Abstract
As medical organizations increasingly adopt the use of electronic health records (EHRs), large volumes of clinical data are being captured on a daily basis. These data provide comprehensive information about patients and have the potential to improve a wide range of application domains in healthcare. Physicians and clinical researchers are interested in finding effective ways to understand this abundance of data. Use of visual analytics to explore healthcare data is one such research direction. Here, we present a visualization and analysis environment to understand patient progression over time. Through the use of optimized data structures and progressive visualization techniques, we allow users to interactively explore how patients and their progression change over time. Compared to existing techniques, our work provides additional flexibility in analyzing patient data and has the potential to be used in a real-time hospital setting. Finally, we demonstrate the utility of our approach using a publicly available intensive care unit (ICU) database.
1. Introduction
The US healthcare system is producing hundreds of thousands of patient records detailing a wide range of information from admission times and dates, to symptoms and outcomes. Until recently, this data has been difficult to access, especially in bulk, often lacked a useful organization, and thus has been generally underutilized for clinical research. With the increasing use of EHRs, this paradigm changes, allowing researchers easy access to a large collection of information. If used effectively, this data may lead to better predictions of patient outcomes, personalized medication, and more targeted interventions. However, to realize this potential requires the ability to understand the clinical data in detail. Given the massive amounts of available data, for example, ICUs may collect real-time data streams of all patients1, which implies automatic or semiautomatic techniques to identify and explore interesting patterns and underlying trends. In this context, visualizing and exploring patient progression over time can provide valuable insights and facilitate the decision-making of physicians and clinical researchers.
Several factors need to be taken into consideration when analyzing this type of data: First, given the large number of patients, an individual, per-patient analysis is time-consuming and does not lend itself to finding commonalities and trends. Instead, patients should be grouped according to various criteria, such as symptoms, outcomes, etc. Second, to compare groups of patients who arrive at different times, their records must be aligned, for example, by their time of admission, time of major procedures or other common factors. Third, patient progression over time needs to be presented in a concise manner to allow simultaneous exploration of large numbers of patients. Finally, to utilize such a system in a hospital setting, the analysis must be interactive, allowing users to quickly explore different hypotheses.
The ideal system described above presents a number of practical challenges, especially for the large databases of interest. First, there exist a number of potentially interesting metrics by which to group patients and thus any analysis must be flexible and efficient enough to change the metric on-the-fly. Furthermore, whereas some metrics are easy to apply and absolute (e.g., splits by gender), others depend on specifying a similarity threshold that determines when two patients are considered to be in the same group. However, in practice this threshold is typically not known a priori, and in fact understanding how patient distributions and progression change with different thresholds may provide important insights. Most existing approaches focus on a single metric and a preselected threshold2,3; we present a system that allows users to freely explore patient grouping metrics and thresholds in an interactive setting.
Another challenge is the size and complexity of the data. Given a large number of patients and high temporal resolution, it is often difficult to grasp the progression of certain groups, let alone identify salient ones. Therefore, presenting data in a concise manner and providing support for various parameter selections and simplifications is crucial to provide the necessary insights.
From an analysis perspective, providing an effective exploration of patient progression requires three abilities: first, grouping patients within a time step at different similarity thresholds; second, correlating patient groups over time; and third, interactively visualizing and exploring patient progression to understand how different similarities affect their behavior. In this paper, we extract patient groups across multiple patient similarities and explore their progression with the aid of tracking graphs, where a concise representation of feature evolution is captured as a collection of feature tracks, see Figure 1. We provide clinical researchers with a visualization and analysis environment that is developed based on our earlier research in the scientific domain4,5. This prior system couples feature grouping and correlation components with visualization techniques to explore the temporal evolution of features in combustion data sets.
Figure 1:
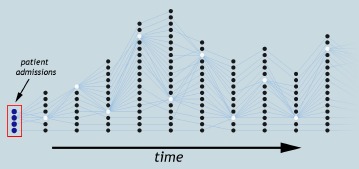
An example tracking graph showing patient progression over time. Each node represents a patient group and its “track” shows how that group progresses over time.
Our system process data in several steps. First, we use the patient similarity metric introduced by Lee et al.6 to group patients across multiple similarity thresholds. Here, it is important to note that any other patient similarity metric can be used within this system as well. Second, patient groups are correlated over time by tracking the individual patients within a group. In order to allow interactive extraction of data, our system uses optimized data structures to store these patient group and correlation details. Within the system, tracking graphs are used to present a global concise view of patient progression, and progressive visualization techniques are employed to enable interactive exploration of data. Finally, in collaboration with clinical researchers, we apply our visualization and analysis environment to a publicly available ICU database, the clinical database of Multiparameter Intelligent Monitoring in Intensive Care (MIMIC II) databases7, and explore the temporal progression of patients for varying similarity thresholds.
2. Related Work
A subset of the relevant related work is presented here to provide context and background for our research work. Analyzing time-varying data sets usually involves feature extraction and tracking steps. For healthcare data, tracking the progression of patient groups, i.e., the features-of-interest, is relevant to clinical researchers. Among the many feature definitions and their computation techniques found in the literature, techniques that extract feature information for all or a large range of values in a single pass are particularly useful. These techniques often result in hierarchical representations. For instance, hierarchical clustering8,9 and various other topological techniques10–12 have been used to effectively capture flexible feature hierarchies.
Hierarchical clustering is considered to be one of the most popular methods for creating a feature hierarchy. It partitions data into homogeneous groups based on a measure of similarity through the use of clustering. Depending on the similarity measure used, the results can lead to very different hierarchies. Moreover, many sequential and parallel algorithms for hierarchical clustering are available in the literature8,9. This type of clustering imposes a hierarchical structure on the underlying data irrespective of whether such a structure is appropriate. However, due to its simplicity, many applications have used this method to explore the clustering hierarchy of features. In this work, we also make use of hierarchical clustering to group patients within a time step at different similarity thresholds. In topological analysis, techniques exist that are able to efficiently extract and encode entire feature families in a single analysis pass. Reeb graphs13, contour trees10, merge trees12, and Morse-Smale complexes11 are several such techniques. Among them, Reeb graph, contour tree, and merge tree are contour-based and the Morse-Smale complex is gradient-based. As a result, the Morse-Smale complex captures very different structural information.
Visualizing the temporal evolution of features has long been a problem of interest within the visualization community. Depending on the subject area, many different techniques have been developed to address this problem. Traditionally, abstraction, illustration, morphing or animation-based techniques have been used to visualize temporal evolution of features14,15. Tracking graphs that show the feature evolution as a collection of feature tracks that split or merge over time are considered to be an effective representation for visualizing feature evolution4. These graphs provide conciseglobal views of feature evolution and are more amenable to filtering and simplifications. As clinical researchers are particularly interested in concise representations, we make use of tracking graphs to visualize patient progression over time.
Rind et al.16 present a comprehensive survey of information visualization systems used to visualize, explore, and query EHRs. These approaches related to EHRs can be broadly categorized into two categories: those that focus on a single patient record17 and those concerned with a collection of patient records2. Approaches in the first category focus on providing comprehensive information about a single patient (e.g., patient history, significant events, medication, and treatment), and the second category aims at presenting an overview from multiple patients. The latter provides less detail on each individual patient and focuses more on recognizing patterns and outliers within patient groups. Among these approaches that fall in the second category, LifeFlow3 and OutFlow2 are particularly interesting as they visualize event sequences in EHRs. LifeFlow uses color for a compact view and OutFlow uses a graph-based representation. In contrast, we do not visualize the progression of patient groups as an event. At a particular time step, the current event of a patient is one of the parameters considered within the patient similarity metric used. Also, within our system any similarity metric can be used to define patient similarities, providing more flexibility.
3. System Components
An interactive visualization and analysis environment is essential to gain an in-depth understanding of patient progression. In this paper, we refine a prior system that relies on dynamically constructed tracking graphs to enable feature extraction, tracking, and simplification4,5. This system is designed to study general time-varying features. However, so far it has only been applied to analyze features in scientific simulations. Here, we extend its functionality to effectively visualize patient progression in healthcare data. This section describes our system partitioned into several subsections dealing with: patient grouping, patient correlation, visualization, exploration, and implementation.
3.1. Grouping Patients Within a Time Step
The first step towards understanding clinical data is defining its features and a time step size based on which subsequent analysis is to be conducted. For our intended research, the feature-of-interest is a patient group (i.e., similar set of patients), and a day is considered to be the appropriate time step size. Next, for each time step in the data set, these patient groups need to be extracted and aligned. In this work, to ensure all patients’ hospital stays start at the same time, we align data based on a patient’s admission time.
Once features are extracted and aligned, they should be grouped based on an appropriate grouping method. By maintaining a notion of scale, this feature grouping naturally approximates a meaningful hierarchy. The naive approach of creating this hierarchy is by exhaustively precomputing all possible features at all possible scales. Many popular grouping algorithms also produce nested sets of features for varying scale, which in turn create feature hierarchies (e.g., hierarchical clustering techniques progressively merge elements8 and threshold-based segmentation creates increasingly larger regions11). In this case, we use hierarchical clustering. P atient groups in each time step are clustered based on their similarity to generate a hierarchical representation in the form of a tree. During clustering, we use the similarity metric by Lee et al.6 to define patient similarities but any other similarity metric could be used as well.
Figure 2(a) shows an example where such a hierarchy is constructed by progressively merging individual patients, with the most similar ones clustered first. Each leaf in the hierarchy represents a patient and each branch a patient group. Along with the hierarchy, various patient group-based attributes such as patient count, mean age and mean heart rate are computed and stored on a per-branch basis. For a given data set, an offline preprocessing step is used to compute these patient hierarchies, and the results are stored in a look-up structure to allow interactive exploration of patient groups. Within this look-up structure, for each patient group, its parent details and patient group-based attributes are stored. To determine correspondences across patients later, each patient is marked with a unique ID. This hierarchy is computed for each time step in the data, and is stored in a separate file to allow interactive exploration of patient groups. Once the hierarchy is computed, patient groups and their attributes can be quickly and easily extracted for any similarity threshold within its range, see Figure 2(b). Given a similarity threshold s within the full range of r, the corresponding patient groups can be extracted by “cutting” the hierarchy at s. This creates a forest of subtrees, where each subtree represents a patient group existing at s.
Figure 2:
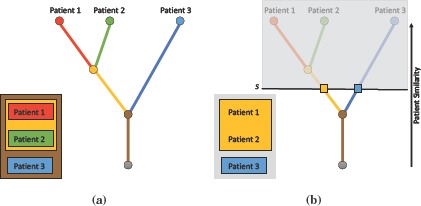
Patient hierarchy construction. (a) A patient hierarchy constructed by progressively merging similar patients, with the most similar ones clustered first. (b) To extract patient groups, the hierarchy is cut at a fixed threshold, resulting in a forest of subtrees, where each subtree represents a patient group.
3.2. Correlating Groups of Patients Over Time
Once patient groups are identified, the next step is to correlate them over time by tracking individual patients within a group. Two patient groups in consecutive time steps are considered to be correlated if they share at least one patient. All such correlations are extracted for each time step. To efficiently store and interactively extract these patient group correlations, we utilize the meta-graph structure of Widanagamaachchi et al.4. Similar to the patient hierarchy, this meta-graph structure is able to encode patient group correlations and their attributes for a range of similarity thresholds.
The meta-graph is generated in two steps. First, per-patient correlations are computed using the patient IDs computed above. For example, two patients in consecutive time steps are considered to be correlated if they have the same ID. As individual patients are represented by leaf branches in the patient hierarchy, this step results in correlations across leaf branches in consecutive time steps. If a correlation exists, we assign an edge with the weight of 1 across the two corresponding leaf branches, (bit, bjt+1), 1). Second, these per-patient correlations are accumulated along the patient hierarchy to compute the per-patient group correlations. At the accumulation time, if a correspondence already exists, we accumulate only the edge weights.
Just as in the patient hierarchy, various correlation-based attributes such as the amount of patient overlap are computed and stored within the meta-graph structure. Again, once the meta-graph is computed, patient group correlations and their attributes can be quickly extracted for any similarity threshold within the full parameter range. For a selected similarity threshold s, first, patient groups existing at s for each time step in the data set are obtained using the precomputed patient hierarchies. Then, correlations that exist across those extracted patient groups are obtained from the meta-graph structure. Together, these extracted patient groups and their correlations form the tracking graph at f, see Figure 3. This meta-graph structure is also created in an offline preprocessing step and the resulting structure is stored in multiple files (i.e., one file per time step), each containing a set of edges representing its correlations to patient groups in the next time step.
Figure 3:
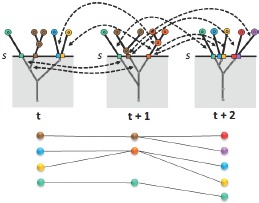
Tracking graph construction. At similarity threshold s, first, patient groups existing at that value are obtained from corresponding patient hierarchies. Then, the meta-graph is used to extract correlation details. Here, the correlations extracted are indicated with black arrows. The resulting tracking graph is displayed at the bottom.
3.3. Visualizing and Exploring Patient Progression
Our system for exploring patient progression over time contains three views: patient grouping view, patient progression view, and patient view, see Figure 4. Within each view, various progressive visualization techniques are employed to achieve interactivity. For instance, data is always presented with respect to a focus time step that is processed first.Data for the neighboring time steps is then extracted and presented in order of increasing distance. All views designed for only a single time step (i.e., patient grouping view and patient view), use the focus to determine their time step. The parameters such as hierarchy parameters and other filter parameters are coordinated across all views to provide a fully linked analysis environment.
Figure 4:
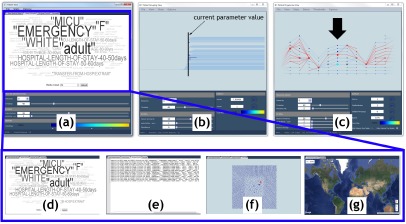
Our system contains three views: (a) patient view, (b) patient grouping view, and (c) patient progression view. The patient view consists of several subcomponents: (d) a word cloud, (e) textual, (f) geometric embedding, and (g) geospatial views. The patient grouping view shows the patient hierarchy for the focus time step and the patient progression view displays the tracking graph for the current focus and time window. Within the patient progression view, nodes are scaled based on the patient group size, and the focus time step is indicated with a black arrow. Here, a patient group is selected, which results in its progression being highlighted (indicated in red in the patient progression view).
3.3.1. Patient Grouping View
To enable researchers to gain a quick visual understanding of how patients group together for varying similarity thresholds, the patient hierarchy of the focus time step is visualized within this view. As the similarity threshold is changed, active patient groups within the hierarchy are also highlighted. In Figure 4(b), the selected similarity threshold within the hierarchy is displayed in a brown vertical line, and the active patient groups are highlighted in prominent colors.
3.3.2. Patient Progression View
This view visualizes the temporal progression of patients using tracking graphs. Starting from the user-defined focus time step, nodes and edges are iteratively added both forward and backward in time up to the user-defined time window to create the tracking graph, see Figure 4(c). Each node in the graph represents a patient group. A set of nodes in the same x coordinate indicates groups in one time step and edges across them indicate their correlations. For visual clarity, nodes in the focus time step are always displayed in prominent colors. Progressive techniques as in4, specifically, a fast initial graph layout and a slower greedy one, are used to visualize these tracking graphs.
3.3.3. Patient View
Several visualization techniques are combined here to present a specialized view of patients. Specifically, we integrate word cloud, textual, geometric embeddings, and geospatial visualizations, see Figure 4(a).
The word cloud visualization is dedicated to providing a quick overview of textual information regarding patients. For a selected patient group, a word cloud is constructed from the patient group-based attributes stored within the patient hierarchy, see Figure 4(d). Here, to obtain more intuitive overviews, the numerical attributes are converted into ranges. This visualization displays high-frequency words using bigger fonts and brighter colors, and others in faded and smaller fonts. As the name suggests, textual visualization presents textual details of patients in their native domain (i.e., as text), see Figure 4(e). For a selected patient group, this component displays its attributes such as hospital admission ID, patient ID, care unit and age.
Regardless of the data type, visualizing geometric embedding reveals interesting details and trends about data. The geometric embedding visualization displays the geometric embedding of patients in either 2D or 3D, see Figure 4(f). As geometric embedding details are not very obvious for the clinical data, for each time step, the GraphViz18 ‘neato’ layout algorithm together with patient similarity details is used to compute the 2D embedding of patients. In the geospatial visualization, when relevant information is available, we allow data exploration to be augmented with geospatial visualizations, see Figure 4(g). For instance, if a patient has his physical location details available for each moment in time (both during and/or prior to his hospital stay), this information will be visualized within this view. In addition to visualizing patient geospatial locations, their trajectories can also be displayed to easily identify data trends related to geographic locations.
3.3.4. Interactive Exploration
As tracking graphs can easily become complex and difficult to understand, various simplifications have to be performed on them to successfully understand their underlying trends. Specifically, we enable several simplification options. Through the linked-view interface, researchers are allowed to explore data sets by changing the focus time step and time window. They can select a particular day within a patient’s hospital stay, expand and contract its neighboring days to view progression both forward and backward in time. Within our system, the similarity threshold within the patient hierarchy, correlation amount within the meta-graph and other attribute values available (i.e., patient group-based and correlation-based), can all be explored. We also allow tracking graphs to be filtered by the length of stay of a patient group, which enables small patient stays to be eliminated from the analysis. Valence two and zero nodes of a tracking graph can be hidden to prevent visual clutter, nodes can be scaled based on their size, and progressions of certain patient groups can be highlighted. To help researchers maintain context across systems’ views, we also make use of correlated color maps and allow nodes to be colored using various patient group-based attributes. All these options combined enable researchers to interactively simplify tracking graphs, isolate interesting patient progressions and explore their parameter space.
3.4. Dataflow
Our system is implemented using the ViSUS framework19, which provides the basic building blocks for designing a streaming, asynchronous dataflow. Figure 5 shows the dataflow utilized within our system. The Data Reader module is dedicated to reading data into the system. It checks whether all data required for the current tracking graph has been loaded. If needed, it loads the required data and passes it to the Filter module. This module filters the received patient group and correlation details for the current parameter and attributes values. This filtered information is then simultaneously sent to layout and view modules.
Figure 5:
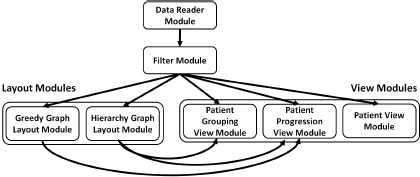
Our system’s dataflow contains several modules. The data is read into the system using the Data Reader module. Then, this node and edge information is filtered according to the current parameters within the Filter module. This module sends the resultant data to Layout and View modules simultaneously. The two Layout modules within the dataflow compute the relevant graph layouts and the View modules render the information received.
Each of the two layout modules computes a graph layout and sends those layout details to the relevant view modules for rendering. The Hierarchy Graph Layout module computes the initial layout for the tracking graph and sends this information to the Patient Grouping View and Patient Progression View modules. This hierarchy graph layout is computed only once for each time step as the data is read for the first time. The second layout module, Greedy Graph Layout, computes a greedy layout for the tracking graph each time its parameters change and passes them to the Patient Progression View module. This greedy layout minimizes the edge crossings within the tracking graph.
Our dataflow contains three view modules. The first view module, Patient Grouping View, visualizes the patient hierarchy of the focus time step. Once the module receivesthe necessary node and hierarchy details from Filter module and the layout details from Hierarchy Graph Layout module, it renders the patient hierarchy. The Patient Progression View module initially renders the tracking graph using the hierarchy graph layout. Then, as the greedy layout becomes available, it is integrated with the current graph. The third view module, Patient View, provides more specific views of patients (geometric embedding, geospatial, word cloud, and textual visualizations). Once this module receives the required data, depending on which visualization mode is selected, the corresponding computations and renderings are triggered. Each time parameters and/or selections are changed, the current processing within the dataflow is interrupted and restarted. However, rendering within the views maintains the current state for visual continuity.
4. Results
We enable clinical researchers to study the progression of patients via interactive exploration of dynamically constructed tracking graphs. The effectiveness of our framework is demonstrated with the use of a publicly available ICU database. The clinical database of Multiparameter Intelligent Monitoring in Intensive Care (MIMIC II) databases7 contains comprehensive EHR data collected from hospital medical information systems (both patient bedside workstations and hospital archives). This data is obtained from a set of ICUs including medical, surgical, coronary care, and neonatal in a single tertiary teaching hospital in the 2001 to 2008 time period. It includes patient information that falls into various categories such as general, physiological, medications, fluid balance, notes, and reports, see Table 1. The entire database totals to about ≈ 27GB and contains information about tens of thousands of ICU patients.
Table 1:
An overview of the data categories within MIMIC II clinical database
| General | Patient demographics, hospital admissions, discharge dates, room tracking, death dates (in or out of the hospital), ICD-9 codes, unique code for healthcare provider, and type (RN, MD, RT, etc). |
| Physiological | Hourly vital sign metrics, SAPS, SOFA, ventilator settings, etc. |
| Medications | IV meds, provider order entry data, etc. |
| Lab Tests | Chemistry, hematology, ABGs, imaging, etc. |
| Fluid Balance | Intake (solutions, blood, etc), output (urine, estimated blood loss, etc). |
| Notes & Reports | Discharge summary, nursing progress notes, etc; cardiac catheterization, ECG, radiology, and echo reports. |
In order to visualize MIMIC II clinical data within our framework, the relevant patient hierarchies and meta-graph structures need to be computed and stored. This is done in an offline preprocessing step. First, for each day in a patient’s hospital stay, patient details available in the database (e.g., admission ID, age, gender, race, ICD-9 code, drug code, hospital stay length, mean heart rate, mean temperature, and max urine output) are extracted, which results in 38291 patient admissions from 32536 patients. These details are then aligned to make sure all admissions fall on the first time step of the resultant data set. The resulting data set after aligning contains 174 time steps (i.e., 174 days).
Patients in each time step are then clustered together using the metric by Lee et al.6. This patient similarity metric was previously applied to the same MIMIC II clinical database to identify patient similarities within the first day of the ICU stay. In order to apply the metric to our research, the required clinical, administrative, and categorical variables are extracted from the database for each day within a patient’s hospital stay. Next, correlations across patient groups are computed by tracking individual patients within the groups. Once the patient groups and their correlation details are stored in our data format, the total data size is reduced to ≈ 680MB. By precomputing the patient hierarchies and meta-graph structures and storing them using optimized data structures, we allow interactive exploration of patient progression over time for several gigabytes of data.
Researchers are provided with the flexibility to vary the patient similarity thresholds and explore the entire parameter space interactively. Such interaction provides an understanding of how patients group together for varying similarity thresholds within a particular day in their hospital stay. In a hospital setting, such interaction provides the flexibility to explore patient progression details to better predict patient outcomes for a specific patient. For example, as new patients come in, for each day in their hospital stay, a patient’s outcome for the next day can be predicted based on their similarity to the existing patient groups. Figure 6 shows several examples of patient groups and their progression for 30, 34, 35 and 38 similarity thresholds. As the similarity thresholds decrease, more patients are grouped together, reducing the complexity of the tracking graph. For a specific similarity metric, exploring the full range of similarities enables researchers to gain insights on that metric’s range of values. In our case, upon exploration we realized that for this particular patient similarity metric, the appropriate similarity threshold range is 30-38. Any similarity threshold below or above that range either grouped all patients into one group or divided each patient to be in a separate group.
Figure 6:
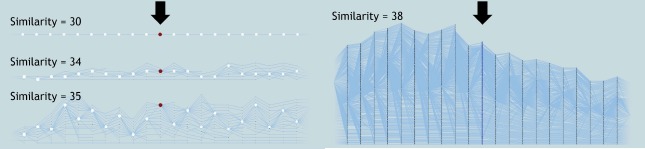
Effects of varying the similarity threshold to explore the temporal progression of patients. Here, patient groups and a portion of their corresponding tracking graphs are shown at 30, 34, 35, and 38 similarity thresholds. The focus time step of the tracking graphs is indicated with a black arrow, and the nodes are scaled based on the patient group’s size. In each graph, patient progression for 10 time steps both forward and backward in time from the focus time step is displayed.
Our system presents a global concise view of patient progression over time using tracking graphs. The full tracking graph showing the patient progression over time at 36 similarity threshold is displayed in Figure 7. By observing these tracking graphs, specifically feature track length indicating the hospital length of stay of patients, it is clear that although many of the patient stays are less than 90 days (i.e., 3 months), our data set also contains several longer patient stays. Of 32536 patients, we found 6 patients with hospital lengths of stay greater than 90 days.
Figure 7:
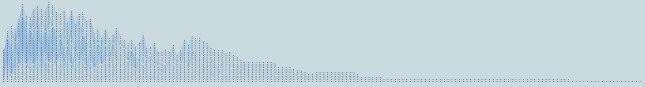
The entire tracking graph showing the complete patient progression for the MIMIC II clinical database. The graph contains 1110 nodes and 1288 edges for a total of 174 time steps. Here, 36 similarity threshold is used.
More importantly, various simplification options available in our system allow researchers to further simplify the tracking graphs. For example, filtering the tracking graph by correlation amount allows removal of the least frequent patient progression paths from the tracking graph, making frequent patterns more prominent. If an analysis is to be conducted only on longer hospital stays of patients, filtering options available within our system, specifically filtering tracking graphs by the length of a feature track, are useful. Furthermore, various patient group based attributes stored along with the patient hierarchy such as mean age and mean heart rate can also be used to filter patient groups. Figure 8 illustrates several such simplification results.
Figure 8:
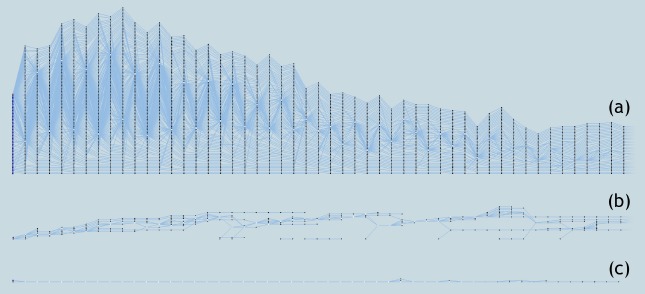
Simplifications of tracking graph. (a) A tracking graph showing the patient progression for the first 50 days within the hospital stay at 37 similarity threshold. (b) Tracking graph in (a) filtered to contain only correlations with overlap≥2. (c) Tracking graph in (a) filtered to contain only patient groups with size≥5.
Additionally, the patient view of our system is useful for obtaining an overview of patient groups. As the user selects a certain patient group, this view displays the details of its patients. The word cloud visualization provides a quick visual overview of the information within a selected patient group, see Figure 4(d). The numerical attributes such as age and hospital length of stay are converted to ranges to obtain more intuitive results. The exact patient details are also presented in the textual visualization within the system, see Figure 4(e).
Due to the generality of its design, this proposed framework provides new avenues for exploring healthcare data. Depending on the domain question we wish to answer, different options can be used within the visual analysis process. Instead of the patient similarity metric used, any other patient similarity based metric (e.g. age based, symptom based) can be used to group patients within time steps. Different patient attributes can be stored along with the patient hierarchies. For example, say we wish to study whether patients with diabetes stay longer in the ICU than other patients. We can use a patient similarity metric based on diabetes-specific symptoms to group patients for each time step. It then allows the users to explore the length of stay of patient groups where the patients are grouped based on their diabetes-specific symptoms.
5. Conclusion and Future Work
In this work, we present a visualization and analysis environment for understanding patient progression over time. The system’s interactive abilities to explore patient progression for different similarity metrics and for varying similarities are a distinct advantage over existing techniques used in healthcare. Using our system, researchers are able to explore how patients group together and progress over time, identify frequent progression paths, and also refer back to the native space of data for a visual understanding. By combining optimized data structures and progressive visualization techniques, we enable interactive exploration of terabytes size data, which provides the platform to use this type of analysis in a hospital setting. Within this work, an existing patient similarity metric is utilized for defining patient similarities. At each moment in time, patient similarities are computed by looking at a patient’s current clinical, administrative, and categorical information. A better similarity metric would be one that considers both the current information of the patient and the entire history starting from the hospital admission time. In order to obtain better results, we hope to utilize such a similarity metric in the future. In this work, we demonstrate the applicability of our approach using a publicly available ICU database. We are looking into obtaining additional healthcare databases to use within our system, specifically, databases with geospatial information for which the patient view within our system would prove to be more beneficial. Finally, we aspire to use our visualization and analysis environment in a real-time setting to assist the decision-making process of our collaborating physicians and clinical researchers.
References
- [1].Celi Leo Anthony, G Mark Roger, J Stone David, A Montgomery Robert. “big data” in the intensive care unit. closing the data loop. American journal of respiratory and critical care medicine. 2013;187(11):1157–1160. doi: 10.1164/rccm.201212-2311ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wongsuphasawat Krist, Gotz David. Outflow: Visualizing patient flow by symptoms and outcome; In IEEE VisWeek Workshop on Visual Analytics in Healthcare, Providence; Rhode Island, USA: American Medical Informatics Association; 2011. pp. 25–28. [Google Scholar]
- [3].Wongsuphasawat Krist, Guerra Gómez John Alexis, Plaisant Catherine, David Wang Taowei, Taieb-Maimon Meirav, Shneiderman Ben. In Proceedings of the SIGCHI conference on human factors in computing systems. ACM; 2011. Lifeflow: visualizing an overview of event sequences; pp. 1747–1756. [Google Scholar]
- [4].Widanagamaachchi Wathsala, Christensen Cameron, Bremer Peer-Timo, Pascucci Valerio. Interactive exploration of large-scale time-varying data using dynamic tracking graphs; In IEEE Symposium on Large Data Analysis and Visualization (LDAV); 2012. pp. 9–17. [Google Scholar]
- [5].Widanagamaachchi W., Klacansky P., Kolla H., Bhagatwala A., Chen J., Pascucci V., Bremer P.-T. Tracking features in embedded surfaces: Understanding extinction in turbulent combustion; IEEE 4th Symposium on Large Data Analysis and Visualization (LDAV); 2015. [Google Scholar]
- [6].Lee Joon, Maslove David M, Dubin Joel A. Personalized mortality prediction driven by electronic medical data and a patient similarity metric. PloS one. 2015;10(5):e0127428. doi: 10.1371/journal.pone.0127428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Saeed Mohammed, Villarroel Mauricio, Reisner Andrew T, Clifford Gari, Lehman Li-Wei, Moody George, Heldt Thomas, Kyaw Tin H, Moody Benjamin, Mark Roger G. Multiparameter intelligent monitoring in intensive care ii (mimic-ii): a public-access intensive care unit database. Critical care medicine. 2011;39(5):952. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Murtagh Fionn. A survey of recent advances in hierarchical clustering algorithms. The Computer Journal. 1983;26(4):354–359. [Google Scholar]
- [9].F Olson Clark. Parallel algorithms for hierarchical clustering. Parallel computing. 1995;21(8):1313–1325. [Google Scholar]
- [10].Carr Hamish. Topological manipulation of isosurfaces. PhD thesis, Citeseer. 2004 [Google Scholar]
- [11].Bremer Peer-Timo, Weber Gunther, Pascucci Valerio, Day Marc, Bell John. Analyzing and tracking burning structures in lean premixed hydrogen flames. IEEE Trans. Vis.Comp. Graph. 2010 Mar;16(2):248–260. doi: 10.1109/TVCG.2009.69. [DOI] [PubMed] [Google Scholar]
- [12].Bremer P-T, Weber G., Tierny J., Pascucci V., Day M., Bell J. Interactive exploration and analysis of large-scale simulations using topology-based data segmentation. IEEE Trans. Vis.Comp. Graph. 2011;17(9):1307–1324. doi: 10.1109/TVCG.2010.253. [DOI] [PubMed] [Google Scholar]
- [13].Pascucci Valerio, Scorzelli Giorgio, Bremer Peer-Timo, Mascarenhas Ajith. ACM Transactions on Graphics (TOG) Vol. 26. ACM; 2007. Robust on-line computation of reeb graphs: simplicity and speed; p. 58. [Google Scholar]
- [14].Muelder Chris, Ma Kwan-Liu. Rapid feature extraction and tracking through region morphing. Technical report, Citeseer. 2007 [Google Scholar]
- [15].Joshi Alark, Caban Jesus, Rheingans Penny, Sparling Lynn. Case study on visualizing hurricanes using illustration-inspired techniques. IEEE Trans. Vis.Comp. Graph. 2009;15(5):709–718. doi: 10.1109/TVCG.2008.105. [DOI] [PubMed] [Google Scholar]
- [16].Rind Alexander, Wang Taowei David, Aigner Wolfgang, Miksch Silvia, Wongsuphasawat Krist, Plaisant Catherine, Shneiderman Ben. Interactive information visualization to explore and query electronic health records. Foundations and Trends in Human-Computer Interaction. 2011;5(3):207–298. [Google Scholar]
- [17].S Pieczkiewicz David, M Finkelstein Stanley, I Hertz Marshall. Design and evaluation of a web-based interactive visualization system for lung transplant home monitoring data; In AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2007. p. 598. [PMC free article] [PubMed] [Google Scholar]
- [18].Ellson John, Gansner Emden, Koutsofios Lefteris, North Stephen, Woodhull Gordon. (Short Description, and Lucent Technologies). Lecture Notes in Computer Science. Springer-Verlag; 2001. Graphviz — open source graph drawing tools; pp. 483–484. [Google Scholar]
- [19].Pascucci V., Scorzelli G., Summa B., Bremer P.-T., Gyulassy A., Christensen C., Philip S., Kumar S. Chapman & Hall/Crc Computational Science. High Performance Visualization: Enabling Extreme-Scale Scientific Insight, chapter The ViSUS Visualization Framework. 2012 [Google Scholar]


