Abstract
A critical issue in the usage of cancer drugs is its association with various adverse events (AEs) in some, but not all, patients. The National Cancer Institute (NCI) Common Terminology Criteria for Adverse Events (CTCAE) is a controlled terminology for AE classification and analysis in cancer clinical trials. The Ontology of Adverse Events (OAE) is a community-based ontology in the domain of AEs. In this study, OAE was first updated by including AE severity grading and OAE-CTCAE mapping. An OAE subset containing CTCAE-related terms and their associated OAE terms was generated to facilitate term usage. A use case study based on a published cancer drug clinical trial demonstrates that OAE provides better hierarchical representation, includes semantic relations, and supports automated reasoning. Demonstrated with a single patient analysis, the OAE framework supports precision informatics for representing AEs and related genetic and clinical conditions in individual patients treated with cancer drugs.
Introduction
While cancer drugs play a critical role in treating cancer patients, the adverse events (AEs) associated with cancer drugs can be very detrimental. As a result, the surveillance and analysis of cancer drug AEs are important to improve public health. To support such surveillance and analysis, it is critical to maintain proper terminology and documentation. A foundation of AE reporting and analysis is the controlled terminology of various AEs. MedDRA [1], WHO-ART [2], and the Common Terminology Criteria for Adverse Events (CTCAE) [3] are commonly used AE controlled terminology systems. These controlled terminologies tend to have drawbacks, such as missing term definitions, poorly defined hierarchies, and lack of semantic relations among terms across hierarchies [4]. Ontologies have been proposed and developed to address these drawbacks [4-6].
As a product of the US National Cancer Institute (NCI), CTCAE is a specialized controlled terminology that provides the standardized classification of AEs of drugs used in cancer therapy. CTCAE has substantially evolved since its inception in 1983. Initially published in 2009, CTCAE v4.0 lists 790 AE terms, and each of these AE terms may be associated with a range of severity grades [7]. Most US and UK cancer drug trials encode their observations based on CTCAE.
The Ontology of Adverse Events (OAE) is a community-based open source ontology that logically represents various AEs [6]. OAE represents various AEs based on patient anatomic regions and clinical outcomes, including symptoms, signs, and abnormal processes. In OAE, an AE is defined as a pathological bodily process that occurs after a medical intervention (e.g., drug treatment). By logically linking the medical intervention, patient, patient conditions, and AE outcomes, OAE provides a robust framework that supports systematic analysis of AE case classification and analysis. With over 5,000 terms, OAE has been used in many scenarios [6, 8-10]. Figure 1 shows the general OAE model of an adverse event of a human patient treated with a drug. Specifically, a human patient is administered a drug in order to treat a specific disease (e.g., cancer). The drug has its active chemical ingredient, dose, and it is administered through a specific route (e.g., oral). After an incubation time, the patient will experience a specific adverse event(s) that occur in a specific anatomic location. The patient conditions (e.g., age, gender, and disease) may affect the drug treatment effectiveness as well as AE outcomes. Even for the same type of AE, there exist different severity grades (as detailed later in this paper).
Figure 1.
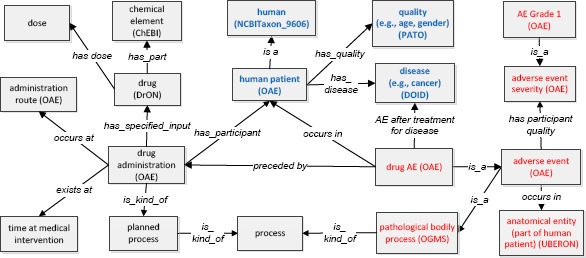
OAE modeling of drug AE and related terms in a systematic framework. The terms inside boxes are ontology classes, and terms in the middle of arrows are relations (i.e., object properties). The boxes with blue text indicate patient information, and the boxes with red text represent AE-related terms. See more explanation in the text.
To support ontology interoperability, in addition to its own terms, OAE imports many terms from existing ontologies such as Basic Formal Ontology (BFO) [11], Ontology for Biomedical Investigations (OBI) [12], and Ontology for General Medical Science (OGMS) [13] (Figure 1). For example, OAE imports many terms from the upper level BFO [11] and aligns with BFO [6]. Both administration and drug AE are classified as BFO:‘process’. On the other hand, the drug administration is subclass type of OBI:‘planned process’ (i.e., a process intentionally planned by human), and an AE is a subclass of OGMS:‘pathological bodily process’ (Figure 1). The disease, drug, quality, and anatomical entity can be represented by Disease Ontology (DOID) [14], Drug Ontology (DrON) [15], Phenotypic Quality Ontology (PATO) [16], UBERON [17], and NCBITaxon [18], respectively (Figure 1). Terms from these ontologies are semantically linked to provide a comprehensive picture of a drug AE and its associated drug, drug administration, anatomical location where the AE occurs, and detailed patient conditions (e.g., gender, age, and health condition).
In the Figure 1 modeling, the relation ‘preceded by’ is used to link drug administration and drug AE. This relation is simply a temporal relation that indicates the drug AE occurs after the drug administration. The usage of this relation is aligned with the FDA definition that a drug AE does not have to be causal. It means that the drug administration does not have to be the cause of the AE. An AE may be just part of a disease process that started with the patient treated with the drug. For example, a patient who has gastric cancer and is treated with a drug has gastrointestinal bleeding. The bleeding AE may be caused by the gastric cancer or the drug usage. If we do not know the cause, we can use the relation ‘preceded by’. To represent a defined causal relation, OAE uses the relation ‘induced by’, which is a special subclass of the ‘preceded by’ relation [6]. If we know a causal relation between the drug administration and the bleeding, the relation ‘induced by’ can be used.
In addition, OAE and CTCAE may assign term hierarchies differently. For example, In CTCAE, ‘Rash pustular’ (CTCAE code E11545) is asserted as a subclass of ‘Infections and infestations’ in CTCAE (Figure 2A). This assertion is questionable since not every rash pustular is associated with an infection. CTCAE includes three other types of rash where ‘Rash acneiform’ and ‘Rash maculo-papular’ are under ‘Skin and subcutaneous tissue disorders’, and ‘Papulopustular rash’ is under ‘Infections and infestations’. However, CTCAE does not have a class called ‘rash’ (Figure 2A). In contrast, OAE has ‘rash AE’ (OAE_0000528) and over ten specific subclasses of ‘rash AE’ (Figure 2B). Such comparisons suggest possible issues with CTCAE classifications and broader coverage of OAE.
Figure 2.
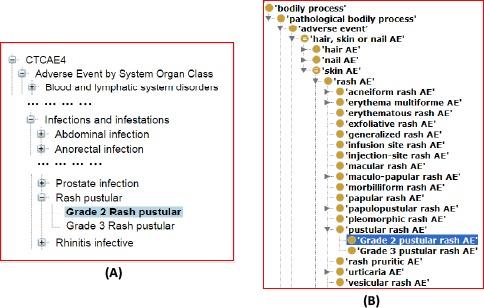
CTCAE and OAE term hierarchy comparison. This comparison uses the Grade 2 pustular rash AE as the example. (A) CTCAE hierarchy. Rash pustular is under ‘Infection and infestation’. (B) OAE hierarchy. OAE includes the term ‘rash AE’ while CTCAE does not have a matching term.
Since CTCAE is commonly used in cancer drug AE reporting, we hypothesized that the CTCAE-OAE mapping followed by OAE-based analysis would improve cancer drug AE classification and analysis and support personalized pharmacovigilance investigation. Not all drug-treated patients will experience AEs. For patients under different conditions, the AE patterns may vary greatly, and should be investigated by considering different variables including personal conditions and drug usage. To address this hypothesis, we updated OAE with a systematic CTCAE-OAE mapping strategy and applied the updated OAE to two different use cases to demonstrate how OAE supports better cancer drug AE classifications and personalized drug-AE case representation.
Methods
OAE ontology visualization and editing. The Protégé OWL editor (http://protege.stanford.edu/) was used for ontology visualization and editing.
OAE modeling of AE severity and semi-automatic OAE-CTCAE mapping. We first generated an OAE design pattern that represents AE severity. Our OAE severity representation strategy is similar to the CTCAE severity grading in terms of severity definitions, but also differs in terms of their classification and usage. The CTCAE source studied in this project primarily comes from the CTCAE 4.0.3, which is the version available in NCBO Bioportal (http://bioportal.bioontology.org/ontologies/CTCAE). To ensure high quality of the OAE-CTCAE mapping, manual curation was applied to annotate individual CTCAE records and map the records to OAE. The annotations, such as, definition, definition source, MedDRA ID, etc., were manually obtained from either CTCAE or online resources, and the mapped results along with the annotated information were initially represented in a pre-defined Excel template. After second review, the Excel data were then automatically converted to an OWL file using the Ontorat software program [19], and merged to OAE. A Java-based program was developed to automatically assign severity axioms to OAE terms to indicate possible grades for specific OAE AE terms.
An important feature of CTCAE is its classification of AE severity. Specifically, the CTCAE v4.0 includes Grades 0 through 5 with unique clinical descriptions of severity for each AE based on the general guideline: Grade 0 – signs and symptoms within normal limits; Grade 1 - Mild AE; Grade 2 - Moderate AE; Grade 3 - Severe AE; Grade 4: life-threatening or disabling AE; and Grade 5 - Death related to AE [7].
Corresponding to the six grades of severity in CTCAE, OAE also has 6 grades of ‘AE severity’. However, OAE and CTCAE implement the severity grades differently. The OAE ‘AE severity’ is a subclass under the term ‘process quality’ (PATO_0001236). The term ‘process quality’, imported from the PATO, represents a quality which inheres in a process. For example, ‘skin ulceration’ includes five grades (Figure 3A).
Figure 3.
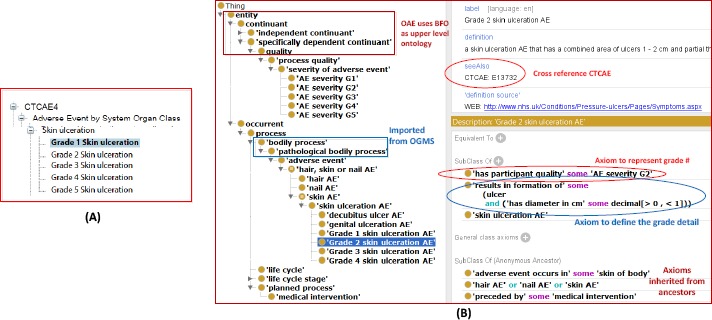
Comparison of AE severity definitions in CTCAE and OAE. (A) CTCAE term ‘Skin ulceration’ and its five grades. (B) OAE term ‘Skin ulceration’, all its child terms, and related axioms. Ontofox was used to extract all children under ‘skin ulceration AE’ and all their related terms and axioms from OAE. The Ontofox output was displayed in Protégé OWL editor, and the screenshot was taken from the display. The OAE annotation includes its mapping to CTCAE ontology term ID. The linkage of the OAE term to the AE severity grade is logically defined using the new relation ‘has participant quality’. The ulcer diameter is included in another axiom to specify the Grade 2 severity. OAE does not include ‘Grade 5 skin ulceration AE’ as explained in the text.
To represent a specific AE severity, OAE uses two approaches. First, for an AE with a defined severity range, OAE uses an axiom with the relation term ‘has participant quality’ to link the AE and a severity grade of the participant undergoing the AE process. For example, OAE defines an axiom of ‘skin ulceration AE’:
‘has participant quality’ some (‘AE severity G1’ or ‘AE severity G2’ or ‘AE severity G3’ or ‘AE severity G4’ or ‘AE severity G5‘)
The second approach in OAE for severity representation is to generate specific subclasses with severity specifically defined. For example, OAE represents 4 different subclasses of ‘skin ulceration AE’ corresponding to the Grade 1-4 of this AE (Figure 3B).
Note that we did not generate a class ‘Grade 5 skin ulceration AE’ because Grade 5 always means death. Since our ‘skin ulceration AE’ axiom has shown Grade 5 is possible, we do not need to generate a specific term for Grade 5. However, OAE represents Grade 1-4 individually (Figure 3B) since each of the grades has its specific criterion other than the general grade definitions. For example, according to CTCAE definition, Grade 2 skin ulceration AE shows a combined area of ulcers <1 cm and nonblanchable erythema of intact skin with associated warmth or edema. In align with the CTCAE definition, we represent such a case in OAE with a specific class “Grade 2 skin ulceration AE’ (OAE_0002618) (Figure 3B). In addition, OAE includes an ontological axiom to logically represent the ulcer size criterion of “Grade 2 skin ulceration AE’:
‘results in formation of’ some (ulcer and ‘has diameter in cm’ some decimal[>0, <1])
With the above axiom, when the ulcer diameter is measured, an OAE-based grading system will be able to automatically classify the ulceration with a Grade 2 severity. The CTCAE does not include such an axiom definition, and its usage requires manual interpretation.
Generation, deposition, and query of CTCAE-related OAE subset view (CTCAE-OAEview). Ontofox [20] was used to generate an OAE subset view that contained all CTCAE-matched OAE terms and their associated OAE terms. The CTCAE-OAEview was then deposited to the Ontobee linked ontology server [21] at http://www.ontobee.org/ontology/CTCAE-OAEview. Ontobee SPARQL (http://www.ontobee.org/sparql) was used to query CTCAE-OAEview for addressing various questions.
OAE application use case studies. Two use cases were analyzed. First, a randomized, multicenter, placebo-controlled phase 2 study of a cancer drug clinical trial was used for testing the application of the newly updated OAE in cancer clinical trial AE analysis [22]. This case study demonstrated how OAE could be used to support AE and AE grading hierarchical classification from a cancer clinical trial. The second use case reported a single patient treated with selumetinib [23] and illustrated the application of the OAE framework for a single patient, personalized pharmacovigilance study.
Results
OAE-CTCAE mapping and annotation results
Using our semi-automatic annotation methods, we have established in OAE the OAE-CTCAE mapping for all the CTCAE terms. Many CTCAE terms like “Blood and lymphatic system disorders, Other, specify” are not included in OAE because this term is not specific and does not meet the requirement of generating a new ontology class term. According to our OAE modeling design of AE severity as described earlier, only those grade-specific terms with details more than the general grade definitions were mapped and included in OAE as OAE classes. Those grade-specific CTCAE terms that do not contain additional information than the common Grade definitions (e.g., those terms with Grade 5) were not included in OAE. Overall, we added 1,140 grade level terms to OAE.
We also generated a CTCAE-related OAE subset view (CTCAE-OAEview) that includes all CTCAE-related OAE terms and the OAE terms associated with these CTCAE-related terms. The generation of such OAE subset (or “view”) supports easy usage of the CTCAE-mapped OAE system for cancer drug clinical trials and cancer drug AE case reporting and analysis. In addition, we can better study the CTCAE-related OAE subset to clarify the coverage of the CTCAE system. The CTCAE-OAEview has been deposited on Ontobee (http://www.ontobee.org/ontology/CTCAE-OAEview). CTCAE-OAEview includes 3,267 CTCAE-associated OAE terms. Among these terms, there are 2,169 terms with “OAE_” prefix, 854 terms with “UBERON_” prefix (for anatomic location), and other terms from different ontologies (Figure 4A).
Figure 4.
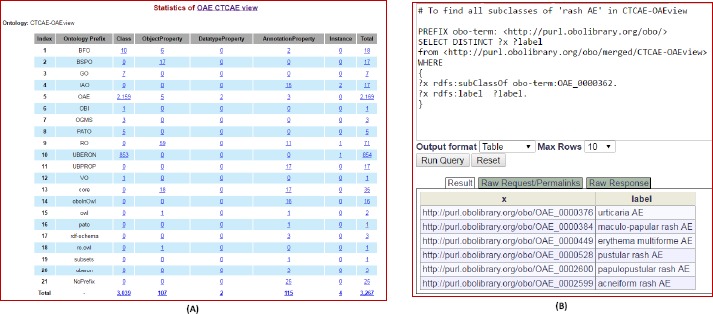
CTCAE-OAEview information and its SPARQL query demonstration. (A) CTCAE-OAEview statistics page information on its Ontobee website (http://www.ontobee.org/ontostat/CTCAE-OAEview). (B) SPARQL query of the CTCAE-OAEview using Ontobee SPARQL website (http://www.ontobee.org/sparql). This query identified all the terms directly under ‘rash AE’ in CTCAE-OAEview.
Use case demonstrating the advantages of OAE-based hierarchical classification
We applied our OAE-CTCAE mapping method to study the AEs reported in a paper published in The Lancet Oncology [22]. Since there was no targeted therapy for KRAS-mutant non-small-cell lung cancer (NSCLC), this clinical study tested whether selumetinib plus docetaxel had a good effect on KRAS-mutant NSCLC. KRAS, encoding for the protein K-Ras that regulates cell division, is the most frequently mutated oncogene in NSCLC. Both selumetinib and docetaxel are antineoplastic drugs. Selumetinib is a potent, selective, orally available, ATP-independent inhibitor of mitogen-activated protein kinase kinase 1 and 2 (MEK1/MEK2), two essential mediators in the activation of the RAS/RAF/MEK/ERK pathway [24]. Inhibition of both MEK1 and 2 by selumetinib prevents cellular proliferation in various cancers. Docetaxel binds to and stabilizes tubulin, thereby inhibiting microtubule disassembly and resulting in cell death. This study found that selumetinib plus docetaxel had promising efficacy but was associated with a higher number of AEs compared to docetaxel alone. Overall, 26 AEs associated with two groups of cancer treatments: selumetinib plus docetaxel, and placebo + docetaxel [22].
After the OAE-CTCAE mapping, we could use the CTCAE and OAE hierarchical structures to classify the 26 AEs and their top-level AEs (Figure 5). Interestingly, CTCAE4 only includes 20 of the 26 terms reported in the paper (Figure 5A) [22], while OAE includes all these 26 AE terms. In addition, our OAE-based AE analysis system demonstrated that OAE at least has the following advantages compared to the CTCAE alone:
Figure 5.
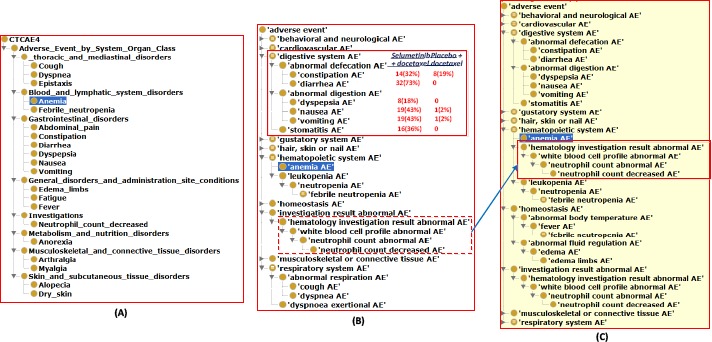
OAE-based classification of adverse events shown in a cancer drug clinical trial. The 26 CTCAE-defined AEs came from Table 8 of the paper [22] were classified using CTCAE or OAE. (A) CTCAE hierarchy of 20 AEs reported in the paper. Note that 6 out of 26 terms in the paper could not be found from CTCAE. (B) Asserted hierarchy of the 26 terms and their associated terms in OAE. (C) Inferred hierarchy using a reasoner in the Protégé OWL editor. See the text for detailed explanation. Note that for (B) and (C), the Ontofox tool (http://ontofox.hegroup.org/) was then used to generate the hierarchical structure of the subset of OAE that contains all the 26 AE terms and the other OAE terms related to these 26 AE terms.
First, OAE provides additional intermediate layers in the AE hierarchy that supports enhanced AE classification. For example, OAE includes an additional layer ‘abnormal defecation AE’ that covers two subclasses - constipation and diarrhea AEs. The combination of these two subclasses in the treatment group makes up to (32% + 73% =) 105% of AE occurrence rates (Figure 5B), suggesting the high susceptibility of this treatment to the abnormal defecation AE. The 105% AE occurrence rate was likely because some patients might have these two AEs simultaneously or at different points in time during the trial. To handle the temporal occurrence of AEs, OAE provides a mechanism to record the time and time period during which an AE occurs (Figure 1) [6].
Second, unlike CTCAE, OAE supports inferred hierarchy generation based on semantic axiom definitions. For example, although OAE does not assert the term ‘hematology investigation result abnormal AE’ as a subclass of ‘hematopoietic system AE’ (Figure 5A), such a subclass relation is inferred using a semantic reasoner (Figure 4C). Note that this term ‘hematology investigation result abnormal AE’ is not an AE term reported in Table 8 of the paper. However, its usage allows the clear classification of this term in OAE.
Third, unlike CTCAE, OAE includes many semantic relations that logically link different AE terms. For example, OAE provides the following axiom: ‘neutrophil count decreased AE’: ‘is evidence of’ some ‘neutropenia AE’. This relation closely links the laboratory count to the neutropenia AE.
In addition, the updated OAE ontology represents different grades of severity. While the contents of defining different grades of severity are the same between OAE and CTCAE, OAE uses its own strategy in representing different grades of severity as detailed earlier in this article. The use case paper summarizes the numbers of cases in Grade 3-4 of 26 different AEs [22]. Such grading results could have been done easily using the OAE grading strategy. As exemplified earlier, one advantage of the OAE grading system is that by measuring some parameter values (e.g., ulcer diameter), OAE will be able to automatically identify the grading level of an AE (e.g., ulceration).
Personal informatics use case demonstrating OAE-based modeling of patient AE after cancer drug treatment
As shown in Figure 1, OAE provides a logical framework that semantically links different variables (e.g., drug features, drug treatment detail, and patient conditions) with the AE outcomes. Each of these variables may affect the final AE outcome. For example, the drug ingredient, the usage of drug, the human quality (e.g., age, gender, concurrent disease, and disease severity) may all affect the outcome of the adverse event.
To show how the OAE-based framework supports practical use case, we used OAE to model a use case of selumetinib treatment on a patient with recurrent low-grade serous ovarian carcinoma (LGSOC) with KRAS mutation [23]. After treatment with selumetinib for more than 7 years, this patient’s tumor has not progressed and the patient has maintained a good general condition without severe toxicities. A next-generation sequencing study showed that her tumor included a G12V mutation in KRAS [23]. Overall, the patient’s LGSOC carcinoma had not progressed, and the patient experienced several mild AEs over the many years of therapy. The most notable AEs were intermittent rashes (grade 2 at the maximum) at anatomically disparate areas including the trunk, chest, and eyelids. She also intermittently experienced grade 1 stomatitis. Figure 6 demonstrates the OAE modeling of the AE process of this LGSOC patient treated with selumetinib.
Figure 6.
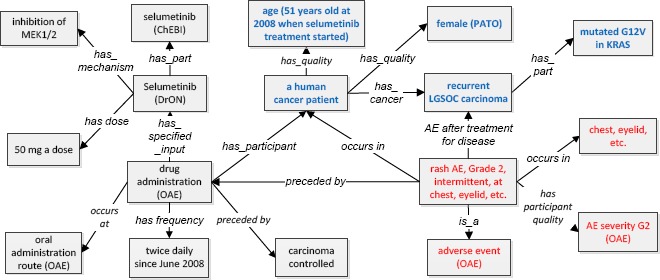
OAE-based instance level modeling of a drug AE from a selumetinib-treated patient. The boxes with blue text indicate patient information. The boxes with red text represent AE-related terms.
In this use case, the patient’s drug AEs were linked with the patient’s background, health condition, drug treatment details, and treatment efficiency (Figure 6). This comprehensive view under the OAE framework provides a systematic understanding of the patient treatment outcomes and possible variable associations. Note that in this model, the “intermittent” in the Grade 2 intermittent rash AE case is not modelled in detail here. This paper does not provide such detail either. For better representation of “intermittent” rash AE, we will need to specify when the rash occurred and then stopped over the course of drug exposure and over the course of the disease process.
Discussion
The major contributions of this study are multiple. First, OAE was updated to include terms representing different grades of AE severity. While such severity definition references the CTCAE definitions, the two definition strategies and their usage still differ in many aspects. Second, we achieved the task of OAE-CTCAE mapping by primarily using manual curation to obtain high accuracy and also using software tools such as Ontorat to facilitate ontology editing process. Note that it is possible to use a natural language processing (NLP) method to achieve part of the manual curation work. However, given the relatively small number of CTCAE terms, we did not perform such a strategy. Third, an OAE view of the CTCAE-related terms (CTCAE-OAEview) was generated, which provides a simplified OAE version to support CTCAE-related cancer drug AE data analysis. Finally, two use cases were applied to demonstrate how the updated OAE system can support AE hierarchical classification and personalized pharmacovigilance research. Future directions of the OAE-CTCAE mapping project include the completion of an OAE-based automatic AE grading classification system.
Since a large number of cancer drug AEs are represented using CTCAE, the CTCAE-OAE mapping allows easy conversion of the CTCAE-reported AE terms to OAE terms, which can then be utilized to support AE hierarchical classification (Figure 5). Based on the first use case study, we demonstrate the advantages of applying OAE to study CTCAE-annotated cancer clinical trial results. Our case study shows that after OAE-CTCAE mapping, OAE can be used to better support AE classification and analysis for cancer clinical trials. Furthermore, since OAE covers more terms than CTCAE, it is possible to apply OAE to leverage the coverage of AEs in cancer clinical trials. In the future, we will investigate more use cases, and provide solutions on best usage of both CTCAE and OAE to support cancer clinical trials.
A major difference between CTCAE and OAE is that CTCAE is a controlled terminology and OAE is an AE-specific ontology. As a controlled terminology, CTCAE includes only simple and loose ‘is a’ hierarchies. In comparison, as an ontology, OAE not only includes ‘is a’ hierarchies, but it also allows the generation of robust axioms that link different terms under different hierarchies [4]. Therefore, unlike CTCAE, OAE provides a platform to support automatic reasoning. As exemplified by the size-based grading ulcerations, Grade 1 ulceration means that the formed ulcer diameter is greater than 0 cm but less than 1 cm. The inclusion of this knowledge as a logical axiom in OAE makes it possible for computers to utilize this and other axioms to automatically reason the Grade 1 ulceration. A reasoning engine can also be developed based on such an ontological system. In the future, we will generate more axioms and based on them to develop an automatic AE grading classification system and semantic reasoning engine.
Personalized informatics, which includes systematic individual data analysis, is critical to personalized medicine. The second use case provided in this report demonstrates that the OAE modeling offers a logic linkage of different variables of this cancer patient. Such a modeling provides the ability to integrate various types of data, such as general patient information (sex, gender), medical condition (e.g., concurrent disease, and disease severity), drug name, drug dose, treatment route, time of drug treatment, or AE symptom occurrence, into a single comprehensive picture. Such personalized information illustration is different from the traditional method of summarizing results from pooled patient records. When many patients’ records are available, it is possible to conduct such personalized record organization for each patient, and develop new statistical methods to better analyze the relations among variables in different patients. Such a strategy would promote personalized medicine and public health.
Conclusion
The OAE ontology was updated to ontologically represent different AE severity grades and mapped to CTCAE. An OAE view of the CTCAE-related terms (CTCAE-OAEview) was generated to support better usage of the mapped information in OAE. Two use cases were applied to demonstrate that the OAE ontology can support cancer drug AE hierarchical classification and provide a semantic framework for personalized AE information representation and analysis.
Disclaimer
This article reflects the views of the authors and should not be construed to represent FDA’s views or policies.
Acknowledgements
We appreciate Dr. Darrell Abernethy’s insightful edits and comments on this manuscript. We also appreciate Dr. Sirarat Sarntivijai’s discussion and comments.
References
- 1.Brown E.G., Wood L., Wood S. ‘The medical dictionary for regulatory activities (MedDRA)’. Drug safety. 1999;20(2):109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 2. https://www.umc-products.com/graphics/28010.pdf.
- 3. [access on October 27, 2016]. http://evs.nci.nih.gov/ftp1/CTCAE/About.html.
- 4.He Y. ‘Ontology-based vaccine and drug adverse event representation and theory-guided systematic causal network analysis toward integrative pharmacovigilance research’. Curr Pharmacol Rep. 2016;2(3):113128. doi: 10.1007/s40495-016-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhichkin P.E., Athey B.D., Avigan M.I., Abernethy D.R. ‘Needs for an expanded ontology-based classification of adverse drug reactions and related mechanisms’. Clin Pharmacol Ther. 2012;91(6):963–965. doi: 10.1038/clpt.2012.41. [DOI] [PubMed] [Google Scholar]
- 6.He Y., Sarntivijai S., Lin Y., Xiang Z., Guo A., Zhang S., Jagannathan D., Toldo L., Tao C., Smith B. ‘OAE: The Ontology of Adverse Events’. Journal of biomedical semantics. 2014;5:29. doi: 10.1186/2041-1480-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen A.P., Setser A., Anadkat M.J., Cotliar J., Olsen E.A., Garden B.C., Lacouture M.E. ‘Grading dermatologic adverse events of cancer treatments: the Common Terminology Criteria for Adverse Events Version 4.0’. Journal of the American Academy of Dermatology. 2012;67(5):1025–1039. doi: 10.1016/j.jaad.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Sarntivijai S., Xiang Z., Shedden K.A., Markel H., Omenn G.S., Athey B.D., He Y. ‘Ontology- based combinatorial comparative analysis of adverse events associated with killed and live influenza vaccines’. PloS one. 2012;7(11):e49941. doi: 10.1371/journal.pone.0049941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie J., Codd C., Mo K., He Y. ‘Differential adverse event profiles associated with BCG as a preventive tuberculosis vaccine or therapeutic bladder cancer vaccine identified by comparative ontology-based VAERS and literature meta-analysis’. PloS one. 2016;11(10):e0164792. doi: 10.1371/journal.pone.0164792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie J., Zhao L., Zhou S., He Y. ‘Statistical and ontological analysis of adverse events associated with monovalent and combination vaccines against hepatitis A and B diseases’. Scientific reports. 2016;6:34318. doi: 10.1038/srep34318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arp R., Smith B., Spear A.D. 2015. ‘Building Ontologies Using Basic Formal Ontology’. Cambridge, MA, USA 2015. [Google Scholar]
- 12.Bandrowski A., Brinkman R., Brochhausen M., Brush M.H., Bug B., Chibucos M.C., Clancy K., Courtot M., Derom D., Dumontier M., Fan L., Fostel J., Fragoso G., Gibson F., Gonzalez-Beltran A., Haendel M.A., He Y., Heiskanen M., Hernandez-Boussard T., Jensen M., Lin Y., Lister A.L., Lord P., Malone J., Manduchi E., McGee M., Morrison N., Overton J.A., Parkinson H., Peters B., Rocca-Serra P., Ruttenberg A., Sansone S.A., Scheuermann R.H., Schober D., Smith B., Soldatova L.N., Stoeckert C.J., Jr., Taylor C.F., Torniai C., Turner J.A., Vita R., Whetzel P.L., Zheng J. ‘The Ontology for Biomedical Investigations’. PloS one. 2016;11(4):e0154556. doi: 10.1371/journal.pone.0154556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. https://github.com/OGMS.
- 14.Schriml L.M., Arze C., Nadendla S., Chang Y.W., Mazaitis M., Felix V., Feng G., Kibbe W.A. ‘Disease Ontology: a backbone for disease semantic integration’. Nucleic acids research. 2012;40(Database issue):D940–946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanna J., Joseph E., Brochhausen M., Hogan W.R. ‘Building a drug ontology based on RxNo rm and other sources’. Journal of biomedical semantics. 2013;4(1):44. doi: 10.1186/2041-1480-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. https://github.com/pato-ontology/pato/
- 17.Mungall C.J., Torniai C., Gkoutos G.V., Lewis S.E., Haendel M.A. ‘Uberon, an integrative multi species anatomy ontology’. Genome biology. 2012;13(1):R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. http://obofoundry.org/ontology/ncbitaxon.html.
- 19.Xiang Z., Zheng J., Lin Y., He Y. ‘Ontorat: Automatic generation of new ontology terms, annotations, and axioms based on ontology design patterns’. Journal of biomedical semantics. 2015;6(1):4. doi: 10.1186/2041-1480-6-4. 10 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang Z., Courtot M., Brinkman R.R., Ruttenberg A., He Y. ‘OntoFox: web-based support for ontology reuse’. BMC research notes. 2010;3(175):1–12. doi: 10.1186/1756-0500-3-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong E., Xiang Z., Zhao B., Liu Y., Lin Y., Zheng J., Mungall C., Courtot M., Ruttenberg A., He Y. ‘Ontobee: A linked ontology data server to support ontology term dereferencing, linkage, query and integration’. Nucleic acids research. 2017;45(D1):D347–D352. doi: 10.1093/nar/gkw918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janne P.A., Shaw A.T., Pereira J.R., Jeannin G., Vansteenkiste J., Barrios C., Franke F.A., Grinsted L., Zazulina V., Smith P., Smith I., Crino L. ‘Selumetinib plus docetaxel for KRAS-mutant advanced nonsmall-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study’. The Lancet. Oncology. 2013;14(1):38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 23.Takekuma M., Wong K.K., Coleman R.L. ‘A long-term surviving patient with recurrent low-grade serous ovarian carcinoma treated with the MEK1/2 inhibitor, selumetinib’. Gynecologic oncology research and practice. 2016;3:5. doi: 10.1186/s40661-016-0026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller C.R., Oliver K.E., Farley J.H. ‘MEK1/2 inhibitors in the treatment of gynecologic malignancies’. Gynecologic oncology. 2014;133(1):128–137. doi: 10.1016/j.ygyno.2014.01.008. [DOI] [PubMed] [Google Scholar]


