technique (LSA). The proposed hybrid approach is supervised and benefits from training data while LSA is unsupervised and does not have any training. Tweets, like other informal texts, required heavy pre-processing and cleaning. The errors of the sys-tem could be reduced by applying more advanced pre-processing like spelling correction. This is the first effort towards the ADR normalization from social media or other noisy text sources, and can provide a baseline for future work.
In the future, we will utilize our tool to perform normalization on a larger set of mentions. We will also incorporate more complex NLP preprocessing techniques, such as negation detection, and perform comparisons of our approach with a larger number of semantic similarity measurement approaches. In the recent past, approaches using distributed representations of words have become very popular, and the use of such representations along with deep neural networks have outperformed past benchmark systems in a variety of natural language processing tasks. Therefore, we will attempt to utilize annotated data to develop and evaluate such neural network based systems against ours. Because of the modular implementation of our system, if such methods provide promising results, we will incorporate them as a module in our concept normalization pipeline.
Table 2.
Illustration of the evaluation technique.
| Mention | Expected Class | Predicted Class | Evaluated class | |
|---|---|---|---|---|
| Class 1 | Class 2 | |||
| M1 | Class1 | Class1 | TP | TN |
| M2 | Class1 | Class2 | FN | FP |
| M3 | Class2 | Class1 | FP | FN |
| M4 | Class2 | Class2 | TN | TP |
References
- 1.Lindquist M. The need for definitions in pharmacovigilance. Drug Saf. 2007;30(10):825–830. doi: 10.2165/00002018-200730100-00001. [DOI] [PubMed] [Google Scholar]
- 2.Harpaz R, DuMouchel W, Shah NH, Madigan D, Ryan P, Friedman C. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Ther. 2012;91(6):1010–1021. doi: 10.1038/clpt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarker A, Ginn R, Nikfarjam A, et al. Utilizing social media data for pharmacovigilance: A review. J Biomed Inform. 2015;54:202–212. doi: 10.1016/j.jbi.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Struik LL, Baskerville NB. The Role of Facebook in Crush the Crave, a Mobile- and Social Media-Based Smoking Cessation Intervention: Qualitative Framework Analysis of Posts. J Med Internet Res. 2014;16(7):e170. doi: 10.2196/jmir.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson CL, Cannon B, Burton S, Giraud-Carrier C. An exploration of social circles and prescription drug abuse through Twitter. J Med Internet Res. 2013;15(9):e189. doi: 10.2196/jmir.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakhasi A, Passarella RJ, Bell SG, Paul MJ, Dredze M, Pronovost PJ. Malpractice and Malcontent: Analyzing Medical Complaints in Twitter. AAAI Tech report, Information Retr Knowl Discov Biomed text,. 2010:1–2. [Google Scholar]
- 7.Lardon J, Abdellaoui R, Bellet F, et al. Adverse drug reaction identification and extraction in social media: A scoping review. J Med Internet Res. 2015;17(7) doi: 10.2196/jmir.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikfarjam A, Sarker A, O’Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Informatics Assoc. 2015;22(3):671–681. doi: 10.1093/jamia/ocu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimeno A, Jimenez-Ruiz E, Lee V, Gaudan S, Berlanga R, Rebholz-Schuhmann D. Assessment of disease named entity recognition on a corpus of annotated sentences. BMC Bioinformatics. 2008;9(Suppl 3):S3. doi: 10.1186/1471-2105-9-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leaman R, Miller C. Enabling Recognition of Diseases in Biomedical Text with Machine Learning: Corpus and Benchmark. In: Proceedings of the 3rd International Symposium on Languages in Biology and Medicine (LBM). 2009:82–89. [Google Scholar]
- 11.Kang N, Singh B, Afzal Z, van Mulligen EM, Kors JA. Using rule-based natural language processing to improve disease normalization in biomedical text. J Am Med Inform Assoc. 2013;20(5):876–881. doi: 10.1136/amiajnl-2012-001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of BioCreAtlvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6(Suppl 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J-D, Ohta T, Pyysalo S, Kano Y, Tsujii J. In: Proceedings of the Workshop on BioNLP Shared Task - BioNLP’09. Morristown, NJ, USA: Association for Computational Linguistics; 2009. Overview of BioNLP’09 shared task on event extraction; p. 1. [DOI] [Google Scholar]
- 14.Clarke CLA, Craswell N, Voorhees EM. Overview of the TREC 2012 Web Track. In: TREC. 2012:1–8. [Google Scholar]
- 15.Uzuner O, South BR, Shen S, Duvall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011 Jun;:552–557. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leaman R, Islamaj Dogan R, Lu Z. DNormml: disease name normalization with pairwise learning to rank. Bioinformatics. 2013;29(22):2909–2917. doi: 10.1093/bioinformatics/btt474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashyam V, Divita G, Bennett DB, Browne AC, Taira RK. A normalized lexical lookup approach to identifying UMLS concepts in free text. Stud Health Technol Inform. 2007;129(Pt 1):545–549. [PubMed] [Google Scholar]
- 18.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001 Jan;:17–21. doi:D010001275 [pii] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang M, Névéol A, Lu Z. Recommending MeSH terms for annotating biomedical articles. J Am Med Inform Assoc. 18(5):660–667. doi: 10.1136/amiajnl-2010-000055. doi:10.1136/amiajnl-2010-000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang M, Liu J, Zhu X. GeneTUKit: a software for document-level gene normalization. Bioinformatics. 2011;27(7):1032–1033. doi: 10.1093/bioinformatics/btr042. doi: 10.1093/bioinformatics/btr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan R, Leaman R, Gonzalez G. In: 2011 10th International Conference on Machine Learning and Applications and Workshops. Vol. 2. IEEE; 2011. The DIEGO Lab Graph Based Gene Normalization System; pp. 78–83. doi: 10.1109/ICMLA.2011.140. [Google Scholar]
- 22.Buyko E, Tomanek K, Hahn U. Resolution of Coordination Ellipses in Complex Biological Named Entity Mentions Using Conditional Random Fields. ISMB BioLink SIG. 2007:163–171. [Google Scholar]
- 23.Tsuruoka Y, McNaught J, Tsujii J, Ananiadou S. Learning string similarity measures for gene/protein name dictionary look-up using logistic regression. Bioinformatics. 2007;23(20):2768–2774. doi: 10.1093/bioinformatics/btm393. doi:10.1093/bioinformatics/btm393. [DOI] [PubMed] [Google Scholar]
- 24.Wermter J, Tomanek K, Hahn U. High-performance gene name normalization with GENO. Bioinformatics. 2009;25(6):815–821. doi: 10.1093/bioinformatics/btp071. doi:10.1093/bioinformatics/btp071. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee S, Pedersen T. An adapted Lesk algorithm for word sense disambiguation using WordNet. Comput LinguistIntell text&. 2002 Feb;:136–145. doi:10.1007/3-540-45715-1_11. [Google Scholar]
- 26.Patwardhan S, Banerjee S, Pedersen T. In: Proceedings of the Fourth International Conference on Intelligent Text Processing and Computational Linguistics. Vol. 4. Springer-Verlag; 2003. Using Measures of Semantic Relatedness for Word Sense Disambiguation; pp. 241–257. doi:10.1007/3-540-36456-0_24. [Google Scholar]
- 27.Han B, Cook P, Baldwin T. Lexical normalization for social media text. ACM Trans Intell Syst Technol. 2013;4(1):1–27. doi:10.1145/2414425.2414430. [Google Scholar]
- 28.Choudhury M, Saraf R, Jain V, Mukherjee A, Sarkar S, Basu A. Investigation and modeling of the structure of texting language. Int J Doc Anal Recognit. 2007;10(3-4):157–174. doi:10.1007/s10032-007-0054-0. [Google Scholar]
- 29.Cook P, Stevenson S. In: Proceedings of the Workshop on Computational Approaches to Linguistic Creativity. Association for Computational Linguistics; 2009. An Unsupervised Model for Text Message Normalization; pp. 71–78. 10.3115/1642011.1642021. [Google Scholar]
- 30.Xue Z, Yin D, Davison B. Normalizing Microtext. Anal Microtext. 2011 Sep;:74–79. [Google Scholar]
- 31.Liu F, Weng F, Jiang X. A Broad-Coverage Normalization System for Social Media Language. Proc 50th Annu Meet Assoc Comput Linguist Vol 1 Long Pap. 2012 Jul;:1035–1044. [Google Scholar]
- 32.Deerwester S, Dumais ST, Furnas GW, Landauer TK, Harshman R. Indexing by latent semantic analysis. J Am Soc Inf Sci. 1990;41(6):391–407. [Google Scholar]
- 33.Sellberg L, Jönsson A. Using Random Indexing to improve Singular Value Decomposition for Latent Semantic Analysis. LREC. 2008:2335–2338. [Google Scholar]
- 34.Wild F, Stahl C, Stermsek G, Neumann G. Parameters Driving Effec-Tiveness of Automated Essay Scoring. Inf Syst J. 2005;80(18):485–494. [Google Scholar]
- 35.Hofmann T. Probabilistic latent semantic analysis. Proc Uncertain Artif Intell. 1999 [Google Scholar]
- 36.Wermter J, Tomanek K, Hahn U. High-performance gene name normalization with GeNo. Bioinformatics. 2009;25(6):815–821. doi: 10.1093/bioinformatics/btp071. doi:10.1093/bioinformatics/btp071. [DOI] [PubMed] [Google Scholar]


