Abstract
Vertebrate cells have evolved two major pathways for repairing DNA double-strand breaks (DSBs), homologous recombination (HR) and nonhomologous DNA end-joining (NHEJ). To investigate the role of DNA ligase IV (Lig4) in DSB repair, we knocked out the Lig4 gene (LIG4) in the DT40 chicken B-lymphocyte cell line. The LIG4−/− cells showed a marked sensitivity to X-rays, bleomycin, and VP-16 and were more x-ray-sensitive in G1 than late S or G2/M, suggesting a critical role of Lig4 in DSB repair by NHEJ. In support of this notion, HR was not impaired in LIG4−/− cells. LIG4−/− cells were more x-ray-sensitive when compared with KU70−/− DT40 cells, particularly at high doses. Strikingly, however, the x-ray sensitivity of KU70−/−/LIG4−/− double-mutant cells was essentially the same as that of KU70−/− cells, showing that Lig4 deficiency has no effect in the absence of Ku. These results indicate that Lig4 is exclusively required for the Ku-dependent NHEJ pathway of DSB repair and that other DNA ligases (I and III) do not substitute for this function. Our data may explain the observed severe phenotype of Lig4-deficient mice as compared with Ku-deficient mice.
DNA double-strand breaks (DSBs) can be caused by a variety of exogenous and endogenous agents, posing a major threat to genome integrity. If left unrepaired, DSBs may cause cell death (1, 2). In vertebrate cells, homologous recombination (HR) and nonhomologous DNA end-joining (NHEJ) are two major pathways for repairing DSBs (2–5). HR allows for accurate repair of the lesion, whereas NHEJ can lead to imprecise joining. NHEJ is the predominant pathway during G0, G1, and early S phases of the cell cycle (6) and relies on Ku (a heterodimer of Ku70 and Ku86), DNA-protein kinase catalytic subunit (PKcs), XRCC4, and DNA ligase IV (Lig4) (3, 5). Extensive biochemical studies have proposed a model for the mechanism of NHEJ (3, 5, 7). First, Ku binds to the ends of a DSB and recruits DNA-PKcs. Nucleases and/or polymerases then trim the ends to make them ligatable. Finally, the Lig4/XRCC4 complex is recruited for ligation.
Consistent with the role of Ku and Lig4 in DSB repair, cells deficient in NHEJ proteins are highly sensitive to ionizing radiation (1, 8–12). Mice deficient in Ku70, Ku86, Lig4, or XRCC4 have also been generated by targeted gene disruption. Lig4 and XRCC4 knockout mice are embryonic lethal, which has been associated with massive neuronal cell death (13–15). However, Ku70- and Ku86-deficient mice are viable, although they are small in size (12, 16, 17). Ku86-deficient mice exhibit premature senescence (18). These observations suggested that Lig4 and XRCC4 have an essential function in the developing central nervous system, whereas Ku is dispensable. Nevertheless, increased neuronal apoptosis has been observed in Ku70- and Ku86-deficient embryos, although it is not as severe as in Lig4- or XRCC4-deficient embryos (19). Thus, the precise roles of these NHEJ proteins are not fully understood. That is, it remains uncertain whether or not the Lig4/XRCC4 complex has a Ku-independent function.
DNA ligase activity is indispensable for the process of HR. In the yeast Saccharomyces cerevisiae, where two DNA ligase genes are present, DNA ligase I is likely the ligase responsible for HR, because cells lacking Lig4 have been shown to possess normal HR activity (20, 21). In vertebrate cells, three DNA ligase genes (LIG1, LIG3, and LIG4) have been reported (22). Although recent studies have clearly established the role of Lig4 in NHEJ (5), it is still unclear which DNA ligase(s) are necessary for HR.
In this article, we generated LIG4−/− and KU70−/−/LIG4−/− clones in a chicken B-lymphocyte DT40 cell line. Consistent with the role of Lig4 in DSB repair, the LIG4−/− cells showed a marked sensitivity to ionizing radiation. HR activity was normal in LIG4−/− cells, supporting the notion that Lig4 is critical for DSB repair by NHEJ. Although LIG4−/− cells were more x-ray-sensitive than KU70−/− DT40 cells, strikingly, the x-ray sensitivity of KU70−/−/LIG4−/− double-mutant cells was essentially the same as that of KU70−/− cells. Our results indicate that Lig4 function is specifically required for the NHEJ pathway of DSB repair and is completely dependent on Ku. Furthermore, our data may explain the observed severe phenotype of Lig4-deficient mice as compared with Ku-deficient mice.
Materials and Methods
Targeting Constructs.
A partial chicken LIG4 cDNA was isolated by reverse transcription–PCR (RT-PCR) and used as a probe to screen a chicken genomic library (Stratagene). A genomic clone containing the LIG4 locus was obtained, partially sequenced (DNA Data Base in Japan/European Molecular Biology Laboratory/GenBank accession no. AB058600), and used to generate targeting constructs by replacing the coding region of the LIG4 gene with the hygromycin or puromycin resistance gene flanked by loxP sequences.
RT-PCR and Cell Cycle Analysis.
RT-PCR was performed as described (23). The primers used were L4–1 (5′-CCCCGGATCCAAGTTGGCTCATGAATCCCTGAGCA-3′) and L4–2 (5′-CATTTGGAGCTGGAGTCTCTGCAATAGCAC-3′). Cell cycle analysis was performed as described (23, 24).
Cell Culture and Transfection.
DT40 cells were cultured in a 5% CO2 incubator at 39°C in ES medium (Nissui Seiyaku, Tokyo) supplemented with 10% FBS and 1% chicken serum (growth medium). For colony formation, cells were grown for 7–11 days in ES medium containing 0.15% agarose, 20% FBS, and 2% chicken serum (soft agarose medium). DNA transfection was performed essentially as described (25, 26). Briefly, 4 × 106 cells were electroporated with 4 μg of DNA construct, and drug-resistant colonies were selected by incubation in soft agarose medium containing 1.5 mg/ml hygromycin B (Wako Pure Chemical, Osaka), 0.5 μg/ml puromycin (Wako Pure Chemical), or 1.6 mg/ml G418 (active; Sigma). Genomic DNA was isolated from drug-resistant clones and subjected to Southern blot analysis as described (25). Before knocking-in of the modified SCneo recombination substrate in LIG4−/− cells, the drug resistance genes were removed by transient expression of Cre recombinase, and the excision was verified by Southern blot analysis (Y.I., N.A., and H.K., unpublished results).
Sensitivity Assays.
To determine bleomycin or VP-16 sensitivity, cells were plated at 102 to 105 cells per dish into 60-mm bacterial dishes containing 5 ml of soft agarose medium with various concentrations of the drug. To determine x-ray sensitivity, cells were plated at 102 to 105 cells per dish into 60-mm dishes containing 5 ml of soft agarose medium and exposed to various doses of x-rays as described (11). In all experiments, cells were incubated for 7–11 days, and resulting, visible colonies were counted. The percent survival was determined by comparing the number of surviving colonies with untreated controls.
Results and Discussion
Construction of a LIG4−/− DT40 Cell Line.
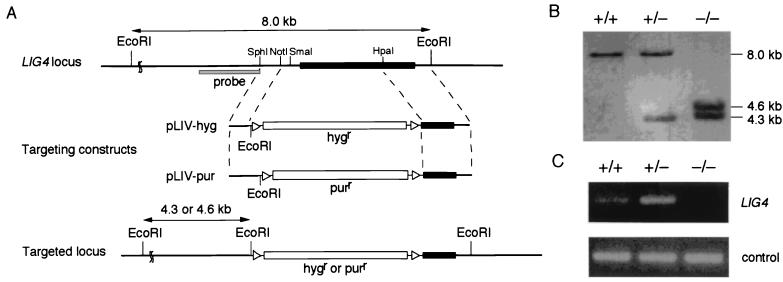
To generate LIG4 targeting constructs, we isolated a genomic clone containing the chicken LIG4 locus. Similar to the mouse and human LIG4 genes (8, 9, 13), inspection of the sequence revealed that the chicken LIG4 gene is encoded by a single exon. An ORF in the genomic clone showed 77% identity to human Lig4 protein. With the use of this genomic clone, we inserted either the hygromycin or puromycin resistance gene in the LIG4 locus as shown in Fig. 1A. Targeted homologous recombination with these constructs was expected to delete amino acids 1–570. The two targeting constructs were sequentially transfected into wild-type DT40 cells. Disruption of the LIG4 gene was verified by Southern blot analysis of EcoRI-digested genomic DNA with the use of an external probe (Fig. 1B) as well as RT-PCR analysis (Fig. 1C).
Figure 1.
Generation of LIG4−/− clones. (A) Schematic representation of targeted disruption of the chicken LIG4 gene. The chicken LIG4 locus, two targeting constructs (pLIV-hyg and pLIV-pur), and the targeted locus are shown. The black box indicates the ORF of the LIG4 gene. The triangles flanking the hygromycin resistance (hygr) or puromycin resistance (purr) gene designate loxP sequences. (B) Southern blot analysis. EcoRI-digested genomic DNA of wild-type (+/+), heterozygous mutant (+/−), and homozygous mutant (−/−) cells was hybridized with the probe shown in A as described. (C) RT-PCR analysis. Total RNA of wild-type, LIG4+/−, and LIG4−/− cells was used as a template to amplify a LIG4 cDNA.
LIG4−/− Cells Are Hypersensitive to DSB-Causing Agents.
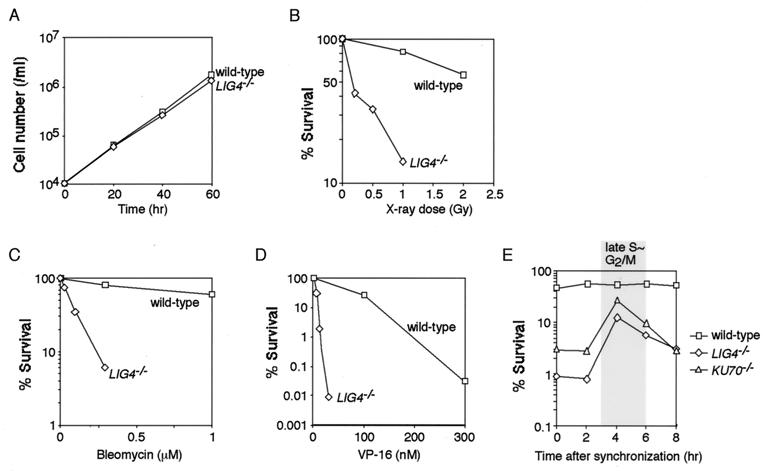
The LIG4−/− cells proliferated at a slightly lower rate than wild-type cells (Fig. 2A). The doubling times of wild-type and LIG4−/− cells were 8.0 and 8.6 h, respectively, at 39°C. We analyzed the cell cycle distribution of asynchronous cells by flow cytometry but observed no significant difference between wild-type and LIG4−/− cells (data not shown). Consistent with previous studies involving Lig4-deficient cells (8–11), LIG4−/− DT40 cells showed a marked sensitivity to x-rays (Fig. 2B). The cells were also hypersensitive to bleomycin (Fig. 2C) and VP-16 (Fig. 2D), a DNA topoisomerase II inhibitor that causes DSBs. The x-ray sensitivity of LIG4−/− cells was nearly restored to wild-type levels by ectopic expression of the chicken LIG4 gene (data not shown). To further characterize the role of Lig4 in DSB repair, we examined the stage(s) of the cell cycle at which the cells were x-ray-sensitive. For this purpose, we synchronized cells in G1 with the use of mimosine (24). After release from mimosine treatment, the cells were treated with 2 Gy x-irradiation at various time points and subsequently examined for their survival. As shown in Fig. 2E, LIG4−/− cells exhibited a higher sensitivity in G1 than in late S or G2/M. These results suggest a critical role for Lig4 in DSB repair by NHEJ, but not by HR. In good agreement with this notion, KU70−/− DT40 cells also showed an elevated sensitivity in G1 (Fig. 2E), although the phenotype was less severe than that of LIG4−/− cells (see below).
Figure 2.
Increased sensitivity of LIG4−/− cells to DSBs. (A) Growth curves of wild-type and LIG4−/− cells. Data are representative of three independent experiments. (B) Sensitivity of cells to ionizing radiation. (C) Sensitivity of cells to bleomycin. (D) Sensitivity of cells to VP-16. (E) X-ray sensitivity of cells throughout the cell cycle. Wild-type, LIG4−/−, and KU70−/− cells were synchronized in G1 with 0.4 mM mimosine. After release from mimosine treatment, the cells were x-irradiated (2 Gy) at the indicated time points and were subjected to a colony formation assay. Data are representative of two independent experiments. (A–D) Data are the mean of three independent experiments.
Lig4 Is Not Involved in HR.
Although the above studies strongly suggest a role for Lig4 in NHEJ, they did not fully exclude the possibility that the hypersensitivity of LIG4−/− cells was caused at least in part by the reduction in HR-mediated DSB repair. To test whether Lig4 participates in HR, we examined the gene-targeting efficiency with the use of the chicken β-actin locus. A β-actin targeting construct (27) was transfected into wild-type or LIG4−/− cells, and genomic DNA of transfectants was analyzed by Southern blot analysis. The targeting frequencies of wild-type and LIG4−/− cells were 34% (14/41) and 36% (24/67), respectively, indicating that gene targeting by HR is not impaired in LIG4−/− cells.
We next wished to examine the chromosomal HR frequency. To accomplish this, we knocked in a recombination substrate at the ovalbumin locus of wild-type and LIG4−/− cells. This recombination substrate was identical to the SCneo substrate composed of two nonfunctional neo genes (28, 29), except that the selection marker was the puromycin resistance gene (T. Fukushima, M. Takata, C. Morrison, R. Araki, A. Fujimori, M. Abe, K. Tatsumi, P. K. Dhar, E. Sonoda, M. Jasin, and S.T., unpublished results). One of the neo genes lacks the 5′ portion of the gene, and the other is mutated by a small internal deletion that destroys an NcoI site and is accompanied by the insertion of the I-SceI rare-cutting endonuclease site. HR between the two nonfunctional neo genes can result in an intact neo gene, giving rise to G418-resistant (neo+) colonies (28, 29). As expected, the frequency of neo+ colonies of LIG4−/− cells was 1.4 × 10−5, which was comparable to that of wild-type cells (1.2 × 10−5). We observed that when a DSB was introduced into the neo substrate by transient I-SceI expression, the HR frequency was at least 100-fold higher in both wild-type and LIG4−/− cells (N.A. and H.K., unpublished results). Taken together, it is highly unlikely that Lig4 is involved in HR, thus indicating that DNA ligase I and/or III are more likely candidates for the ligase function in HR.
Lig4 Is Exclusively Required for the Ku-Dependent DSB Repair Pathway.
Recently, Takata et al. (6) reported that although KU70−/− DT40 cells were hypersensitive to low doses of ionizing radiation, they were more resistant to high doses of ionizing radiation than were wild-type cells. This phenotype can be explained by the intrinsic error-prone nature of NHEJ compared with the precise repair by HR. At higher doses of ionizing radiation, followed by repair, NHEJ generates a large number of detrimental mutations in wild-type cells, which ultimately can result in cell death, whereas HR efficiently repairs DSBs in NHEJ-defective KU70−/− cells.
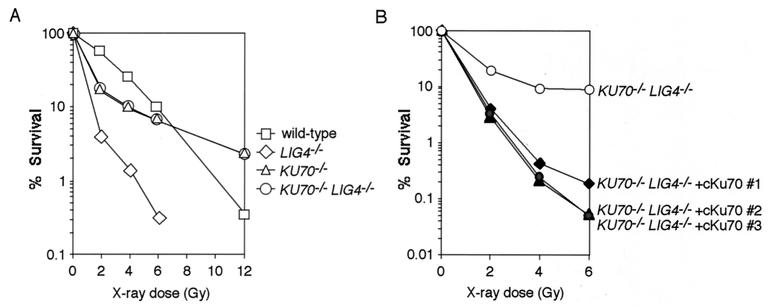
Given that Ku70 and Lig4 are involved in a common NHEJ pathway, it is conceivable that LIG4−/− DT40 cells would be more resistant to high doses of ionizing radiation than wild-type cells would be. However, unlike KU70−/− cells, LIG4−/− cells were consistently more sensitive than wild-type cells (Fig. 3A). Because LIG4−/− cells were more radiosensitive than KU70−/− cells, we speculated that Lig4 possessed a function that is independent of Ku-dependent NHEJ. To test this possibility, we generated KU70−/−/LIG4−/− double-mutant DT40 cells, by disrupting two alleles of the LIG4 gene with the use of KU70−/− cells, as shown in Fig. 1. The gene disruption was verified by Southern blot and RT-PCR analyses (data not shown). Strikingly, the x-ray sensitivity of KU70−/−/LIG4−/− double-mutant cells was essentially the same as that of KU70−/− cells (Fig. 3A). In addition, ectopic expression of chicken Ku70 in KU70−/−/LIG4−/− cells dramatically increased the x-ray sensitivity (Fig. 3B). These results indicate that Lig4 deficiency has no effect in the absence of Ku70, thus eliminating the possibility of any Ku-independent Lig4 function in DSB repair. Furthermore, our data indicate that the Ku-dependent NHEJ pathway specifically requires Lig4, and that other DNA ligases (I and III) cannot completely substitute for this in vivo Lig4 function, which is in good agreement with recent biochemical studies (30, 31).
Figure 3.
LIG4−/− cells are more x-ray-sensitive than KU70−/− and KU70−/−/LIG4−/− cells. (A) X-ray sensitivity of wild-type, LIG4−/−, KU70−/−, and KU70−/−/LIG4−/− cells. Data are representative of two independent experiments. (B) X-ray sensitivity of KU70−/−/LIG4−/− cell lines stably transfected with an expression plasmid for chicken Ku70 (6).
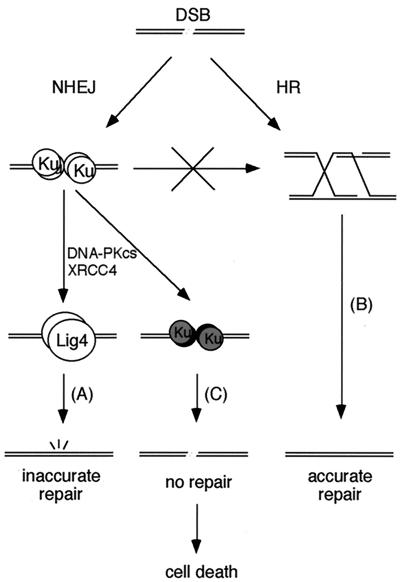
The present study provides insights into the mechanism of DSB repair. NHEJ is the predominant pathway during G0, G1, and early S phases of the cell cycle (6) but appears to be active in the late S and G2 phases as well, when HR can also repair DSBs (32). At least in DT40 cells, Ku deficiency does not result in hypersensitivity to a large amount of DSBs, irrespective of the presence of Lig4 (ref. 6 and this study), presumably because HR is able to compensate for NHEJ in late S and G2. In contrast, in Ku-proficient cells, an NHEJ reaction in the absence of Lig4 is extremely deleterious, owing to an exclusive requirement for Lig4 in NHEJ. We therefore propose that once Ku binds to DSB ends, the DSB is obligated to be repaired by the NHEJ pathway, thereby preventing HR-mediated repair of the lesion (Fig. 4). When Lig4 is absent, such Ku-bound DNA ends would result in unrepaired lesions, leading to cell death (Fig. 4). It has recently been reported that the absence of ataxia telangiectasia mutated (ATM) or p53 rescues the lethal phenotype of Lig4-deficient mice (14, 33), raising the possibility that these proteins are involved in the apoptosis of cells harboring such unrepairable DNA lesions.
Figure 4.
Model for DSB repair pathways. DSBs can be repaired by either NHEJ (A) or HR (B). However, once Ku binds to DSB ends, the DSBs must be repaired by NHEJ, preventing the opportunity to use HR. Because Lig4 is exclusively required for the Ku-dependent NHEJ pathway, its absence results in unrepaired lesions, leading to cell death (C). At least in DT40 cells, Ku deficiency is not so severe, presumably because HR is able to partially compensate for NHEJ.
Our model may provide an explanation for the difference in phenotype between Ku-deficient mice and Lig4- or XRCC4-deficient mice. In mice deficient in Lig4 or XRCC4, cells harboring Ku-bound DSBs eventually die, because these lesions cannot be repaired by HR, as discussed above. In contrast, in the absence of Ku, HR can partially substitute for the lost NHEJ function, resulting in the less severe phenotype observed in Ku-deficient mice. In this regard, it will be intriguing to determine whether mice deficient in both Ku and Lig4 show a phenotype similar to that of Ku-deficient mice.
Acknowledgments
We thank M. Jasin for generously providing us with the SCneo construct. We also thank C. Nishigaki for excellent technical assistance. We acknowledge Z. E. Karanjawala and F. Chedin for critical reading of the manuscript. This work was supported in part by grants-in-aid from the Ministry of Health and Welfare, and the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations
- DSB
double-strand break
- HR
homologous recombination
- Lig4
DNA ligase IV
- NHEJ
nonhomologous DNA end-joining
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB058600).
References
- 1.Chu G. J Biol Chem. 1997;272:24097–24100. doi: 10.1074/jbc.272.39.24097. [DOI] [PubMed] [Google Scholar]
- 2.Kanaar R, Hoeijmakers J H, van Gent D C. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 3.Critchlow S E, Jackson S P. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 4.Liang F, Han M, Romanienko P J, Jasin M. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieber M R. Genes Cells. 1999;4:77–85. doi: 10.1046/j.1365-2443.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- 6.Takata M, Sasaki M S, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee K J, Huang J, Takeda Y, Dynan W S. J Biol Chem. 2000;275:34787–34796. doi: 10.1074/jbc.M004011200. [DOI] [PubMed] [Google Scholar]
- 8.Frank K M, Sekiguchi J M, Seidl K J, Swat W, Rathbun G A, Cheng H L, Davidson L, Kangaloo L, Alt F W. Nature (London) 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 9.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 10.Riballo E, Critchlow S E, Teo S H, Doherty A J, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett C F, et al. Curr Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 11.Sado K, Ayusawa D, Enomoto A, Suganuma T, Oshimura M, Sato K, Koyama H. J Biol Chem. 2001;276:9742–9748. doi: 10.1074/jbc.M010530200. [DOI] [PubMed] [Google Scholar]
- 12.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes D E, Stamp G, Rosewell I, Denzel A, Lindahl T. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 14.Frank K M, Sharpless N E, Gao Y, Sekiguchi J M, Ferguson D O, Zhu C, Manis J P, Horner J, DePinho R A, Alt F W. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, Seidl K J, Rathbun G A, Zhu C, Manis J P, van der Stoep N, Davidson L, Cheng H L, Sekiguchi J M, Frank K, et al. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 17.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 18.Vogel H, Lim D S, Karsenty G, Finegold M, Hasty P. Proc Natl Acad Sci USA. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt F W. Proc Natl Acad Sci USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teo S H, Jackson S P. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson T E, Grawunder U, Lieber M R. Nature (London) 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 22.Tomkinson A E, Mackey Z B. Mutat Res. 1998;407:1–9. doi: 10.1016/s0921-8777(97)00050-5. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi M, Adachi N, Koyama H. Gene. 1998;215:329–337. doi: 10.1016/s0378-1119(98)00283-2. [DOI] [PubMed] [Google Scholar]
- 24.Adachi N, Kobayashi M, Koyama H. Biochem Biophys Res Commun. 1997;230:105–109. doi: 10.1006/bbrc.1996.5893. [DOI] [PubMed] [Google Scholar]
- 25.Aratani Y, Andoh T, Koyama H. Mutat Res. 1996;362:181–191. doi: 10.1016/0921-8777(95)00049-6. [DOI] [PubMed] [Google Scholar]
- 26.Fujimaki K, Aratani Y, Fujisawa S, Motomura S, Okubo T, Koyama H. Somatic Cell Mol Genet. 1996;22:279–290. doi: 10.1007/BF02369567. [DOI] [PubMed] [Google Scholar]
- 27.Buerstedde J M, Takeda S. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 28.Johnson R D, Liu N, Jasin M. Nature (London) 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 29.Johnson R D, Jasin M. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Trujillo K, Sung P, Tomkinson A E. J Biol Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- 31.Nick McElhinny S A, Snowden C M, McCarville J, Ramsden D A. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson C, Jasin M. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Y, Barnes D E, Lindahl T, McKinnon P J. Genes Dev. 2000;14:2576–2580. doi: 10.1101/gad.837100. [DOI] [PMC free article] [PubMed] [Google Scholar]