Abstract
Research data warehouses integrate research and patient data from one or more sources into a single data model that is designed for research. Typically, institutions update their warehouse by fully reloading it periodically. The alternative is to update the warehouse incrementally with new, changed and/or deleted data. Full reloads avoid having to correct and add to a live system, but they can render the data outdated for clinical trial accrual. They place a substantial burden on source systems, involve intermittent work that is challenging to resource, and may involve tight coordination across IT and informatics units. We have implemented daily incremental updating for our i2b2 data warehouse. Incremental updating requires substantial up-front development, and it can expose provisional data to investigators. However, it may support more use cases, it may be a better fit for academic healthcare IT organizational structures, and ongoing support needs appear to be similar or lower.
Introduction
Research data warehouses typically provide access to data from the electronic health record (EHR) and administrative systems, potentially linked to other sources such as a cancer registry or death index.1–3 These systems frequently employ relational database management for data storage and query, and they may be paired with business intelligence software that supports constructing queries and reports through a graphical user interface.4 Research use cases include cohort discovery for assessing study feasibility, identifying potential participants for studies, retrospective analyses, developing predictive models, testing novel healthcare software, and providing data access to multi-institutional research data networks. While trained data analysts usually run queries and retrieve data on behalf of investigators, increasingly, systems such as i2b25 provide self-service interfaces for constructing some queries such as getting patient counts that meet specified inclusion and exclusion criteria.6,7
Research data warehouse approaches have primarily focused on the data models and terms that are used. The National Patient-Centered Clinical Research Network (PCORnet) common data model8,9 supports a limited subset of EHR and administrative data in a standard form for aggregation across large research data networks. The Observational Medical Outcomes Partnership (OMOP) common data model10 supports a broader variety of data and a set of statistical analysis tools that can operate on local data that are loaded into the model. The Accrual to Clinical Trials (ACT) common data model11 is similar to that of PCORnet, with minor modifications to support rapidly finding institutions with potential participants of interest for a clinical trial. In these approaches, the actual process of loading data from source systems into a research data warehouse, called Extract, Transform and Load (ETL), is largely left as an implementation detail. As a consequence, there is a dearth of published best practices on how to implement ETL in an academic health system environment to support the research use cases above. ETL implementations tend to be specific to organizations due to their specific combination of EHR and other data systems, but the architectural choices do not have to be.
In an effort to contribute to best practices in this area, we address a key architectural question for an ETL process: whether to implement an ETL process that fully reloads the entire warehouse weekly, monthly or quarterly; or an incremental loading process that refreshes the warehouse without reloading on a far more frequent basis such as daily. While almost nothing is published on this choice in healthcare, anecdotally our institution appears to be highly unusual in its decision to implement an incremental process for i2b2. Full reload is relatively simple to implement because there is no need to migrate live data from one version of the ETL process to the next, such as when new subject areas are added or data issues are discovered. In our environment, we initially attempted the full reload approach. The process took over a week, and it required ETL developer and database administrator time from multiple parts of the organization, including our Clinical and Translational Science Award (CTSA)12 program’s biomedical informatics core, healthcare IT and research IT. The intermittent nature of the work made freeing up staff from multiple parts of the organization simultaneously to perform the reload a challenge.
As a result of this initial experience, we implemented an incremental updating process that is entirely automated and is designed to require only a basic level of ongoing support from outside of our research IT and informatics units. This choice traded a larger upfront development effort for a far smoother maintenance phase. We describe below our incremental updating methodology, including the types of data changes that it supports during its daily refresh process, and the types of data additions and corrections that it permits. We report on the performance of the system since it was deployed in October 2016, and we describe the effort required to build and maintain it as compared with a full reload process.
Background
Research data warehouses are typically implemented using relatively standard approaches that were adopted from the IT industry. They use a relational database for the storage itself, and a commercial, open source or locally developed set of procedures for the ETL. The data models employed in most warehouses follow standard design patterns. ETL systems are generally designed to support these patterns, and the physical model stores an audit trail of the ETL process’s work. In the case of ETL systems that provide incremental updating, the physical model also supports determining which records need to be altered during the updating process. Eureka! Clinical Analytics, our locally developed open source ETL system for i2b2,13 follows these design patterns, and we recently extended it to support incremental updating as described in Methods. While incremental updating of a data warehouse in general is not novel (see 14 for an example in the clinical domain), its application to i2b2 research data warehouses appears to be unusual, thus the pros and cons of it in an academic healthcare environment are not widely known.
ETL systems15 implement three sequential functions. They extract data from a source system, typically as flat files, or by copying selected tables from a source database into a database with a similar schema, called a staging database. Next, they transform the source system data from a local data model with local codes into a standardized data model with standardized codes. Finally, they load the transformed data into the target warehouse database. ETL solutions typically involve specifying these functions as a flow diagram of procedural steps, or more recently, as declarative metadata that map the source schema and semantics to the target schema and semantics.16 Eureka implements the latter mechanism using an augmented i2b2 metadata schema as its metadata repository.13 As described above, the procedural steps in an ETL process tend to be highly site-specific. The Observational Health Data Sciences and Informatics (OHDSI) community,17 which maintains the OMOP common data model, has published scripts that populate an OMOP schema from some commercial EHR systems. Similarly, the Greater Plains Collaborative, a PCORnet network that uses i2b2, has published many of their ETL scripts as open source.18 While these code resources can be very helpful, they ultimately are a starting point for creating an ETL process that takes local variation into account.
A widely-adopted data warehouse methodology, developed by Kimball,15 prescribes modeling a data warehouse as a star schema, shown in Figure 1. A star schema contains a central fact table and a collection of dimension tables that model attributes of each fact. Foreign key relationships assign each fact a value from each of the dimensions. Facts are uniquely identified by their combination of dimension values. In general, i2b2 adopts this approach.5 The i2b2 fact table contains a row for each diagnosis, procedure, laboratory test, and so forth. Each fact is linked by foreign keys to patients, visits, providers, metadata about the concept represented by the fact (lab test, diagnosis code, and so on), and metadata about attributes associated with the fact called modifiers (for example, a diagnosis’ priority). The dimensions have additional values describing the patients, visits, and providers such as birth date and race. The Kimball approach may be extended by composing a warehouse out of multiple star schemas that share dimension tables. ETL systems, including Eureka, are typically architected to support star schemas.
Figure 1.
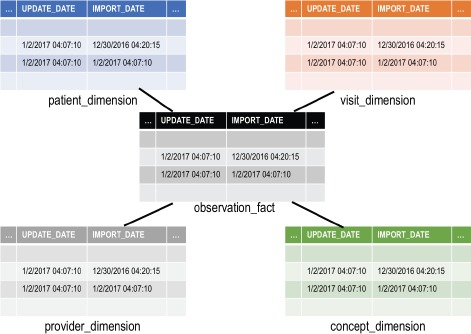
The primary tables of the i2b2 star schema, with their auditing columns shown. Not shown are the modifier_dimension table, which stores metadata on concept modifiers, and the encounter_mapping and patient_mapping tables, which map generated patient and visit numbers to local identifiers. The latter three tables also have the same auditing columns.
The i2b2 star schema has “auditing” columns in each fact and dimension table, illustrated in Figure 1, that can support an ETL process that implements incremental updating. The import_date column specifies when the row was inserted into i2b2, and the update_date column specifies when the row was last updated by the source system. Warehouses sometimes also have an auditing column for specifying when a record was logically deleted, thus keeping a historical record of the last state of the fact. I2b2 lacks this column, thus an ETL process would physically delete a row when it is removed from the source system. Some data warehouses additionally employ an active record indicator column, which contains a numerical value indicating whether a record is not deleted, physically deleted, or logically deleted from the source system. The audit columns, in combination with a row’s unique combination of dimension values, can be compared to a record from a source system to determine if it is a new record or an update to an existing record.
Eureka, our locally developed i2b2 ETL system, is a collection of Java web services that implement loading clinical data from an existing database into i2b2. It is a successor to the Analytic Information Warehouse system, also developed at our institution.19 In addition to ETL services, it provides computing EHR phenotypes20 during the ETL process that reflect sequence, frequency, and threshold patterns in the source system data. Eureka is the central component of a metadata-driven architecture that we are implementing for our CTSA that will make local clinical data sources and EHR phenotyping available to investigators through i2b2 projects, as shown in Figure 2. This architecture is designed to support maintaining our i2b2-based study feasibility service, launched in October 2016, which allows querying for patient counts across most of our organization’s patients. It also is designed to support future creation of study-specific data marts containing data from a cohort of interest that is customized for the study. We have described prototypes of this service elsewhere.21,22
Figure 2.
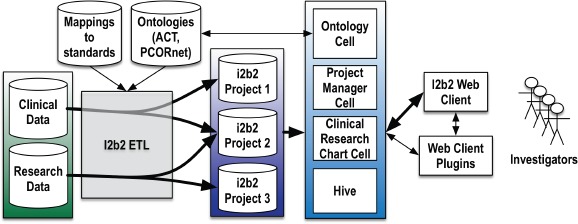
Eureka-based architecture for providing i2b2 clinical databases to investigators using standard tools and data models.
In architecting our i2b2 ETL process, we had concerns about implementing full reload. Each i2b2 project would require its own reload on a schedule appropriate for its users such as weekly, monthly, or quarterly. In the worst case, we could have an almost continuous process of reloading i2b2 projects, which would be unacceptable to the source system managers. In order to avoid significant i2b2 downtime during each reload, we would need two copies of each i2b2 project so that we could reload the inactive copy and swap it with the active copy. Maintaining two copies increases storage requirements, and requires all database optimizations like indexes and statistics gathering to be performed twice for each project.
Several additional factors figured into our decision to implement an incremental updating process. The high data volume involved in full reloads makes them vulnerable to network errors, enterprise-level scheduled maintenance that may involve restarting network services, and other glitches that may halt the process part-way through. Full reload ETL processes typically involve a collection of database and ETL-specific tools for exporting, transforming and importing data, which may have limited support for orchestration into a single automated process that is resilient to these errors. The semi-manual alternative involves close coordination of IT personnel from across the organization, which can be challenging in healthcare because clinical and operational needs typically are higher priority for the personnel managing clinical data systems. Because full reloads can take days to complete for academic health system data volumes (millions of patients), and they can place substantial load on source systems, personnel in healthcare IT may be reluctant to allow them to occur on a regular basis. We believe that an incremental updating approach would be more resilient to errors due to the lower data volume per load, would be easier to schedule during times when systems that are relied on by healthcare operations are lightly used, and thus would be more amenable to an automated approach that would loosely couple the systems managed by healthcare personnel from those managed by research IT and informatics personnel.
Methods
Clinical data flow into i2b2
A locally developed clinical data warehouse serves as an integration point for data from our EHR (Cerner Millennium), administrative systems, and other transactional systems that support healthcare operations. It is implemented using the Oracle 11g relational database management system. PL*SQL stored procedures and scripts, not described here, update it daily from source systems. The warehouse supports access for research, quality improvement and business needs through a commercial business intelligence system (Microstrategy). It also provides data to downstream data marts including our i2b2 deployment. It has median daily volume of 12,258 new encounters (interquartile range [IQR], 824–14,560) for 10,158 patients (IQR, 804–12,000) plus any updates to older data.
Our i2b2 ETL process uses this clinical data warehouse as its source system. Figure 3 illustrates the data flow from the clinical data warehouse into i2b2. The ETL process into i2b2 is implemented in two parts. It copies data from the clinical data warehouse into a relational staging database with a subset of the clinical data warehouse’s schema. The Eureka system queries the staging database, maps codes to standards, and transforms the data into the ACT common data model. It loads the transformed and mapped data into a collection of tables for temporary data storage. Stored procedures then load the data from those tables into i2b2 as a full-reload or as an incremental update as described below.
Figure 3.
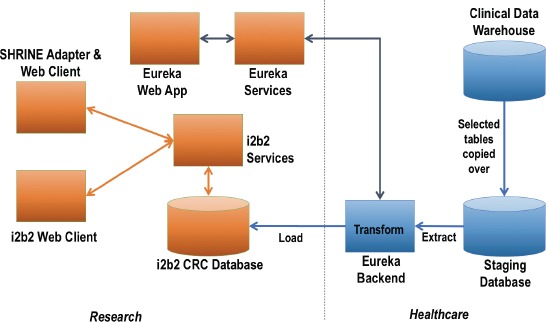
Data flow from our clinical data warehouse into i2b2.
Full reload approach
We described the full reload process previously.13 Briefly, selected tables from our clinical data warehouse (Figure 3) were exported to files using Oracle’s built-in utilities and imported into a staging database. The staging database was indexed and statistics were collected. Next, the Eureka ETL process (Eureka Backend in Figure 3) truncated and reloaded the i2b2 star schema (i2b2 CRC Database in Figure 3) in six month batches by visit end date. The staging database reload requires healthcare IT database administrator time to start and monitor the process and correct any issues. The i2b2 reload requires CTSA engineering time to start, monitor and validate each batch.
Incremental update approach
The incremental updating process depends on auditing columns in the central clinical data warehouse, the staging database and the i2b2 star schema; and an extra “flagging” column in each table of the clinical data warehouse and the staging database called extract_ind_daily. The process is illustrated in Figure 4. The flagging column has the value 1 for records that are changed and 0 for records that are unchanged. When records are inserted, updated or logically deleted in the clinical data warehouse, the value of this column is set to 1 for those records. For logically deleted records, the delete timestamp audit column is set, and the active record indicator column is set to an inactive value.
Figure 4.
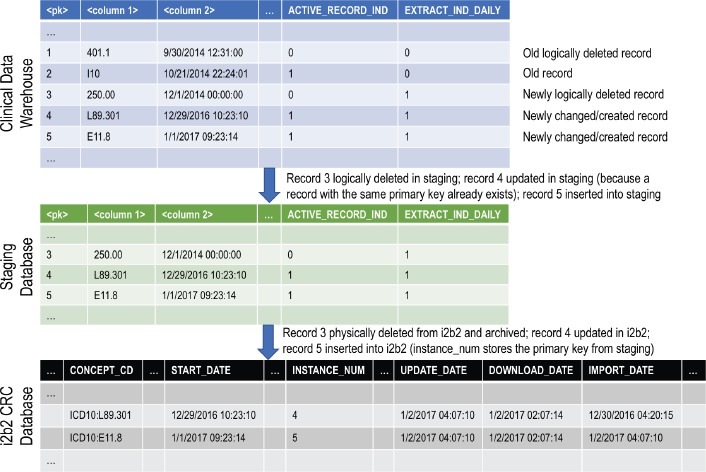
How the auditing and flagging columns flow from the clinical data warehouse through to the i2b2 database during incremental updating. <pk>=primary key column, <column 1>=data column, <column 2>=data column.
The clinical data warehouse’s ETL cycle runs overnight starting at midnight and ending at between 6 and 9am. Locally developed functions allow programmatically checking whether or not the cycle is running. The new work described in this paper is the implementation of incremental updating of the staging database (staging ETL cycle) followed by incremental updating of the i2b2 star schema (i2b2 ETL cycle).
i2b2 ETL registration
There is a simple lock file-based registration process in which each of the two steps of the ETL process (staging ETL cycle, i2b2 ETL cycle) registers itself as running so that the other step cannot run simultaneously. This prevents corrupted data in the i2b2 star schema if the two steps were to run accidently in parallel.
Updating the staging database
A script checks automatically at half-hour intervals every morning whether the clinical data warehouse’s ETL cycle is completed. The script then launches the staging ETL cycle.
A collection of stored procedures selects all records in the clinical data warehouse that are “flagged” (have the extract_ind_daily flag set to 1) in the tables of interest, deletes any existing versions of those records from the staging database based on each record’s primary key, and inserts new copies. The flagging column is copied into the staging database, so the newly inserted records all have the staging database’s flagging column set to 1 as well. Upon successful completion of this process, a stored procedure is invoked in the clinical data warehouse that sets the flagging column to 0 for all previously flagged rows. The number of records inserted into each staging database table is logged. If an error occurs during the updating of the staging database, the unflagging step is skipped, and the whole process can be rerun safely without corrupting the staging database. If the clinical data warehouse’s ETL cycle does not finish by noon, which rarely occurs, the population of the staging database is postponed until the following morning. The next time the staging ETL cycle is run, multiple days of updates will be loaded into the staging database. During beta testing, we introduced a week-long delay in running the staging ETL cycle, and the system performed normally, albeit with a longer runtime.
Updating the i2b2 database
At midnight, the i2b2 ETL cycle commences automatically. If there are flagged records in the staging database, Eureka performs the following steps:
Retrieve all configuration metadata.
Generate and execute SQL to retrieve data from the staging database.
Map local codes to standard codes.
Transform retrieved data into the ACT common data model.
Load mapped and transformed data into temporary tables in the i2b2 schema.
Generate i2b2 patient numbers and encounter numbers for new patients and encounters.
Copy records that violate i2b2 schema constraints into a “failed record” table.
Copy records that are marked for deletion into an archived record table.
Insert and update valid dimension records in the i2b2 dimension tables, and delete records that are marked for deletion.
Insert and update valid fact and modifier records into the i2b2 fact table, and delete records that are marked for deletion.
Log changed record counts and query timings.
Adding new data types/codes to the data warehouse
In addition to the normal operations described above, the process supports adding new codes and subject areas as part of the regular daily cycle. Assuming the appropriate extract_ind_daily columns are implemented in the clinical data warehouse for the tables containing the new data, the schema of the staging database includes any new tables that are needed, and all of the new data is flagged in the clinical data warehouse, the new data will load into the staging database during the next staging ETL cycle. For the i2b2 ETL cycle, if code changes to Eureka are required, we redeploy the new code using Ansible deployment automation software.23 We execute SQL manually in the staging database to set extract_ind_daily to 1 for the new data, if the data was not flagged by the staging ETL cycle, and the new data will load into i2b2 during the next i2b2 ETL cycle.
Addressing data issues
Similarly, Eureka supports correcting data issues in i2b2 by reflagging the data to be corrected in the staging database or the clinical data warehouse, depending on the nature of the data issue. The normal staging and i2b2 ETL cycles will update i2b2 after deployment of any code changes to the staging ETL cycle or Eureka.
Data that is in error in the clinical data warehouse will be updated in i2b2 entirely automatically through the flagging system after the clinical data warehouse team corrects the issue, or the issue is corrected in the clinical data warehouse’s source systems.
If data is inadvertently not loaded into i2b2, but the flags in the staging database and clinical data warehouse are unset, we can send a request to the clinical data warehouse team to reflag all rows that are changed after a specified date. The next staging and i2b2 ETL cycles will load the data into i2b2.
In the i2b2 ETL cycle, facts and their associated modifiers are managed as a unit, so corrections to modifiers involve flagging their associated facts in the staging database tables, after which the facts and their modifiers will all be reloaded during the next cycle.
Data that is deleted from i2b2 in error can be corrected by flagging the corresponding records in the staging database and correcting and redeploying Eureka, after which the data will be reinserted during the next i2b2 ETL cycle.
Errors due to i2b2 metadata issues and incorrect mappings from local codes to standard codes have been the most common data issues that we have encountered. There are three general scenarios:
A local code has a theoretical mapping to a standard code but is not mapped in Eureka: in this scenario, the data with the unmapped local codes would not be loaded. We deploy an update to Eureka with updated mappings, and we flag all records in the staging database that have the affected local code. Those records will be loaded into i2b2 during the next i2b2 ETL cycle.
Data is mapped correctly to a standard code that is present in the i2b2 metadata schema, but a metadata issue causes the data not to be loaded: we correct the i2b2 metadata manually, and flag all records in the staging database with the affected local codes. Those records will be loaded into i2b2 during the next i2b2 ETL cycle.
A local code is mapped to the wrong standard code: if the data with the local codes should not have been loaded into i2b2 at all, those records must be deleted from i2b2 by executing SQL statements manually. Otherwise, we update the mapping in Eureka, redeploy Eureka, and set the flags for the corresponding records in the staging database to 1. The updated records will load into i2b2 during the next i2b2 ETL cycle.
These workflows for correcting data issues support a systematic approach that mostly avoids manual changes to the i2b2 warehouse data. In general, the system is capable of inserting, updating and deleting about 8 million flagged records daily, depending on the database hardware. For higher data volumes, the i2b2 ETL cycle may continue running into normal business hours and impact performance in a user-visible fashion. If more data requires updating, records could be divided by date or other criteria and flagged sequentially over multiple days.
Results
The staging and i2b2 databases are Oracle 11g databases running on Linux. Eureka runs on Oracle Java 8 on Linux. Eureka is available as open source under the Apache 2 software license (source code available at https://github.com/eurekaclinical). The Eureka server executes the staging ETL cycle and the i2b2 ETL cycle. The staging ETL cycle and i2b2 ETL cycle have been operational since October 2016. Initially, the staging and i2b2 databases were populated using the full reload process described above. We then activated the incremental process. As of February 2017, our i2b2 contains 294 million facts from 23 million encounters on 2.7 million patients with discharges since 2011.
Full reload implementation — level of effort and time required
The initial population of the staging database took approximately 72 hours. The population of the i2b2 database took approximately 10 hours for each 6-month increment. In total, the process took more than five days of data processing time, assuming it were running continuously. In practice, the process took almost 1.5 weeks because each step had to be verified manually, and some steps had to be restarted due to network glitches during a long-running query. If we were to fully reload i2b2 periodically, the process would require 24 hours of effort to reload the staging database and 16 hours of effort to reload the i2b2 database.
Incremental updating implementation — level of effort and time required
Implementation of the incremental updating staging ETL cycle and i2b2 ETL cycle took 914 work hours. This level of effort includes project management; oversight from CTSA leadership and the leadership of our central clinical data warehouse; design; implementation of flagging columns in the clinical data warehouse and the staging database; extension of Eureka to support incremental updating; integration testing; and deployment. We have submitted no requests for support from healthcare IT to correct issues or reflag records since the system went into production.
Incremental updating performance
From October 2016 to February 2017, median staging ETL cycle time was 33 minutes (IQR, 13–38 minutes). Median i2b2 ETL cycle time was 51 minutes (IQR, 34–161 minutes). Data volumes by subject area are shown in Table 1. Overall, data from a median of 113,863 patients (IQR, 68,406–125,611) and 116,515 financial encounters (IQR, 66,285–132,174) were updated daily by the ETL cycle, which is ten times as many encounters as occur daily and ten times as many patients as are seen daily (see Methods).
Table 1.
Staging ETL cycle performance by subject area for daily cycles from October 2016 to February 2017.
| Subject Area | Median nightly record count (interquartile range) |
|---|---|
| Demographics | 22,269 (5,994–25,094) |
| Visit details | 53,063 (28,435–58,876) |
| Diagnosis codes | 296,105 (58,220–337,952) |
| Procedure codes | 25,787 (300–28,897) |
| Laboratory test results | 234,433 (97,996–253,424) |
| Medication orders | 205,253 (153,454–221,595) |
Discussion
Our institution has implemented an i2b2 research data warehouse with close to 300 million facts on 2.7 million patients. It is populated from a locally developed clinical data warehouse that itself is populated from our EHR and administrative systems (Figure 3). Both the clinical data warehouse and i2b2 are refreshed daily. The ETL process for i2b2 uses a method based on auditing columns, which is described above (Figure 4), to identify new, updated and logically deleted records for insert, update and delete in i2b2. There is a two-day time lag from when data is recorded in our EHR and administrative systems to when it appears in i2b2 for query by investigators (see Methods). This i2b2 environment has provided an opportunity to characterize the performance of an incremental updating ETL system for i2b2 at a major academic health system in order to assess this approach’s benefits and drawbacks as compared with the more typical full reload ETL approach.
Implementing an incremental updating ETL process is a major undertaking, requiring over 900 hours of one-time effort at our institution. By comparison, our experience indicates that a full reload process would require 40 hours of effort per reload. If we assume a monthly full reload process as the alternative to incremental updating, we will recoup the costs of the incremental updating implementation after approximately 2 years. The full reload process is more difficult to staff, because it requires coordinated bursts of resources potentially from both healthcare IT and academic IT personnel, access to which may compete with healthcare business needs. In our environment, the incremental updating implementation has required no i2b2-specific effort from healthcare IT personnel since it went into production. Maintenance of the process has been performed entirely by research IT and CTSA staff. Because our incremental updating process is in its first year of production, we cannot yet assess the extent to which substantial ETL logic updates may be needed in excess of what might be required as part of a full reload implementation.
Our experience indicates that level of effort and IT organizational structure are important factors in determining whether to use incremental updating or full reload for a research data warehouse. While ETL is often perceived as a decidedly backroom operation, in academic health environments, it sits at the intersection of complex organizational structures that, if not taken into account, can foil the best technical implementation. In our environment, three organizational units are involving in the i2b2 ETL process (research IT, healthcare IT, and CTSA). Due to the challenges of coordinating resources across them, we believe that the larger upfront effort required to implement incremental updating is worth it.
In addition to potential long-term effort savings, incremental updating provides more timely refreshes of the warehouse from source systems that may allow the warehouse to serve more use cases. We suspect that healthcare operations would primarily benefit from data that is refreshed with a one-day time lag, which is provided by our clinical data warehouse but not by our i2b2 at present. Similarly, data that is refreshed with a short time lag would be of substantial benefit to clinical trial accrual efforts that aim to find potential participants who are currently hospitalized. While our i2b2 captures hospitalizations that are in progress at the start of ETL, assuming a typical 48- to 72-hour length of stay, our current time lag is too long for some studies. Refinement of the ETL processes of our data warehouse and i2b2 likely could reduce the overall time lag to one day. Further reducing the time lag may involve adopting alternative database technologies that better support frequent data updates while users are running queries.
The incremental updating process captured a large number of updates that occur to transactional EHR and administrative data after they are recorded initially. We would expect these updates to include coding of encounters for billing purposes, which impacts data from many subject areas (diagnoses, procedures, discharge status, and more); and laboratory testing that may take multiple days to finalize. Most investigators only care about final test results, and the diagnoses, procedures and other codes that are recorded in the final bill. The extent to which these provisional data values might alter query results is unclear. Characterization of the rate at which different subject areas become finalized could help with building into the incremental updating process a subject area-specific lag time that allows data to “mature” before it becomes available for query. There may be tradeoffs between supporting clinical trial accrual and waiting until data likely has stopped being updated.
Incremental updating relies on source systems to provide a unique identifier for records that is consistent across refreshes for the updating and deleting processes. If a unique identifier is not available, the ETL process must implement its own logic for generating and maintaining consistent unique identifiers. As a result, full reload may be the only option for some warehouses that contain fully de-identified data unless the ETL process maintains metadata that supports consistently transforming patient identifiers into the same de-identified values. For example, an i2b2 ETL process could securely maintain a per-patient configuration for hashing algorithms, and a random temporal offset per patient for altering timestamps. Our i2b2 warehouse has a limited dataset as defined by HIPAA and omits identifiers that would disqualify the data from being a limited dataset, so we avoided this issue.
Thus, limitations of incremental updating for an i2b2 research data warehouse include de-identification challenges, exposure of provisional data, and large up-front effort. A potential additional limitation is database fragmentation over time. Full reload processes may more easily take advantage of database management system-specific features that permit optimizing the storage of database rows on disk to avoid wasting disk space or slowing queries as a result of seeks across empty disk blocks. It is possible that fragmentation will result in performance degradation over time. Database management systems may provide tools for addressing this, but they may require downtime. Our i2b2 has not been deployed for long enough to assess the likelihood of any performance degradation becoming noticeable.
Conclusion
Incrementally updating a research data warehouse such as i2b2 has substantial advantages in the academic healthcare environment, including more timely refreshes of the warehouse with new and updated data, greater automation of ETL tasks, and fit with often complex organizational structures around research IT and informatics. While the upfront level of effort required to implement incremental updating is substantial, subsequent ongoing support requirements are anticipated to be similar or smaller. Further characterization is needed of the impact on research use cases of the large volume of updates that occurs to data after initial loading into the warehouse.
Acknowledgements
This work has been supported by PHS grant UL1 TR000454 from the CTSA program, NIH, NCATS; NCI grant U01 CA187013l; NHLBI grant R24 HL085343; and Emory University Library and Information Technology Services.
References
- 1.Chelico JD, Wilcox A, Wajngurt D. Architectural design of a data warehouse to support operational and analytical queries across disparate clinical databases. AMIA Annu Symp Proc. 2007:901. [PubMed] [Google Scholar]
- 2.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010;17(2):131–5. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy SN, Morgan MM, Barnett GO, Chueh HC. Optimizing healthcare research data warehouse design through past COSTAR query analysis. Proc AMIA Symp. 1999:892–6. [PMC free article] [PubMed] [Google Scholar]
- 4.Ferranti JM, Langman MK, Tanaka D, McCall J, Ahmad A. Bridging the gap: leveraging business intelligence tools in support of patient safety and financial effectiveness. J Am Med Inform Assoc. 2010;17(2):136–43. doi: 10.1136/jamia.2009.002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE--An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc. 2009:391–5. [PMC free article] [PubMed] [Google Scholar]
- 7.Lyman JA, Scully K, Harrison JH., Jr The development of health care data warehouses to support data mining. Clin Lab Med. 2008;28(1):55–71. doi: 10.1016/j.cll.2007.10.003. vi. [DOI] [PubMed] [Google Scholar]
- 8.PCORnet. Common Data Model (CDM) Specification, Version 3.0. 2015. [cited 2016 Jan 5]. Available from: http://www.pcornet.org/wp-content/uploads/2014/07/2015-07-29-PCORnet-Common-Data-Model-v3dot0-RELEASE.pdf.
- 9.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21(4):576–7. doi: 10.1136/amiajnl-2014-002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Observational Health Data Sciences and Informatics (OHDSI) OMOP Common Data Model. 2016. [cited 2016 Jan 5]. Available from: http://www.ohdsi.org/data-standardization/the-common-data-model/
- 11.ACT Network. [cited 2016 Apr 22]; Available from: https://www.act-network.org/ [Google Scholar]
- 12.Leshner AI, Terry SF, Schultz AM, Liverman CT, editors. The CTSA Program at NIH: Opportunities for Advancing Clinical and Translational Research. Washington (DC): National Academies Press (US), National Academy of Sciences; 2013. Committee to Review the Clinical and Translational Science Awards Program. [PubMed] [Google Scholar]
- 13.Post AR, Pai AK, Willard R, May BJ, West AC, Agravat S, et al. Metadata-driven Clinical Data Loading into i2b2 for Clinical and Translational Science Institutes. AMIA Summits Transl Sci Proc. 2016:184–93. [PMC free article] [PubMed] [Google Scholar]
- 14.Evans RS, Lloyd JF, Pierce LA. Clinical use of an enterprise data warehouse. AMIA Annu Symp Proc. 2012;2012:189–98. [PMC free article] [PubMed] [Google Scholar]
- 15.Kimball R, Ross M. 2nd ed. New York: Wiley Computer Publishing; 2002. The Data Warehouse Toolkit: The Complete Guide to Dimensional Modeling. [Google Scholar]
- 16.He T, Gudyka M. Build a Metadata-Driven ETL Platform by Extending Microsoft SQL Server Integration Services. 2008. [cited 2016 Jan 5]. Available from: http://download.microsoft.com/download/D/2/0/D20E1C5F-72EA-4505-9F26-FEF9550EFD44/Build a Metadata-Driven ETL Platform by Extending Microsoft SQL Server Integration Services.docx.
- 17.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 18.Waitman LR, Aaronson LS, Nadkarni PM, Connolly DW, Campbell JR. The Greater Plains Collaborative: a PCORnet Clinical Research Data Network. J Am Med Inform Assoc. 2014;21(4):637–41. doi: 10.1136/amiajnl-2014-002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Post AR, Kurc T, Cholleti S, Gao J, Lin X, Bornstein W, et al. The Analytic Information Warehouse (AIW): A platform for analytics using electronic health record data. J Biomed Inform. 2013;46(3):410–24. doi: 10.1016/j.jbi.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richesson RL, Smerek M. Electronic Health Records-Based Phenotyping. Rethinking Clinical Trials: A Living Textbook of Pragmatic Clinical Trials: Duke University. 2014 [Google Scholar]
- 21.Post A, Kurc T, Rathod H, Agravat S, Mansour M, Torian W, et al. Semantic ETL into i2b2 with Eureka! AMIA Summits Transl Sci Proc. 2013:203–7. [PMC free article] [PubMed] [Google Scholar]
- 22.Post A, Kurc T, Overcash M, Cantrell D, Morris T, Eckerson K, et al. A Temporal Abstraction-based Extract, Transform and Load Process for Creating Registry Databases for Research. AMIA Summits Transl Sci Proc. 2011:46–50. [PMC free article] [PubMed] [Google Scholar]
- 23.Red Hat Inc. Ansible. 2017. [cited 2017 Feb 25]. Available from: https://www.ansible.com/


