Abstract
The wide availability of electronic health record (EHR) data for multi-institutional clinical research relies on accurately defined patient cohorts to ensure validity, especially when used in conjunction with open-access research data. There is a growing need to utilize a consensus-driven approach to assess data quality. To achieve this goal, we modified an existing data quality assessment (DQA) framework by re-operationalizing dimensions of quality for a clinical domain of interest - heart failure. We then created an inventory of common phenotype data elements (CPDEs) derived from open-access datasets and evaluated it against the modified DQA framework. We measured our inventory of CPDEs for Conformance, Completeness, and Plausibility. DQA scores were high on Completeness, Value Conformance, and Atemporal and Temporal Plausibility. Our work exhibits a generalizable approach to DQA for clinical research. Future work will 1) map datasets to standard terminologies and 2) create a quantitative DQA tool for research datasets.
Introduction
In recent years, with increasing availability of electronic health record (EHR) data for research, studies frequently integrate EHR data with insurance billing and claims data to study health outcomes and cost-effectiveness.1–3 Large-scale multi-institutional studies, such as pragmatic clinical trials and comparative effectiveness research, rely on accurately defined patient cohorts to ensure that findings are valid. As more and more clinical data derived from EHRs, claims, and other sources are being collected and stored in publicly accessible repositories, such as the Database of Genotypes and Phenotypes (dbGaP)4 and the Biologic Specimen and Data Repository Information Coordinating Center (bioLINCC),5 there is an overwhelming need to harmonize these datasets and assess their quality.6,7
Several studies have looked at quality issues in clinical research data, such as the lack of data standardization, missing or incomplete clinical data, incompatible representations of data types and elements, and identified three primary challenges to evaluating the quality of a research dataset.8–11 First, data quality assessment (DQA) is often subjective and is dependent on the evaluative task or objective, which is particularly problematic because clinical research datasets are often phenotype-specific, requiring a unique patient cohort, or may cater to a specific set of participating medical centers. Moreover, data within the EHRs may be sufficient for clinical purposes, but not for research, which are typically more objective-driven. For example, the clinical concept “History of Cerebrovascular Accident” can be found in the EHR, and would provide actionable information for patient care. A particular research dataset, however, might require the presence of “History of Cerebrovascular Accident Within 3 years of Encounter.” In other words, data quality is context-dependent.12 Second, although there are certain assurance checks that researchers can conduct to ensure data quality, this process is frequently time-consuming and cumbersome, and the results of these assessments may not be meaningful without a thorough understanding of the researcher’s intended goal. Evaluating data missingness, distributions, and accepted values do not entirely paint a full picture of the quality of the dataset as described by the research objective, and manually evaluating quality in this fashion is a resource-intensive task. Finally, there are no consistent evidence-based or community-driven metrics for assessing the quality of research data. Study investigators frequently develop ad-hoc metrics that are specific to the study, and cannot be replicated.13,14
Objective
It is crucial to quantitatively analyze the quality of a dataset to ensure reproducibility in research studies. To encourage implementation of the recommended concepts of quality assessment, rules and conditions are required to formulate an assessment framework that is related to a task or specific phenotype of interest. This is a more challenging task when evaluating quality in research datasets as they are often task- or domain-specific.
The present study sought to promote the development and utilization of large, multi-institutional research datasets based on existing data sources.4,5 Specifically, we inventoried and assessed the quality of common phenotypic data elements (CDPEs) for heart failure research, enabling a reusable framework development process for other researchers looking to use or contribute to this composite dataset. Our first aim is to modify an existing DQA framework by re-operationalizing the definitions of several data quality dimensions. The framework will be dependent on a particular research goal, which in this case are studies identifying novel biomarkers for heart failure diagnosis, prognosis and treatment. The second aim is to create an inventory of CPDEs derived from the research datasets. In this study, we limit our scope to heart failure biomarker studies in the following open-access databases: dbGaP and BioLINCC. The third and final aim is to evaluate the data element inventory using the modified DQA framework. The task-oriented approach will evaluate, based on the necessary data elements for a study design or goal, the Completeness, Conformance, and Plausibility of the CPDE inventory.
Materials and Methods
Prior Work in Quality Assessment Frameworks
Data quality frameworks and harmonized assessment terminologies have been created to evaluate EHR data. Prior research in DQA defined broad dimensions of data quality.15,16 However, in a growing field of data quality research, inconsistent definitions make it difficult to uniformly compare results across data sharing partners and institutions. Efforts to harmonize these concepts are necessary to promote interoperability in the field of data quality research. A harmonized and revised DQA terminology framework was developed to encompass quality concepts that have been defined by other researchers.12 The proposed harmonized framework takes the categories of Conformance, Completeness, and Plausibility and expands on each.
Conformance is defined by whether data values adhered to pre-specified standards or formats. Conformance was separated into three distinct sub-categories: Value Conformance (whether recorded data elements agree with constraint-driven data architectures, such as data models or rules defined in a data dictionary), Relational Conformance (determines if data elements agree with structural constraints of the physical database that stores these values, hinging on the importance of primary key and foreign key interactions within relational databases), and Computational Conformance (focuses on the correctness of the output value of calculations that were made from existing variables, either within the dataset or between datasets). Although Value Conformance can be ascertained at both the data element and data value levels, Computational Conformance can only be properly assessed at the value level. The scope of this study focused only on single datasets, typically in flat files, such as Excel or CSV formats. As a result, Relational Conformance is not applicable at the data element level for this study.
Completeness evaluates data attribute frequency within a dataset without reference to the data values. It does not consider its structure or its plausibility, but instead looks at the absence of data at a specific point in time agreeing with a trusted standard, common expectation, or existing knowledge. This dimension is applicable to both higher-level data element and more granular data value levels of assessment.
Plausibility is defined by whether or not the values of data points are believable when compared to the expected representation of an accepted value range or distribution. Plausibility was separated into Uniqueness Plausibility (values that identify a particular object—person, institution, etc.—are not duplicated), Atemporal Plausibility (data values adhere to common knowledge or are verified by an external source), and Temporal Plausibility (whether time-varying variables also have changing values, based on gold standards or existing knowledge). All sub-categories within Plausibility are applicable to both data element and data value levels.
DQA Framework Modification
The study team used a consensus-driven approach to finalize the framework creation to redefine the harmonized DQA terminology from Kahn et al.12 to the specific research task. In particular, the concepts of Conformance, Completeness, and Plausibility were operationalized to be specific to heart failure biomarker research. Table 1 includes definitions of the harmonized data quality assessment terms and examples.
Table 1.
Harmonized Terminology with Examples Drawn from Heart Failure Research Studies.
| Concept | Applicable Level | Definition | Example |
|---|---|---|---|
| Value conformance | Data Element Data Value | Whether recorded data elements agree with constraint-driven data architectures | The description for the data element, “Body Mass Index (BMI)” is defined by units of kg/m2. |
| Relational conformance | Data Element* Data Value | Whether data elements agree with structural constraints of the physical database that stores these values | The data element for “History of Cerebrovascular Incident” is represented by categorical values, Yes or No. The values are represented as such, and are not in any other form. |
| Computational conformance | Data Value | Whether the correctness of the output value of calculations that were made from existing variables, either within the dataset or between datasets | Calculating patient body weight and height would produce the same value as the value represented in the BMI data element. |
| Completeness | Data Element Data Value | Whether data values or elements are present | The research data elements in the inventory are complete when compared to the aggregated list from literature. |
| Uniqueness plausibility | Data Element Data Value | Whether values that identify a particular object--person, institution, etc.--are not duplicated | Each data element is not duplicated or represented by another data element within the inventory. |
| Atemporal plausibility | Data Element Data Value | Whether or not data values adhere to common knowledge or are verified by an external source | Chronic Kidney Disease (CKD) stage 2 criteria of GFR <60 mL/min/1.73 m2 for >=3 months is in concordance with existing knowledge and guidelines for of CKD diagnosis. |
| Temporal plausibility | Data Element* Data Value | Whether time-varying variables also have changing values, based on gold standards or existing knowledge | Follow-up dates are sequentially collected after the study enrollment date. |
* Rows marked with an asterisk indicate exceptions to the dimensional applicability at that particular data level. Relational Conformance requires a SQL database or relational database structure. While Relational Conformance can be applied at the data element level, it requires a specific structure of tables beyond the scope of the study. Similarly, Temporal Plausibility requires time-varying data elements, which were not included within the ‘Demographics’ and ‘Medications’ categories in the current CPDE inventory.
In our modified framework, Value Conformance measures that assessed the adherence of data values to internal constraints necessitated the evaluation of acceptable ranges for data elements like patient sex, blood pressure, or cholesterol levels. Completeness compared the data element inventory to the aggregated data elements within literature.17–19 These studies compiled cardiovascular EHR data elements, and in particular, key heart failure data elements, that have research utility and maximum clinical impact. Plausibility required the evaluation of data values specific to cardiovascular disease, and understanding that patients with a heart failure diagnosis should have statistically similar distributions for certain data elements compared to patients without a heart failure diagnosis.
Conformance for research data is still defined by whether data values adhered to pre-specified standards or formats. We also separate conformance into three sub-categories. Value conformance in research data would be determining whether elements are represented according to some standard medical terminology or appropriate clinical nomenclature. External standards can be applied as well, such as units of measurement or ranges of accepted values. Relational conformance determines if data elements agree with structural constraints of the physical database that stores these values. Relational conformance deals with how a data model represents reality via metadata descriptions or database rules. Computational conformance within research datasets include validation checks that are consistent with EHR validation checks, such as whether body mass index (BMI) calculations for data elements like, “Patient Body Weight” and “Patient Height” should yield the same values for the data element, “Body Mass Index”.
Completeness evaluates the presence of data elements within a particular dataset. It does not reference data values and it compares the dataset to an existing standard knowledge or common expectation. Because of the subjective nature of compiling a “complete” research dataset for a given disease domain, we compared the data element inventory against cardiovascular data elements aggregated through literature guidelines.17-19 Although these were EHR data elements, they were compiled to create a list of attributes that had research utility in clinical settings. This was in line with our focus for heart failure biomarker research studies.
Plausibility is whether or not the values of data points are believable when compared to the expected representation of an accepted value range or distribution. Uniqueness plausibility ensures that values are not duplicated or represented by another entity within the dataset. At the research data element level, we ensure that data elements are not duplicated or represented by another data element or attribute. At the data value level, we ensure that each data point is not duplicated within another data element. Atemporal plausibility determines whether or not data values adhere to common knowledge or are verified by an external source. This extends to research data elements with metadata descriptions that align with existing knowledge. For example, a data element representing “Chronic Kidney Disease Stage 2” should have metadata descriptions consistent with Chronic Kidney Disease stage 2 criteria, such as appropriate laboratory values for glomerular filtration rate (GFR) and blood urea nitrogen (BUN). Values should be consistent with external standards of acceptable ranges or distributions of values. Temporal plausibility is whether time-varying variables also have changing values or follow sequentially, based on gold standards or existing knowledge. For example, do values vary over time as expected, such is the case for spikes in flu diagnosis in emergency room or outpatient visits during flu season? Similarly, recruitment dates into the research study should not come before patients’ dates of birth. A follow-up date should not precede the recruitment date for the study. These values follow for other variables that require follow-up dates within a study.
Common Phenotype Data Element Inventory Creation
We conducted a retrospective review of open-access cardiovascular disease datasets in order to identify CPDEs related to heart failure. We focused our literature search on studies identifying biomarkers and risk factors for heart failure diagnosis, prognosis and treatment. Several cardiovascular research studies, particularly focusing on heart failure, from dbGaP and BioLINCC were aggregated to compile an inventory (work was led by co-authors KL and JP) of commonly used phenotype data elements. Our inclusion criteria for studies were: 1) focus on heart failure or congestive heart failure biomarker research; 2) use of clinical data in addition to genotype or sequencing data; and 3) inclusion of demographics, diagnostic test, patient history, physical examination, and medications variables. Authorized access was obtained for these research datasets. Data elements for the inventory were selected based on their relevancy to heart failure biomarker research, research utility in clinical settings, and their presence in EHR systems. Figure 1 illustrates the process for collating CPDEs from dbGaP and BioLINCC.
Figure 1.
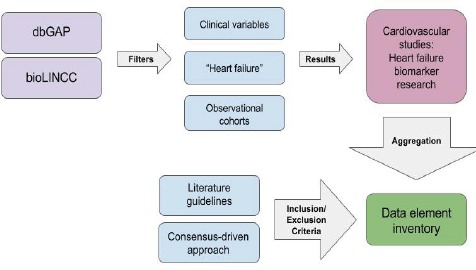
Common Phenotype Data Element (CPDE) Aggregation.
This illustrates the workflow in creating the data element inventory. The first step was to search through open-access databases for research datasets that focused on biomarker discovery for heart failure diagnosis, prognosis, and treatment. Additionally, we narrowed our search to observational cohort studies and projects that included clinical patient data. There were many research datasets with purely genotyping or sequencing data elements, which we excluded for this study. The second step was to use literature guidelines to prioritize a baseline set of standard cardiovascular data elements along with the commonly occurring elements found in our aggregated studies. The final step yielded our
The development of our data element inventory consisted of a two-pronged approach. First, we aggregated CPDEs that we found in heart failure biomarker research studies in dbGaP and BioLINCC. In aggregating appropriate CPDEs from the studies, we focused on those that had greater generalizability for heart failure studies. Elements like indices of social support networks, for example, were not included in our inventory, as they were not deemed to be generalizable to most heart failure research studies that utilized EHR data. We followed guidelines from literature that enumerated relevant data elements present in EHR databases that could be repurposed for clinical research. Based on these guidelines, social elements were excluded from the inventory. Second, following similar data element guidelines from literature, clinical variables with high research utility, such as diagnostic tests and medical history, were prioritized over those that are more attuned to specific research designs, such as insurance, government aid sources, and billing zip codes. Of particular usefulness in our aggregation of data elements were a report from the American College of Cardiology Foundation and the American Heart Association Task Force on Clinical Data Standards17, which focused on harmonizing existing data standards with newly published ones, and to establish terms that are available in every general purpose EHR, and are extendable and reusable in clinical research, a report from the Data Standards Workgroup of the National Cardiovascular Research Infrastructure Project, which attempted to create or identify and harmonize clinical definitions for a general set of cardiovascular data elements18, and a report from the American College of Cardiology and American Heart Association Task Force on Clinical Data Standards reviewed key data elements for heart failure management in clinical research.19
Results
Finalized Data Element Inventory for Heart Failure Research
We created an inventory of 100 data elements from the following studies: the Cardiovascular Health Study (CHS), the Jackson Heart Study, the Framingham Heart Study, the Heart Failure Network (HFN) CARdiorenal Rescue Study in Acute Decompensated Failure (CARRESS), and the Sudden Cardiac Death in Heart Failure Trial (SCDHeFT).20–24 Data elements were selected based on comprehensive coverage of cardiovascular conditions, test results, and clinical presentations that would best reflect the breadth and variety of commonly occurring heart failure biomarker research data elements in clinical research. The final data element inventory is available as an online supplement (https://goo.gl/GNk4Zn). Our review of relevant guidelines led to the identification of six categories of frequently utilized and clinically meaningful phenotypic data elements:
Demographics: All research datasets have at minimum a demographics category for patient records. Commonly occurring data elements within this category included the Date of Birth, Sex, Race, and Ethnicity. Four demographic data elements were included in the inventory.
Physical Examination or Baseline Observation: This category can often be fairly detailed depending on the research study. Data elements related to systemic observations of the body and functions include measurements of height, weight, body mass index, blood pressure, and heart rate. We included ten data elements in this category.
Diagnostic Tests: Diagnostic and therapeutic procedures, as well as laboratory tests are included in this category. Units of measurement in their metadata description often accompany these variables. Data elements resulting from electrocardiograms, bypass graft surgeries, echocardiograms, and stress tests are included, as well as laboratory values like cholesterol, glucose, creatinine, and blood urea nitrogen measurements. Twenty-six data elements were included in the inventory.
Patient Medical History: Typical data elements in this category include prior history of disease or diagnoses, prior surgeries or hospitalizations, history of tobacco use, drug use, and alcohol use, and family history of disease. These data elements are often categorical, such as the data element, “Type of stroke”, which takes on values, “Hemorrhagic”, “Nonhemorrhagic”, and “Unknown.” Twenty-seven medical history items were included in the data element inventory.
Clinical Diagnoses or Presentation: This category is limited to the patient’s current status. In research datasets for heart failure biomarker research, patients are often diagnosed with heart failure, or another cardiovascular disease to be included in the study cohort. Inclusion of clinical presentations was considered for this section as EHR systems record current statuses of patients differently. Data elements, such as Myocardial Infarction, Chest Pain (Angina), Heart Failure, Syncope, and the like, were included. Eight patient assessment data elements were included in the inventory.
Medications: Types of medication, such as beta blockers, ACE inhibitors, and statins were included, as well as data elements describing usage, such as Medications Held or Discontinued, Contraindications, and timepoints of usage were added to the inventory. The inventory contained twenty-five medications-related data elements.
Assessment of Data Element Inventory Against Modified DQA Framework
Having redefined the DQA framework and creating an inventory of accepted data elements for comparison, we were able to evaluate the quality of our collected data element inventory. Table 2 displays the results of the inventory against the framework. The inventory was assessed at the data element variable level. For some DQA concepts that are more suitable for assessing variable values, we evaluated the appropriate variables based on their ability to capture the significant aspect. For instance, to evaluate particular data elements on Temporal Plausibility at the data element variable level, we assessed their ability to capture temporal aspects, such as continuity of data collection and whether or not they were defined by a numeric date, where appropriate.
Table 2.
Data Element Inventory Framework Assessment by Category
| Number of data elements from the inventory adhering to concept criteria | ||||||
|---|---|---|---|---|---|---|
| Data Element Category | N | Value Conformance | Completeness | Uniqueness plausibility | Atemporal plausibility | Temporal plausibility |
| Demographics | 4 | 4 (100%) | 4 (100%) | 4 (100%) | 4 (100%) | N/A |
| Physical Exam | 10 | 10 (100%) | 10 (100%) | 10 (100%) | 10 (100%) | 10 (100%) |
| Diagnostic Test | 26 | 26 (100%) | 23 (88.4%) | 26 (100%) | 26 (100%) | 23 (88.4%) |
| Medical History | 27 | 27 (100%) | 27 (100%) | 23 (85.2%) | 27 (100%) | 27 (100%) |
| Patient Assessment | 8 | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%) | 8 (100%0 |
| Medications | 25 | 25 (100%) | 25 (100%) | 25 (100%) | 25 (100%) | N/A |
Discussion
This work is not an exhaustive list of modifications to a DQA framework for research datasets. Although there are many ways that research data and EHR data coincide in terms of data quality, the applications for verification and validation can be quite different. While we understand that the data element inventory was created to obtain a “model” for accepted variable values that work within our modified DQA framework, we acknowledge the potential limitations for its application as a comparison to other research datasets that may have a broader or narrower focus even within the scope of heart failure biomarker research studies. Our goal was to adapt a harmonized DQA framework to a clinical domain, such as heart failure, and inevitably to compile a working DQA framework that can be reusable in clinical research.
Although harmonized frameworks are typically evaluated at the data value level, we attempted to apply it at a broader element level for this study. All data elements met the criteria for Value Conformance by being thoroughly represented by appropriate units of measurement in their metadata descriptions. However, mapping data elements to a standard terminology, such as the Systematized Nomenclature of Medicine (SNOMED)25,26 would further ensure that data elements adhered to the best practices for research data management. There remains a subjective aspect to our approach. 88.4% of the Diagnostic Test data elements met the criteria for Completeness. Including dates for certain diagnostic procedures that may vary over time might have led to a more complete list. For example, including the date of “Radionuclide Ventriculography Findings” typically helps investigators track a patient’s disease progression over time if more tests are conducted at different time points.
The data elements in the inventory are considered a complete collection of appropriate variables to be included in a research dataset for heart failure biomarker research as it includes elements that encompass demographics, physical examination, tests, patient medical history, and medications. Collating the data elements for the inventory involved consulting guidelines set in literature for high research utility clinical variables in heart failure studies, as well as evaluating commonly occurring data elements that are present in open-access clinical research datasets. Some studies included other lifestyle factors, such as sources of social support, eating habits, or scales for depression and anxiety. We considered these data elements to be not as generalizable in heart failure studies according to literature guidelines previously set for EHR data elements. These guidelines included only common elements that had high clinical research utility, and as a result, the aforementioned social indices were not included in our data element inventory. Uniqueness Plausibility ensures that elements are not duplicated and values are not dually represented within a dataset. 85.2% of Medical History data elements adhered to the concept of Uniqueness Plausibility. Certain data elements, such as Family History of Coronary Arteriosclerosis, Family History of Cardiomyopathy, and Family History of Sudden Cardiac Death shared some overlapping definitions and descriptive criteria.
Because our data element inventory is presented in a single table, and there are no relationships between the data elements themselves (for example, as one would observe in an Entity-Relationship model), it did not qualify to be assessed for Relational Conformance. There are no additional tables in the data element inventory, although some research datasets may have separate tables for categories of measurements. Additionally, we are unable to evaluate Computational Conformance as we are only evaluating at the data element variable level, which provides no output variable values for us to calculate on. Temporal Plausibility was incalculable for Demographics and Medications categories as the elements were not expected to vary over time. Should there have been prescription dates for Medications, perhaps Temporal Plausibility might have been more applicable. In our inventory, however, Medications were only listed as drug classes.
Limitations
The first limitation is that there are certain data quality concepts that cannot fully apply to research data. Relational Conformance, for example, deals with the ability to navigate between different tables, which may not be necessary if the table in question is a research study data dictionary. These are structured differently and are typically not stored in the same manner as EHR systems tables.
Second, to properly assess Value Conformance, it is often necessary that data elements in research datasets are mapped to standardized biomedical terminologies. An underlying data quality issue in research data is the inability to be readily integrated or linked to relevant datasets due to a lack of standardization. This can be addressed by mapping data elements to terminologies or models to further ensure that the data elements are represented appropriately. In addition, string variables present a familiar challenge for researchers working with ontologies as mapping these terms can be difficult when compiling EHR data and when transforming data from one schema to another. We aimed to include values within a string variable that contain more structured response options. For a variable, such as “Lung (pulmonary) examination”, the values could take the form of free-text responses, which would present a challenge for data mapping. Its values, however, include structured responses, like “Clear or normal, Rales (height of rales when patient sitting upright should be noted), Decreased breath sounds or dullness, Rhonchi, or Wheezing”, which can more easily facilitate mapping to standard terminologies.
Third, because data quality assessment is a subjective task, it is necessary to have an external gold standard for comparison. We used literature guidelines that focused on EHR heart failure data elements with high research utility to create our data element inventory. The objective of this study, after all, is to evaluate data quality within open-access research datasets that focus on biomarker discovery, which are often linked to EHR data. Fourth, our methodology for assessing data quality was limited only to the data element variable level: were the data elements complete and in line with a set of external standards or common knowledge? Focusing only on evaluating data quality at the data element variable level, much like assessing a data dictionary, restricts our ability to look at the variable values and understand its accepted distributions, ranges, and completeness or missingness. Further, we were unable to conduct validation checks that may enable us to assess for computational conformance.
Future Work
Assessing data quality at the higher-level data element phase was a necessary first step in determining the ability of clinical research data to adhere to a harmonized DQA framework. To more comprehensively evaluate the utility of our harmonized framework, future projects will adapt the framework at the data value level as well. We anticipate that including data values will produce more criteria with which to better assess quality, most likely producing less adherence than what our current results exhibit. In addition, we anticipate that the addition of data elements, such as social support and insurance and billing procedures may provide us with a richer set of criteria to broaden our DQA dimensions. Incorporating more granular data values may involve either a more comprehensive data element inventory, or data transformation of several datasets to a common model so that they have a similar baseline for comparison. The latter exercise of data transformation into a common data model can also help alleviate the mapping challenges of string variable responses that can be particularly difficult to standardize. Evaluating the framework against differing granularities can more appropriately showcase its ability to be repurposed continuously in research, independent of clinical domain. Our eventual goal is to create an assessment tool to better quantify data quality using this framework. The first step is to create a stepwise script that can run on a statistical program, such as R or SAS. This would enable researchers to go through their appropriate data elements within their data dictionary and check for missingness and completeness, appropriate ranges, and distributions. They can also check Computational Conformance in this way. The second goal is to create a stepwise tool that can assess the overall dataset, much like the National Institute of Standards and Testing (NIST). NIST created a set of methods to test data compliance to meaningful use standards.27 Future work will also include utilizing the Yale University Open Data Access (YODA) platform to obtain research studies that can be evaluated using our modified DQA framework.28
Acknowledgements
This work was funded in part by NIH R01 GM105688 and R01 GM103859.
References
- 1.Reimer AP, Milinovich A, Madigan EA. Data quality assessment framework to assess electronic medical record data for use in research. Int J Med Inform. 2016;90:40–47. doi: 10.1016/j.ijmedinf.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sox HC, Goodman SN. The methods of comparative effectiveness research. Annu Rev Public Health. 2012;33:425–445. doi: 10.1146/annurev-publhealth-031811-124610. [DOI] [PubMed] [Google Scholar]
- 3.Weiner MG, Embi PJ. Toward reuse of clinical data for research and quality improvement: the end of the beginning? Ann Intern Med. 2009;151(5):359–360. doi: 10.7326/0003-4819-151-5-200909010-00141. [DOI] [PubMed] [Google Scholar]
- 4.NCBI. dbGaP. [Accessed February 3, 2017]. https://www.ncbi.nlm.nih.gov/gap.
- 5.NHLBI. BioLINCC. [Accessed February 3, 2017]. https://biolincc.nhlbi.nih.gov/home/
- 6.Min L, Liu J, Lu X, Duan H, Qiao Q. An Implementation of Clinical Data Repository with openEHR Approach: From Data Modeling to Architecture. 2016:100–105. doi: 10.3233/978-1-61499-666-8-100. [DOI] [PubMed] [Google Scholar]
- 7.Chute CG, Ullman-Cullere M, Wood GM, Lin SM, He M, Pathak J. Some experiences and opportunities for big data in translational research. Genet Med. 2013;15(10):802–809. doi: 10.1038/gim.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Field D, Sansone S. A Special Issue on Data Standards. Omi A J Integr Biol. 2006;10(2):84–93. [Google Scholar]
- 9.Richesson RL, Krischer J. Data Standards in Clinical Research: Gaps, Overlaps, Challenges and Future Directions. J Am Med Informatics Assoc. 2007;14(6):687–696. doi: 10.1197/jamia.M2470.Introduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinov ID. Methodological challenges and analytic opportunities for modeling and interpreting Big Healthcare Data. Gigascience. 2016;5(1):12. doi: 10.1186/s13742-016-0117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mead CN. Data interchange standards in healthcare IT--computable semantic interoperability: now possible but still difficult, do we really need a better mousetrap? J Healthc Inf Manag. 2006;20(1):71–78. [PubMed] [Google Scholar]
- 12.Kahn MG, Callahan TJ, Barnard J, et al. A Harmonized Data Quality Assessment Terminology and Framework for the Secondary Use of Electronic Health Record Data. EGEMS (Washington, DC) 2016;4(1):1244. doi: 10.13063/2327-9214.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: Enabling reuse for clinical research. J Am Med Informatics Assoc. 2013;20(1):144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zozus MN, Hammond WE, Green BB, et al. Assessing Data Quality for Healthcare Systems Data Used in Clinical Research. 2014. pp. 1–26. file:///C:/Users/anobles/Downloads/Assessing-data-quality_V 10 (1).pdf.
- 15.Weiskopf NG, Hripcsak G, Swaminathan S, Weng C. Defining and measuring completeness of electronic health records for secondary use. J Biomed Inform. 2013;46(5):830–836. doi: 10.1016/j.jbi.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013;20(1):144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weintraub WS, Karlsberg RP, Tcheng JE, et al. ACCF/AHA 2011 key data elements and definitions of a base cardiovascular vocabulary for electronic health records: A report of the American College of Cardiology Foundation/American Heart Association Task Force on clinical data standards. J Am Coll Cardiol. 2011;58(2):202–222. doi: 10.1016/j.jacc.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Anderson VH, Weintraub WS, Radfod MJ, et al. Standardized Cardiovascular Data for Clinical Research, Registries, and Patient Care: A Report from the Data Standards Workgroup of the National Cardiovascular Research Infrastructure Project. A collaboration of the Duke Clinical Research Institute and th. 2014;61(18):1835–1846. doi: 10.1016/j.jacc.2012.12.047. Standardized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radford MJ. ACC/AHA Key Data Elements and Definitions for Measuring the Clinical Management and Outcomes of Patients With Chronic Heart Failure: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Data Standards. Circulation. 2005;112(6):1888–1916. doi: 10.1161/CIRCULATIONAHA.105.170073. [DOI] [PubMed] [Google Scholar]
- 20.dbGaP. Cardiovascular Health Study (CHS) Cohort: an NHLBI-funded observational study of risk factors for cardiovascular disease in adults 65 years or older. [Accessed January 3, 2017]. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000287.v6.p1.
- 21.dbGaP. Jackson Heart Study (JHS) Cohort. [Accessed January 3, 2017]. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000286.v5.p1.
- 22.BioLINCC. Heart Failure Network (HFN) CARdiorenal REScue Study in Acute Decompensated Heart Failure (CARRESS) [Accessed January 3, 2017]. https://biolincc.nhlbi.nih.gov/studies/carress/?q=heart failure.
- 23.BioLINCC. Sudden Cardiac Death in Heart Failure Trial (SCD-HeFT) [Accessed January 3, 2017]. https://biolincc.nhlbi.nih.gov/studies/scd_heft/
- 24.dbGaP. Framingham Cohort
- 25.Cornet R, de Keizer N. Forty years of SNOMED: a literature review. BMC Med Inform Decis Mak. 2008;8(Suppl 1):S2–S2. doi: 10.1186/1472-6947-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randorff Højen A, Gøeg KR. SNOMED CT Implementation. Methods Inf Med. 2012;51(6):529–538. doi: 10.3414/ME11-02-0023. [DOI] [PubMed] [Google Scholar]
- 27.National Institute of Standards and Technology. Health IT Testing Infrastructure
- 28.Yale University. The YODA Project


