Abstract
Gleason grading of histological images is important in risk assessment and treatment planning for prostate cancer patients. Much research has been done in classifying small homogeneous cancer regions within histological images. However, semi-supervised methods published to date depend on pre-selected regions and cannot be easily extended to an image of heterogeneous tissue composition. In this paper, we propose a multi-scale U-Net model to classify images at the pixel-level using 224 histological image tiles from radical prostatectomies of 20 patients. Our model was evaluated by a patient-based 10-fold cross validation, and achieved a mean Jaccard index of 65.8% across 4 classes (stroma, Gleason 3, Gleason 4 and benign glands), and 75.5% for 3 classes (stroma, benign glands, prostate cancer), outperforming other methods.
Introduction
Prostate cancer is the most common and second most deadly cancer in men in the United States [1]. A key component of prostate cancer staging and treatment selection is the Gleason grading system, in which histopathological slides of prostate tissue are assigned grades that represent the aggressiveness of the cancer. The Gleason scale ranges from Gleason 1 (G1) - Gleason 5 (G5), with a score of G1 indicating cancer that closely resembles normal prostate glands and a score of G5 indicating the most abnormal histopathology, which is associated with the highest mortality risk. Final Gleason scores are generated by summing the first- and second-most prevalent patterns in the tissue sections. Currently, Gleason score is the best biomarker in predicting long term outcome of prostate cancer [2–4]. Yet, a recent clinical trial found no significant difference in mortality at 10 years between patients on active surveillance and immediate surgery [5], underscoring the need for more effective risk stratification tools to improve the outcomes of treatment deferral. Studies have also demonstrated the prognostic value of quantitative pathology features, such as the percent of Gleason 4, for diagnosis and treatment planning [3,6,7]. In addition, Gleason scores are assigned manually through pathologist review, a process that has been shown to have low inter-observer agreement across pathologists, especially when differentiating Gleason 3 (G3) vs Gleason 4 (G4), a distinction that may have substantial impact on further care [8–10].
A computer aided diagnosis (CAD) tool for performing Gleason scoring would provide a repeatable method for grading cancers and may be used as a pre-step for quantitative pathology feature extraction, thus providing a more precise assessment of cancer stage and treatment planning. In this paper, we propose a multi-scale U-Net CNN model for pixel-wise Gleason score prediction. We also compare the performance of several machine learning approaches to Gleason score assignment, including a pixel-wise deep convolutional neural network (CNN), a standard U-Net, the proposed multi-scale U-Net, and the previous work by Gertych, et.al. [11] on 224 histological image tiles from radical prostatectomies of 20 patients.
Previous Work
Previous work has been done in developing an automatic Gleason grading system to help improve diagnosis accuracy and achieve quantitative histological image analysis. A commonly used approach is to extract tissue features and apply classifiers on pre-selected small image tiles, each of which only contains one tissue class. Farjam, et.al. [12] developed a method to segment prostate glands with texture-based features, and then used the size and shape features of glands to classify image tiles into benign or malignant glands. Nguyen, et.al. [13] used structural features of prostate glands to classify pre-extracted regions of interest (ROIs) into benign, G3, and G4, achieving an overall accuracy of 85.6%. In the work by Gorelick, et.al. [14], a two stage Adaboost model was applied to classify around 991 sub-images extracted from 50 whole-mount sections of 15 patients. They achieved 85% accuracy for distinguishing high-grade (G4) cancer from low-grade cancer (G3).
However, the above algorithms require a set of pre-extracted image tiles with homogeneous tissue content, which may not be generalizable to larger and more heterogeneous images. Moreover, accurate localization of such small image tiles is a non-trivial problem [15]. Rather than attempting to classify the entire image tile, some efforts focus on segmenting and classifying glands with glandular features such as lumen shape and nuclear density [16,17], but these efforts require well-defined gland boundaries, and may not be applicable for high-grade prostate cancer with few recognizable glands.
Instead of using features from segmented glands, Gertych, et.al. [11] used intensity and texture features from joint histograms of local binary patterns and local variance to segment stroma (ST), prostate cancer (PCa), and benign glands (BN). In their two-stage classifier, a support vector machine (SVM) was trained with local intensity histogram to separate ST and epithelium (EP) areas, and then a random forest (RF) classifier was trained to segment BN and PCa. They obtained an average Jaccard index (J) of 59.5% for segmenting ST and EP areas. For separating BN and PCa, they achieved a JBN of 35.2% ± 24.9%, and a JPCa of 49.5% ± 18.5% in the test set. Although their model was able to do pixel-wise classification on heterogeneous image tiles, they did not address the problem of differentiating high-grade (G4) versus low-grade cancer (G3).
Additional previous work has explored the use of neural network models to learn features directly, rather than using handcrafted features. Deep convolutional neural network (CNN) models have demonstrated high performance in a variety of natural image analysis tasks [18–21]. Litjens, et.al. [22] implemented a deep convolutional neural network (CNN) to detect cancerous areas on prostate biopsy slide images at 5x magnification. They achieved around 0.90 AUC, but did not address the challenge of distinguish high-grade versus low-grade cancer.
Pixel-wise deep convolutional networks are difficult to apply to pathology image analysis due to the high resolution of digital pathology slides [23,24], direct analysis of which would generally require more memory than is available on a graphics processing unit (GPU). Two approaches to handling this challenge are resolution downsampling and patch extraction. In down-sampling, high resolution images are scaled down to more manageable sizes, at the cost of the loss of potentially discriminative fine details. In patch extraction, images are divided into (possibly overlapping) sub-patches that are then treated as independent training samples. This approach allows for the analysis of full resolution data, but may lead to an intractable number of potential patches, requiring subsampling of the dataset. In both of these methods, an overall prediction is created for the image or the patch.
Shelhamer and Long, et.al. [24,25] proposed a fully convolutional network (FCN) that can be trained from end to end to output pixel-wise predictions for an entire input image patch (rather than a single prediction for the patch). In order to get dense predictions for each pixel, they used up-sampling operations, and replaced the final fully connected layer with an Nx1x1 convolution layer, which output probabilities for N classes. Shallow layer features were fused with deep layer features to mitigate the challenge that intensive up-sampling can lead to coarse segmentation results. The model obtained a mean J of 67.5% and showed 30% improvement on PASCAL VOC 2011 test datasets.
The U-Net architecture proposed by Ronneberger, et.al. [26] extended the FCN by adding a relative symmetric upsampling path to down-sampling path, creating a U-shaped network architecture. Another important modification of U-Net was the use of an overlap-tile strategy for large image segmentation, in which a slightly larger tile is used as input and predictions are produced for the centered small tile. This method achieved an average J of 77% on a cell segmentation task [26].
While much work has been done in histological image analysis of prostate cancer, few addressed the problem of differentiating high-grade versus low-grade cancer. In this paper, we developed a multi-scale U-Net to predict four tissue classes at once (ST, BN, G3, and G4). We compared the proposed method with a pixel-wise CNN, a standard U-Net and a previously developed model by Gertych et.al. [11] that uses a combined SVM and RF classifier. Our multi-scale U-Net outperformed all other models and achieved the highest mean J of 65.8% across four classes.
Methods
Dataset
Radical prostatectomy specimens from 20 patients with a diagnosis of G3 or G4 prostate cancer according to the contemporary grading criteria [27,28] were retrieved from archives in the Pathology Department at Cedars-Sinai Medical Center (IRB approval no. Pro00029960). The specimens were previously stained with hematoxylin and eosin (H&E) for histological evaluation of the tumor. Slides were digitized by a high resolution whole slide scanner SCN400F (Leica Biosystems, Buffalo Grove, IL). The scanning objective was set to x20. The output was a color RGB image with the pixel size of 0.5pm x 0.5pm and 8 bit intensity depth for each color channel. Areas with tumor previously identified by the pathologist were extracted from whole slide images (WSIs) and then saved as 1200 x 1200 pixel tiles for analysis. 224 tiles were selected by three collaborating pathologists [11] who identified stroma (ST), benign glands (BN), G3 cancer, and G4 cancer containing cribriform and non-cribriform growth patterns. Individual glands and stroma in each tile were annotated manually using a custom graphical user interface [11]. All annotated image tiles were cross-evaluated by the pathologists, and corrections made if there was no consensus. This collection contains: BN (n=32), G3 (n=24), G4 (n=22), G3 and BN (n=29), G4 and BN (n=6), G3 and G4 (n=80), and G3 and G4 and BN (n=31) image tiles. All tiles were normalized to account for stain variability [29].
Semantic Image Segmentation with a Deep Convolutional Neural Network
For baseline comparison, a deep CNN model was trained to produce pixel-wise class predictions. The tile dataset was split into a training set containing 187 tiles and a testing set containing 37 tiles. In order to avoid correlations between data in the training and tests sets, tiles belonging to the same patient were restricted to either the training set or the testing set, yielding 17 unique patients in the training set, and 3 unique patients in the testing set (cross-validation was not used due to the large time requirements for evaluating the model).
The Inception V3 CNN model [30] was used with an input size of 299x299 pixels. Patches of size 299x299 were extracted from the pathology image tiles and then used for training and evaluation of the network. The label for any given patch was set to be the true label of the central pixel of the patch. Because there are a large number of possible 299x299 patches (each tile has over 800,000 possible 299x299 patches), it is impractical to train a network on every patch that exists in the dataset. Instead, patches were sampled (with replacement) from the training set using balanced random sampling. In this approach, patches were sampled with equal probability for each class. Within a given class, every potential patch that would fall into the class had equal probability of being sampled. Because of the class imbalance of the dataset, in this methodology, individual potential patches from different classes would have unequal probability of being sampled. Training was performed using an RMSProp (LR = 0.001, p = 0.9, e = 10–8) [31] optimizer using Keras [32] with Tensorflow [33] on two NVIDIA Titan X GPUs with synchronous gradient updates and a batch size per GPU of 50 patches. In order to saturate the GPUs during training, patch sampling was run in threads with separate state; one sampler thread was used per GPU. Training was performed over 25 “epochs” of 100,000 patches.
For evaluation, every possible patch was extracted from tiles in the testing set, and any patches that would have extended outside of the bounds of the original tile were discarded. Class predictions were obtained for these patches from the network, and each pixel was assigned a class based on the maximum prediction probability for that pixel.
Semantic Image Segmentation with U-Nets
Convnets produce dense predictions by extracting patches around every pixel, which can be inefficient even for images with moderate size. The FCN proposed by Shelhamer and Long, et.al. [24,25] uses up-sampling and fully convolutional layers to generate pixel-wise predictions efficiently in a single pass. The pooling operation makes CNNs relatively invariant to spatial transformations and also reduces spatial resolution of feature maps. To enable making local predictions with global context, the U-Net [26] extends an FCN with a U-shape architecture, which allows features from shallower layers to combine with those from deeper layers [24,26].
One intuitive way of performing semantic segmentation with FCN is to use the entire image as the input. However, training FCN with large images may require a huge number of samples and also can cause high GPU memory requirement. To solve those problems, large images are divided into several relatively smaller patches, and the overlap- tile strategy is used for seamless segmentation [26]. This requires the size of the patch to be carefully chosen so that the patch can be segmented with sufficient contextual information. Yet, the size of cellular structures such as glands may vary greatly as shown in (Figure 1). To better segment tissue structures with variable size, we propose a multiscale U-Net architecture that incorporates patches (subtiles) of three different sizes: 400x400, 200x200, and 100x100 to explicitly provide contextual information at multiple scales [34]. To handle border patches that cause one of these patch sizes to extend past the boundary of a given image tile, the tile is padded with reflection of the border [26]. A detailed overview of our multi-scale U-Net architecture is shown in (Figure 2). Instead of taking the whole 1200 x1200 image tile as input, we divided images into 100x100 subtiles and extracted the three patches of varying size around each of these image subtiles. Features from different sizes of patches were then concatenated together and used as inputs for the multi-scale U-Net model. The commonly used fully connected layer was replaced by a 4x1x1 convolutional layer that output pixel-wise probabilities for four classes (G3, G3, ST, and BN).
Figure 1.
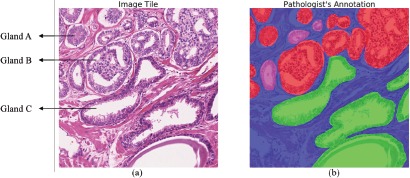
Variations in gland size. (a) shows a tile with heterogeneous Gleason grades (G3, G4 and benign glands). Pathologist annotation mask is shown in (b). The high-grade cancer (G4) areas are shown in red, low- grade cancer (G3) areas are denoted as pink, benign glands are indicated by green, and stroma areas are represented by blue. These images demonstrate the heterogeneity of glands both between grades (e.g. glands A and C) and within the same grade (e.g. glands A and B).
Figure 2.
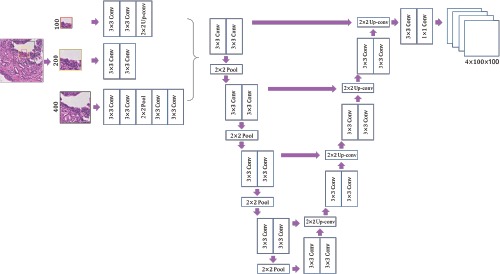
Architecture of the multi-scale patch-based U-Net. The whole image was divided into multiple nonoverlapping 100x 100 subtiles. To capture contextual information, a 200x200 patch (framed in yellow) and a 400x400 patch (framed in black) were extracted around each centered 100x100 patch (framed in red). Features of different sizes were either down-sampled or up-sampled to 200x200, and concatenated into 64x200x200 feature maps that were input to a U-Net model. The final layer output a 4x 100x 100 probability map, each channel of which corresponded to a probability map of one class.
In this experiment, we trained two FCN models. The first was the baseline U-Net model that followed an existing work [26]. The other is the multi-scale U-Net. Both models were trained with batch gradient descent (batch size: 25) and backpropagation. A momentum of 0.9 and a learning rate of 0.05 were used. A heuristic was followed to improve the learning of deep neural network model [19], where the learning rate was decreased by 10x when validation errors stopped decreasing. Models were implemented in Torch7 [35], and the training was done on two NVIDIA Titan X GPUs. The dataset of 20 patients was divided into 10 folds resulting in two patients in each fold. This patient-based cross validation ensured independence of training and testing data.
Evaluation Metrics
Overall pixel accuracy, mean accuracy for each class, and Jaccard index are three commonly used evaluation metrics for multi-class semantic image segmentation. Overall pixel accuracy measures the proportion of correctly classified pixels, however, it can be biased by imbalanced datasets. Mean single-class accuracy calculates the average proportion of correctly classified pixels in each class, which can also be biased by imbalanced datasets and overestimates the true accuracy due to combining multiple negative classes into one inference class [36–38]. Jaccard index, also known as intersection-over-union, overcomes the limitations of overall pixel accuracy and mean accuracy since it considers both false positives and negatives.
Here, we report Jaccard index and overall pixel-wise accuracy for our models, which can be obtained from a pixel- wise confusion matrix C. Cij is the number of pixels labeled as i and predicted as j. The total number of pixels with label i is denoted as where N is the number of classes. The number of pixels predicted as j is represented as Pj = Σi Ci,j, [36]. The Jaccard index for class i is then defined as follows:
| 1 |
The overall pixel-wise accuracy (OP) is defined as
| 2 |
Results and Discussion
Model Comparison
For the pixel-wise deep CNN model, class predictions were produced for a testing set comprising 30,170,133 pixels in 37 tiles across 3 patients. For the standard and multi-scale U-Net models, pixel-wise confusion matrices were summed across all 10 folds. In the first evaluation, true positive, true negative, false positive, and false negative rates for each class were calculated for all pixels in the dataset. Gleason 3 and Gleason 4 predictions were summed into a single inference class (PCa) for evaluation. For comparison, results from a baseline SVM + RF model by Gertych et.al. [11] are also included. The Jaccard index and overall pixel accuracy of each model are reported in (Table 1). The analysis was also performed without combining Gleason 3 and Gleason 4 into a single class, with performance shown in (Table 2). In both cases, the same network (trained on separate classes) was used for prediction. The multiscale U-Net architecture achieved the highest Jaccard index in both segmentation tasks: mean J = 75.5% for 3 class segmentation and mean J = 65.8% for 4 class segmentation. Both the U-Net and multi-scale U-Net models outperformed the pixel-wise CNN and the SVM-RF model by Gertych et.al. [11].
Table 1.
Model performances on segmenting prostate cancer (PCa), benign glands (BN) and stroma (ST).
| JPCa | JBN | JST | Mean J | OP | |
|---|---|---|---|---|---|
| U-Net | 74.3% | 70.6% | 80.1% | 75.0% | 86.6% |
| Multi-scale U-Net | 74.7% | 72.6% | 79.3% | 75.5% | 86.7% |
| Pixel-wise CNN | 66.0% | 59.0% | 71.0% | 65.0% | 63.9% |
| Gertych, et.al. [11] | 49.5% | 35.2% | 59.5% | 48.1% | n/a |
Table 2.
Model performances on segmenting Gleason 4 (G4), Gleason 3 (G3), benign glands (BN), and stroma (ST).
| JG3 | JQ4 | JBN | JST | Mean J | |
|---|---|---|---|---|---|
| U-Net | 45.8% | 60.9% | 70.6% | 80.1% | 64.4% |
| Multi-scale U-Net | 49.8% | 61.5% | 72.6% | 79.3% | 65.8% |
| Pixel-wise CNN | 23.0% | 25.0% | 59.0% | 71.0% | 45.0% |
| Gertych, et.al. [1l]a | n/a | n/a | 35.2% | 59.5% | 47.4% |
The previous model (SVM+RF) by Gertych, et.al. only addressed three class segmentation by combining G3 and G4 to PCa.
The approximated inference time for each model is also measured. It took about 2 hours for pixel-wise deep CNN model, around 3 seconds for U-Net model, and 9 seconds for multi-scale U-Net to generate predictions for a 1200X 1200 tile on one NVIDIA Titan X GPU. Dense predictions can be much more efficiently produced by FCN.
Segmentation results generated by U-Net and multi-scale U-Net for two representative image tiles are shown in (Figure 3). Our models performed well in segmenting different tissue types on image tiles with heterogeneous content, but both models struggled with some border areas due to a lack of contextual information. The small high-grade gland marked by a white arrow in the second row in (Figure 3), for example, was segmented as low-grade gland by both models.
Figure 3.
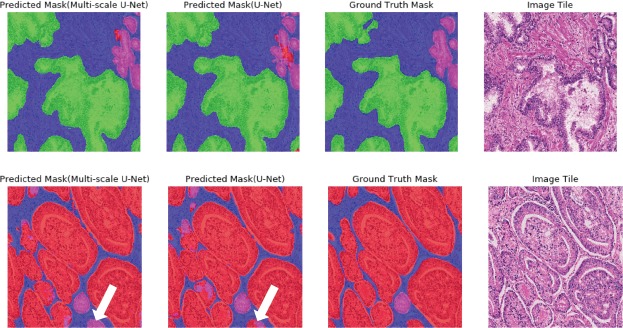
Segmentation masks generated by the U-Net and the multi-scale U-Net. Both ground truth masks and predictions are overlaid on original image tiles for easy interpretation. Colors follow the same schema illustrated in (Figure 1). The first row shows segmentation results for an image tile with three tissue types (benign, stroma, and G3 cancer). The second row shows a representative image tile with three tissue types (stroma, G3 and G4 cancer). White arrows point to border areas that both models struggle with.
In cases in which global information may be more important for class prediction, the multi-scale U-Net showed superior performance. As shown in (Figure 4), the single input U-Net misclassified areas with dense nuclei on a large benign gland. However, the multi-scale U-Net was able to segment this area correctly. Though both models could segment large irregular high-grade glands very well (Figure 3), they had limited power in segmenting poorly-formed high-grade areas, as shown in the first row of (Figure 5). Models could detect the approximate location of high-grade cancer, but failed to segment the exact areas. Segmentation performance of both models decreased on tiles with a mixture of small high-grade glands and small low-grade glands. The highest Jaccard indices for G3 and G4 achieved by the multi-scale U-Net were 49.8% and 61.5%, respectively. This reflects the reality that differentiating G3 and G4 is a challenging task, even for pathologists. The inter-observer agreement of clinical pathologists for distinguishing G3 from G4 is between 25% to 47% [11,39]. A larger training dataset that represents more of the natural variance of these cancer grades could allow for improving the models’ ability to discriminate between these classes.
Figure 4.
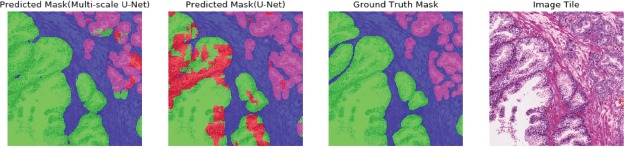
Segmentation results comparison for the multi-scale U-Net and the U-Net. Colors follow the same schema illustrated in (Figure 1). The multi-scale U-Net successfully segmented the large irregular benign gland, while the U- Net with single scale input did not.
Figure 5.
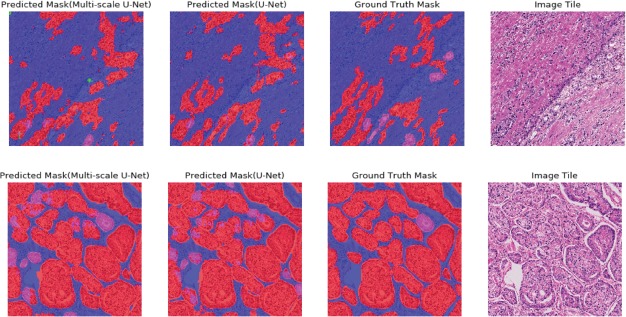
Segmentation results for two challenging tiles. Colors follow the same schema illustrated in (Figure 1). The first row shows an image tile containing G4 cancer with poorly-formed glands. Glands were less differentiated on that tile, likely increasing segmentation difficulty. The second row presents a tile with a mixture of small high-grade glands and small low-grade glands.
Conclusion
In this paper, we addressed the challenge of segmenting different tissue types on heterogeneous histological image tiles by using deep learning techniques. The performance of three different deep learning models (pixel-wise CNN, U-Net, multi-scale U-Net) were evaluated and compared using the Jaccard index and overall pixel accuracy. All three models outperformed a reference algorithm on three-class (ST, BN, PCa) segmentation. Both the U-Net and multiscale U-Net models achieved a higher Jaccard index than the pixel-wise model. The multi-scale model with three types of inputs (400x400, 200x200, 100x100) showed superior performance as compared with the original U-Net, likely due to its ability to explicitly make use of more global information without overly increasing memory requirements during model training.
There are some limitations in our work. Models were only trained on image tiles, rather than whole histological images. Though our method can be extended to whole image segmentation by splitting these images into nonoverlapping tiles, the prediction accuracy for boundary patches could be influenced by lack of contextual information and changes in class balance. Also, our model did not perform as well in segmenting G4 cancer with less differentiated glands. Exploring other approaches, such as the use of two separated models with two scales of inputs [40], could improve performance in the future. We also plan to investigate the influence of global versus local features on predicting dense labels, and will perform further evaluations of our models with whole histological images and extend our algorithm to a computerized tool which can be used to extract reliable and reproducible quantitative features from histological images.
Acknowledgments
The authors would like to acknowledge support from the UCLA Radiology Department Exploratory Research Grant Program (16-0003). KVS acknowledges support from an AMA Foundation Seed Grant, NIH NCI F30CA210329, NIH NIGMS GM08042, and the UCLA-Caltech Medical Scientist Training Program.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015 Jan;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Epstein JI, Hoshino K, Sasaki T, Al. E. An Update of the Gleason Grading System. J Urol. 2010 Feb 1;183(2):433–40. doi: 10.1016/j.juro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 3.Cole AI, Morgan TM, Spratt DE, Palapattu GS, He C, Tomlins SA, et al. Prognostic Value of Percent Gleason Grade 4 at Prostate Biopsy in Predicting Prostatectomy Pathology and Recurrence. J Urol. 2016 Aug;196(2):405–11. doi: 10.1016/j.juro.2016.01.120. [DOI] [PubMed] [Google Scholar]
- 4.Lee G, Sparks R, Ali S, Shih NNC, Feldman MD, Spangler E, et al. Zuo Z, editor. Co-Occurring Gland Angularity in Localized Subgraphs: Predicting Biochemical Recurrence in Intermediate-Risk Prostate Cancer Patients. PLoS One. 2014 May 29;9(5):e97954. doi: 10.1371/journal.pone.0097954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamdy FC, Donovan JL, Lane JA, Mason M, Metcalfe C, Holding P, et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N Engl J Med. 2016 Oct 13;375(15):1415–24. doi: 10.1056/NEJMoa1606220. [DOI] [PubMed] [Google Scholar]
- 6.Sauter G, Clauditz T, Steurer S, Wittmer C, Büscheck F, Krech T, et al. Integrating Tertiary Gleason 5 Patterns into Quantitative Gleason Grading in Prostate Biopsies and Prostatectomy Specimens. Eur Urol. 2017 Jan;137(0):1740–6. doi: 10.1016/j.eururo.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Perlis N, Sayyid R, Evans A, Van Der Kwast T, Toi A, Finelli A, et al. Limitations in Predicting Organ Confined Prostate Cancer in Patients with Gleason Pattern 4 on Biopsy: Implications for Active Surveillance. J Urol. 2017;197(1):75–83. doi: 10.1016/j.juro.2016.07.076. [DOI] [PubMed] [Google Scholar]
- 8.Lavery HJ, Droller MJ, Michalski J, Al. E. Do Gleason Patterns 3 and 4 Prostate Cancer Represent Separate Disease States? J Urol. 2012 Nov;188(5):1667–75. doi: 10.1016/j.juro.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 9.Huang CC, Kong MX, Zhou M, Rosenkrantz AB, Taneja SS, Melamed J, et al. Gleason Score 3 + 4=7 Prostate Cancer With Minimal Quantity of Gleason Pattern 4 on Needle Biopsy Is Associated With Low-risk Tumor in Radical Prostatectomy Specimen. Am J Surg Pathol. 2014 May;38(8):1. doi: 10.1097/PAS.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 10.Humphrey PA. Gleason grading and prognostic factors in carcinoma of the prostate. Mod Pathol. 2004 Mar 13;17(3):292–306. doi: 10.1038/modpathol.3800054. [DOI] [PubMed] [Google Scholar]
- 11.Gertych A, Ing N, Ma Z, Fuchs TJ, Salman S, Mohanty S, et al. Machine learning approaches to analyze histological images of tissues from radical prostatectomies. Comput Med Imaging Graph. 2015 Dec;46:197–208. doi: 10.1016/j.compmedimag.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farjam R, Soltanian-Zadeh H, Jafari-Khouzani K, Zoroofi RA. An image analysis approach for automatic malignancy determination of prostate pathological images. Cytom Part B Clin Cytom. 2007 Jul;72B(4):227–40. doi: 10.1002/cyto.b.20162. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen K, Sabata B, Jain AK. Prostate cancer grading: Gland segmentation and structural features. Pattern Recognit Lett. 2012 May;33(7):951–61. [Google Scholar]
- 14.Gorelick L, Veksler O, Gaed M, Gomez JA, Moussa M, Bauman G, et al. Prostate histopathology: Learning tissue component histograms for cancer detection and classification. IEEE Trans. 2013 Oct;32(10):1804–18. doi: 10.1109/TMI.2013.2265334. [DOI] [PubMed] [Google Scholar]
- 15.Doyle S, Feldman M, Tomaszewski J, Madabhushi A. A Boosted Bayesian Multiresolution Classifier for Prostate Cancer Detection From Digitized Needle Biopsies. IEEE Trans Biomed Eng. 2012 May;59(5):1205–18. doi: 10.1109/TBME.2010.2053540. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen K, Sarkar A, Jain AK. Berlin, Heidelberg: In Springer; 2012. Structure and Context in Prostatic Gland Segmentation and Classification; pp. 115–23. [DOI] [PubMed] [Google Scholar]
- 17.Peng Y, Jiang Y, Eisengart L, Healy MA, Straus FH, Yang XJ. In: 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE; 2011. Segmentation of prostatic glands in histology images; pp. 2091–4. [Google Scholar]
- 18.Razavian AS, Azizpour H, Sullivan J, Carlsson S. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops. IEEE; 2014. CNN Features Off-the-Shelf: An Astounding Baseline for Recognition; pp. 512–9. [Google Scholar]
- 19.Krizhevsky A, Sutskever I, Hinton GEGE, Sulskever Ii, Hinton GEGE. ImageNet Classification with Deep Convolutional Neural Networks. In: Advances in Neural Information Processing Systems. 2012:1097–105. [Google Scholar]
- 20.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2016:770–8. [Google Scholar]
- 21.Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y. OverFeat: Integrated Recognition, Localization and Detection using Convolutional Networks. 2013 Dec 21; [Google Scholar]
- 22.Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016 Sep 23;6(1):26286. doi: 10.1038/srep26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH. Patch-based Convolutional Neural Network for Whole Slide Tissue Image Classification. Proceedings IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2016;2016:2424–33. doi: 10.1109/CVPR.2016.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long J, Shelhamer E, Darrell T. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2015. Fully convolutional networks for semantic segmentation; pp. 3431–40. [DOI] [PubMed] [Google Scholar]
- 25.Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2017:1–1. doi: 10.1109/TPAMI.2017.2699184. [DOI] [PubMed] [Google Scholar]
- 26.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Med Image Comput Comput Interv. 2015 May;9351:234–241. [Google Scholar]
- 27.Fine SW, Amin MB, Berney DM, Bjartell A, Egevad L, Epstein JI, et al. A Contemporary Update on Pathology Reporting for Prostate Cancer: Biopsy and Radical Prostatectomy Specimens. Eur Urol. 2012 Jul;62(1):20–39. doi: 10.1016/j.eururo.2012.02.055. [DOI] [PubMed] [Google Scholar]
- 28.Brimo F, Montironi R, Egevad L, Erbersdobler A, Lin DW, Nelson JB, et al. Contemporary grading for prostate cancer: implications for patient care. Eur Urol. 2013 May;63(5):892–901. doi: 10.1016/j.eururo.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Reinhard E, Adhikhmin M, Gooch B, Shirley P. Color transfer between images. IEEE Comput Graph Appl. 2001;21(4):34–41. [Google Scholar]
- 30.Szegedy C, Wei Liu, Yangqing Jia, Sermanet P, Reed S, Anguelov D, et al. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 31.Tieleman T, Hinton G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. COURSERA Neural networks Mach Learn. 2012;4(2) [Google Scholar]
- 32.Chollet F. Keras. [Google Scholar]
- 33.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. 2016 Mar [Google Scholar]
- 34.Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Collobert R, Kavukcuoglu K, Farabet C. Torch7: A Matlab-like Environment for Machine Learning. BigLearn, NIPS Work. 2011:1–6. [Google Scholar]
- 36.Csurka G, Larlus D, Perronnin F, Meylan F. What is a good evaluation measure for semantic segmentation? IEEE PAMI. 2004 [Google Scholar]
- 37.Everingham M, Van Gool L, Williams CKI, Winn J, Zisserman A. The Pascal Visual Object Classes (VOC) Challenge. Int J Comput Vis. 2010 Jun 9;88(2):303–38. [Google Scholar]
- 38.Everingham M, Eslami SMA, Van Gool L, Williams CKI, Winn J, Zisserman A. The Pascal Visual Object Classes Challenge: A Retrospective. Int J Comput Vis. 2015 Jan 25;111(1):98–136. [Google Scholar]
- 39.Allsbrook WC, Mangold KA, Johnson MH, Lane RB, Lane CG, Epstein JI. Interobserver reproducibility of Gleason grading of prostatic carcinoma: General pathologist. Hum Pathol. 2001;32(1):81–8. doi: 10.1053/hupa.2001.21135. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, MacKenzie JJD, Ramachandran R, Chen DZ. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2016. A Deep Learning Approach for Semantic Segmentation in Histology Tissue Images; pp. 176–84. [Google Scholar]


