Abstract
Healthcare quality research is a fundamental task that involves assessing treatment patterns and measuring the associated patient outcomes to identify potential areas for improving healthcare. While both qualitative and quantitative approaches are used, a major obstacle for the quantitative approach is that many useful healthcare quality indicators are buried within provider narrative notes, requiring expensive and laborious manual chart review to identify and measure them. Information extraction is a key Natural Language Processing (NLP) task for discovering and mining critical knowledge buried in unstructured clinical data. Nevertheless, widespread adoption of NLP has yet to materialize; the technical skills required for the development or use of such software present a major barrier for medical researchers wishing to employ these methods. In this paper we introduce Canary, a free and open source solution designed for users without NLP and technical expertise and apply it to four tasks, aiming to measure the frequency of: (1) insulin decline; (2) statin medication decline; (3) adverse reactions to statins; and (3) bariatric surgery counselling. Our results demonstrate that this approach facilitates mining of unstructured data with high accuracy, enabling the extraction of actionable healthcare quality insights from free-text data sources.
1. Introduction
Improving the quality of healthcare is a fundamental but difficult task in any healthcare system. The first step to such improvements is the identification of specific performance deficits. This can be achieved through measuring healthcare quality by using and/or developing performance measures that can serve as indicators of quality. Such metrics range from simple measures (e.g., patient waiting time) to more sophisticated ones (e.g., time to administration of antibiotics to a patient with pneumonia).
Research methods in healthcare quality can be broadly categorized as qualitative and quantitative in nature. “Quality” is not an easily defined concept, but is a much more complicated and sophisticated issue. Consequently, the research questions that may arise do not always have quantifiable answers. Qualitative research has proven to be an effective way to answer some of these complex questions using approaches based on interviews, observation and data analysis.
Quantitative methods, on the other hand, aim to measure phenomena and statistically evaluate them. Common measures include prevalence, incidence, frequency and severity. This research generates numeric data by applying specific methods to preselected data. It also bears mentioning that mixed methods – those combining both qualitative and quantitative approaches – continue to gain wider currency in the research community.1 Regardless of the research paradigm, informatics plays an important role in obtaining the right data.
One issue with quantitative research is that beyond statistics derived from structured data (e.g., patient records or laboratory results), the development of measures for more sophisticated questions has proven to be more difficult.2 A major hindrance is that while some of the target information is readily available as structured data, the great majority of it is stored as unstructured data, such as free text written by care providers. This use of free text is borne out of the flexibility required by professionals in describing their observations, diagnoses and treatment strategies. However, the unstructured natural language data that they produce is not directly usable in large-scale quantitative analyses. To use this narrative data, it would need to be manually abstracted via labor-intensive chart review. Indeed, retrospective chart review is a commonly used method for identifying issues not well documented by other methods.3 While it is widely considered to be a laborious process, it has also been said that “chart review is more difficult than it appears on the surface.”4 A demonstrative example of these issues is seen in the work of Pivovarov et al.,5 who state:
“Indicators used to assess quality of care are often buried within patient records. To accurately abstract these quality indicators, specially trained nurses manually comb through patient records to locate relevant information. Our 2,600-bed institution employs 35 full-time data abstraction specialists dedicated to reporting quality metrics for 30 databases covering 13 disease states and processes of care.”
Given that these free-text notes contain vital clinical data and that their manual review is a costly process, this has led to the development of computational methods to process and mine them for information of interest.6 This is often done via information extraction (IE), the task of identifying and extracting relevant fragments of text from a larger, unstructured document. Researchers have been developing such methods to extract the information that they require from their own data sources. Once extracted, this information is used for quantitative research, clinical decision support, evidence-based medicine or further processing.
The overarching aim of the present paper is to highlight how unstructured data can be utilized by researchers for quantitative healthcare research. We approach this by introducing a new platform for extracting information from free-text data, and demonstrate its application in four different projects focused on healthcare quality.
The introduction of information extraction software is an important facet of facilitating more sophisticated quantitative healthcare quality research. This is because despite the importance, cost and laborious nature of this task, as we highlighted above, no readily usable tools for biomedical researchers are available. Ideally, such tools should be easy to use for researchers, even those without any NLP or software development experience. However, no such free or open-source solution exists. In this paper we present Canary, an NLP-based information extraction platform designed to meet these criteria. Canary has been developed for processing clinical documents to support the extraction of data using user-defined information discovery parameters and vocabulary. The various components that form the Canary NLP pipeline and the system for capturing text fragments are described, followed by an empirical evaluation on clinical data from four projects.
2. Background: Approaches to Information Extraction
In this section we describe a number of different approaches that have been used for information extraction, highlighting some of their advantages as well as disadvantages. In doing so we aim to position Canary within the wider context of the information extraction literature.
Simple Text Matching: The most elementary and straightforward approach to IE is based on defining specific words or sequences of words to be matched and output. While this can work in the simplest of cases, such as identifying particular drugs, it is impractical for more complex tasks.
Pattern Matching: Some shortcomings of the string matching method can be addressed by using more expressive and powerful pattern matching methods. Regular expressions are a common technique commonly employed for this purpose. They can be useful in extending patterns to match variations (e.g,. different patterns of expressing drugs and dosage) or to account for other patterns like typographical errors. Disadvantages include the large number of rules required to capture all possible variations as well as difficulties in maintaining and updating the rules. They are also unable to capture structure, as we will discuss in the next section.
Language Parsing The methods described thus far can be considered as “shallow” text processing techniques, relying solely on the words as they appear in text. Another approach involves using a “deeper” understanding of the text by parsing it to produce syntactic representations of the data; this can include constituency or dependency parsing. The parsing process can add linguistic information such as part-of-speech tags for each word as well as structural information such as noun phrases or prepositional phrases. This information can then be incorporated into the IE rules to produce more accurate and generalizable rules. These methods have been successfully applied in recent studies. For example, Wang et al. showed that such parser-based methods can be useful for automatic extraction of substance use information from clinical notes.7 One disadvantage of this approach is that the parsing process can be slow.
Supervised Learning A more recently developed family of techniques based on statistical analysis of text is supervised machine learning. This involves the use of labelled training data to train a learning algorithm to identify elements of interest. While it does not involve manually engineering information extraction rules like the previous approaches, the costs of creating labelled data for supervised training are also significant. Moreover, learning algorithms require large amounts of data for effective training, along with the relevant expertise in tuning them. Nevertheless, a number of successful and popular machine learning-based toolkits for clinical information processing have been developed. The Automated Retrieval Console (ARC) is one such tool that attempts to eliminate rule creation via supervised learning.8 The Apache cTAKES system has also gained wide currency among clinical researchers in recent years.9 However, there is a population of clinical researchers without the prerequisite NLP and/or computer science skills that are not able to make use of these solutions. In this regard, the solution that we present here is complementary and designed to aid the aforementioned researchers in conducting their investigations in a self-sufficient manner.
3. Methods
3.1. Design
We designed a method for information extraction and developed NLP software implementing it. This platform was evaluated by conducting four quantitative healthcare quality experiments using unstructured clinical notes.
3.2. A Hybrid Information Extraction Method Based On User-Defined Parameters
The Canary software employs a hybrid approach for the IE task, combining the pattern matching and parsing approaches in order to address their shortcomings. The approaches are not mutually exclusive and can be complementary, as we show.
Regular expressions can be very useful for recognizing words or chunks of text, but they are not designed for capturing structure within text. While they can be helpful for identifying text in a pre-specified format, such as dates, numbers, email addresses or measurements,10 they are not suitable for more sophisticated structures with greater variance in their composition. For example, writing a regular expression to match text fragments describing a body part can be a cumbersome and error-prone task, given the large number of possible variations. Let us consider the following example phrases of interest:
| (1)left hand | (2)anterior cruciate ligament |
| (3)lower back region | (4)lateral ankle ligament |
| (5)right hand’s index finger | (6)upper left abdominal quadrant |
A key shortcoming of regular expressions is their inability to capture recursive structures, such as nested components in a tree structure. For example, they are unable to match nested brackets within a string.
Phrase (5) above is an example of such a nested structure in which a single entity is composed of two smaller entities: right hand and index finger. Capturing such nested constructs requires the definition of recursive rules, something of which regular expressions are inherently incapable.
On the other hand, detecting nested constructs can be achieved through a parsing-based approach in which recursive grammatical rules can be defined. While all aspects of regular expressions can be implemented through parsing, only some of the parsing functionality can be performed using regular expressions.
Another disadvantage of using regular expressions is that the rules encompass both words and their possible ordering. However, given the size of the target vocabulary that researchers use (particularly in the medical domain), it would be advantageous to separate the lexical entries and the ways in which they can be combined. That is to say, split the rules into a vocabulary (a set of recognized words and their categories) and a grammar (rules defining how words can be combined). This can be easily addressed by a parse-based solution by defining a vocabulary of terms and grammatical rules that define how they can be combined.
A related issue with regular expressions is that they can quickly grow in complexity, resulting in an unwieldy set of cryptic rules that can be very hard to comprehend. Updating such rules is also fraught with difficulties. Researchers have noted that the modification and documentation of regular expressions are a source of difficulty in their work.11 The isolation of vocabulary and grammar rules can help address these issues, as we will demonstrate.
The use of a complete language parser would require that users define their rules on top of the linguistic rules governing the target language (i.e., first categorize words into their grammatical categories and then create subsets of interest). An alternative approach, employed by Canary, is to allow users to create a simple, customizable grammar that enables them to model their target information, e.g., body parts or something broader that includes body parts. Complete language parsing is also a more computationally intensive task.
We now turn to a concrete example to illustrate how this approach works. The first step is to define the vocabulary or lexicon, which is a set of words organized into word classes. A class is a grouping of words from the same semantic category. For our body part example, we can define two classes referring to body parts and anatomical adjectives, which could be defined as:
ANATOMICAL → (bi)?-?lat(eral)?, anterior, caudal, upper, lower, left, right, […]
BODYPART → (gastro)?intestinal, (gastro)?o?esophageal, (musculo)?skeletal, abdom.?, abdomen, […]
Users may define as many classes as needed, and the words belonging to each class can be matched using a regular expression, as seen in the above example. This allows users to create customizable ontologies to meet their needs.
The second step involves defining grammatical rules that define how these word classes can be combined for form phrases. A phrase can be a single word or a combination of words, as allowed by the grammar.
BODYPARTPHRASE → BODYPART BODYPARTPHRASE
BODYPARTPHRASE → ANA TOMICAL BODYPART
The above rules state that a body part phrase can be a single body part or an anatomical adjective followed by a body part. These rules are then processed by a parser to match all fragments of a text that match any of the supplied rules. Words that are not found in the vocabulary are ignored. Some examples matched by this simple grammar are shown in Figure 1.
Figure 1.
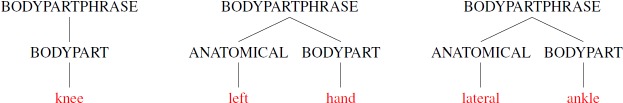
Example fragments of text describing body parts matched by our simple grammar.
We can also extend the grammar to match nested body parts by simply adding a recursive rule:
BODYPARTPHRASE → BODYPARTPHRASE BODYPARTPHRASE
A recursive rule expresses that a phrase may include a sub-phrase of the same type as its own constituent, allowing us to capture the recursive property of natural language. This is important because, for example, English nouns and sentences can be infinitely recursive. In the above example, this extension allows the capture of one or more adjacent body part phrases. We illustrate this with the examples in Figure 2.
Figure 2.
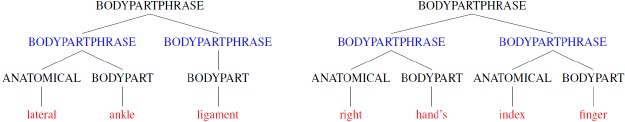
Examples of recursive body part phrases that contain other body parts (marked in blue) as constituents.
We can see that this simple extension allows the capture of the more complex phrases. This is something not possible with regular expressions. A parser, on the other hand, can find an arbitrary number of such nested elements.
Furthermore, these constituency grammar rules can be used to build more sophisticated phrases that contain multiple recursive elements, as we show in the next section.
3.2.1. Creating Longer Phrases
Researchers often need to extract complex phrases that capture more information than just a body part. To this end, the simple rules we have seen so far can serve as the building blocks for forming longer, more sophisticated phrases. Building on the previous example, we now extend the rules to detect a medical condition involving a body part. We first add additional entries to our vocabulary:
ARTICLE → the, a, an PREPOSITION → of, in, […]
POSSPRONOUN → his, her, your, my CONDITION → (hepato)?(to)?xicity, aches?, aching, […]
We also add some grammar rules to capture a phrase indicating a condition involving a body part:
CONDITIONPHRASE → BODYPARTPHRASE CONDITION
CONDITIONPHRASE → CONDITION PREPOSITION BODYPARTPHRASE
Applying these rules, we can match the phrases shown in Figure 3. We can also observe that the body part phrase in the left example has three levels of nested phrases. This example demonstrates that an advantage of the grammar-based approach is that the items in our vocabulary can be easily referenced and used to build larger and more meaningful phrases, something that is not possible with regular expressions.
Figure 3.
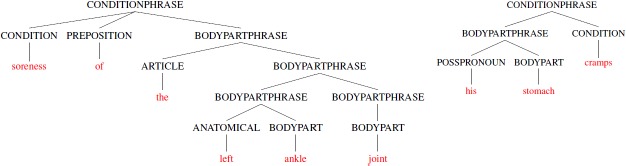
Two parse trees for condition phrases that include recursive body part phrases.
3.3. Canary: An NLP-based Information Extraction Platform
In order to facilitate the mining of text documents by researchers, we created a free software program called Canary,12 as shown in Figure 4 below. The software, which can be downloaded for free (http://canary.bwh.harvard.edu/), was developed to enable information extraction using the approach discussed in section 3.2. The software also includes numerous sample projects that demonstrate how the grammar-based approach described above is used in practice. A number of factors were considered in designing the Canary platform, some of which we highlight here.
Figure 4.
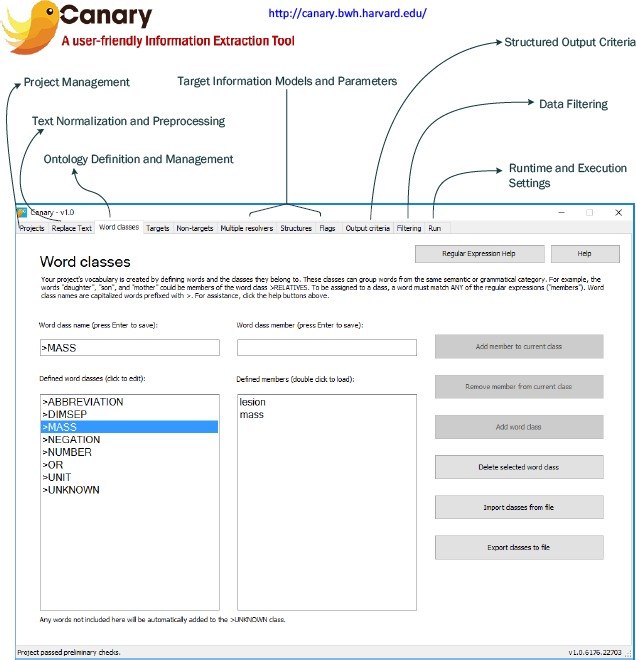
An overview of the Canary software.
Ease of Use: Canary was designed for users without software development or engineering experience, allowing the vocabulary and grammar rules to be defined through a unified graphical user interface (GUI). The output is generated in plain text format for easy analysis.
Easy Setup: Canary is made available as an off-the-shelf software solution. It can be installed in a local folder without administrator privileges and can even run off a flash drive. It was designed to work out of the box.
Security and Privacy Concerns: Researchers in health informatics often work with data that includes protected health information, requiring compliance with the appropriate legal and security measures to guard this information (e.g., HIPAA compliance). These data may be stored on local networks and behind firewalls, with strict rules governing their transmission. Although a number of cloud-based data processing and information extraction solutions have been proposed, the abovementioned restrictions may rule out their use for many clinical researchers. In such cases, the use of self-contained software packages that can be run on local machines may be a better option. This is one of the motivating reasons that underlies Canary’s design.
4. Insulin Decline
The first task we demonstrate is the quantitative assessment of insulin decline. It is anecdotally known that patients frequently decline medications that are recommended by their healthcare providers. However, little systematic data are available on this phenomenon. It is not known how commonly patients decline medications and how frequently they ultimately receive medications they initially declined. This information, if extracted, would be of great value to healthcare quality and outcome research.
Insulin is thought to be one of the medications that are especially frequently declined by patients. Many patients are reluctant to start injectable medications; others express fear that “once you start insulin, you can’t get off it.” Studies show that patients whose diabetes is poorly controlled on oral medications take a very long time to be started oninsulin;13 insulin decline by patients could be one of the reasons for this. However, data on insulin decline remains extremely limited.14
One reason for the paucity of research in this area is that information on patients declining medications is not easily available. As these patients declined the medication before any prescription was written, no trail is generated in the data sources that are typically used to study medication prescribing, such as pharmacy insurance claims or EMR medication records. Instead, medication decline is primarily recorded in narrative notes, requiring labor-intensive chart review. To this end, NLP software like Canary holds great promise for allowing clinical researchers to access the valuable pieces of relevant information locked away among millions of unstructured health records. Accordingly, we evaluate our software on data for this task, using it to extract this information and conduct a study to assess the prevalence of patients who decline insulin.
4.1. Data Collection
Data for this study comes from the clinical records of all adult patients with diabetes treated in primary care practices affiliated with Massachusetts General Hospital and Brigham and Women’s Hospital between 2000 and 2014. The number of notes reviewed for each task varied between 600 and 50,000.
4.2. Insulin Decline Language Model
A manual review of a subset of the collected data containing 50,000 notes was conducted to identify instances of insulin decline by patients. This task required the largest set of notes due to the extremely low prevalence of the information. Next, Canary vocabulary and information extraction criteria were created by a clinical researcher with no formal training in NLP or software development. They were designed to detect the language used to document insulin decline in manually identified instances with maximum accuracy and generalizability. After iteratively refining the language model, it resulted in a set of 148 word classes and 284 rules.
4.3. Evaluation
The primary aim of our evaluation is to assess the accuracy with which instances of insulin decline can be detected using Canary. This evaluation was conducted against a held-out gold standard dataset of 1,501 provider notes which were randomly selected and independently annotated by trained pharmacy and medical students. The reviewers marked all sentences describing patients who refuse to take insulin,. We then compare the reviewers’ annotation to output generated by Canary. We conducted our evaluation against this annotated gold standard at two levels of granularity:
Note level: detecting notes that contain any mention of insulin decline anywhere in the document.
Sentence level: detecting sentences that mention insulin decline, across all notes.
The sentence-level assessment is the more challenging task. At the note level, sensitivity (recall), specificity and positive predictive value (PPV/precision) were calculated. Specificity has no meaning at the sentence level because of the arbitrary nature of the tokenization process (i.e. token boundaries in free text are implementation specific), so only sensitivity and PPV were calculated. Results from this experiment will be used to estimate the prevalence of insulin decline in the unstructured data belonging to patients with diabetes.
4.4. Results
The manually annotated test set included a total of 19 sentences mentioning insulin decline across only 14 notes. This is a note-level prevalence of 0.93%, highlighting the difficulty associated with identifying this information through manual review. It should also be noted that given the length of the average note, this prevalence is significantly lower at the sentence level, i.e., less than 1% of sentences.
We next applied Canary to the same data. The results for this evaluation are listed in Table 1. We observe that at the note level, Canary achieved a sensitivity of 100.0% and a PPV of 93.3%, with even better results at the sentence level.
Table 1.
Evaluation results on the test set of 1,501 manually annotated gold-standard notes. The 95% confidence interval is provided in parentheses.
| Sensitivity | Specificity | PPV | |
|---|---|---|---|
| Note-level | 100.0% (76.8–100.0) | 99.9% (99.6–100) | 93.3% (68.0–99.8) |
| Sentence-level | 100.0% (82.4–100.0) | N/A | 95.0% (74.4–99.9) |
5. Statin Decline
The study of medication decline, as outlined in the previous section, can be extended to other classes of drugs. Cardiovascular disease is the number one cause of death both in the United States and worldwide, and hypercholesterolemia is the most common risk factor. HMG-CoA reductase inhibitors (statins) reduce the risk of cardiovascular events in patients with hypercholesterolemia. Nevertheless, many patients at high cardiovascular risk are not taking statin therapy, likely leading to thousands of preventable deaths. The reasons for this are not fully understood, but it is thought that medication decline may play a role.
Our methodology can be used to empirically study this phenomenon and analyze initial statin decline rates, how frequently patients ultimately start statin therapy after initially declining it, and whether the provider who eventually prescribes statin is likely to be different from the one whose statin recommendation was initially refused in the population of patients at high cardiovascular risk.
To this end we conducted a preliminary study to assess the specificity and PPV of our method in detecting instances of statin decline.
5.1. Data Collection
Data for this study comes from the clinical records of all adult patients with coronary artery disease treated in primary care practices affiliated with Massachusetts General Hospital and Brigham and Women’s Hospital between 2000 and 2013.
5.2. Statin Decline Language Model
A set of 8,800 notes were used by a clinical researcher with no formal training in NLP or software development to create a Canary model to detect instances of statin decline. Canary vocabulary and grammar rules were created to match the text fragments with maximum accuracy and generalizability. This resulted in a model with 97 semantic word classes and 88 structures.
5.3. Evaluation and Results
A set of 4,000 held-out notes were used for evaluation. The model achieved sensitivity (true positive rate) of 88% and PPV (precision) of 92%. These preliminary results highlight the utility of our approach to studying this healthcare issue. We are now in the process of creating additional resources to measure the specificity of our models, in order to apply them for answering the research questions laid out in section 5.
6. Adverse Reactions to Statins
EMRs are widely used in documenting adverse reactions to medications. Clinical decision support systems that analyze previous allergies and reactions have been shown to dramatically decrease prescription errors, making this a widely-researched area. However, a substantial portion of these reactions are not documented in a structured format but instead persist as free text. To this end, we applied our methodology to quantitatively extract this information and conduct a study to assess the prevalence of adverse reaction information that is not present in a structured format. More specifically, we studied this for 3-hydroxy-3-methyl-glutaryl-CoA reductase inhibitors (i.e., statins), as they are thought to have high rates of adverse reactions reported by patients.
6.1. Data Collection
This study was conducted at Partners HealthCare System, a healthcare delivery network in eastern Massachusetts. Partners Healthcare maintains a network-wide repository of medication allergies called the Partners Enterprise Allergy Repository (PEAR). The EMR system used at Partners allows data entry into PEAR. Data for our study came from all patients who were recorded as having been prescribed a statin between 2000 and 2010. This resulted in a set of 4.7 million provider notes.
6.2. Statin Side Effects Language Model
Trained pharmacy students manually reviewed a set of 3,175 narrative provider notes, annotating instances of adverse statin reactions. These notes were randomly selected from a set of all notes written on the data a statin was noted as being discontinued in the EMR system. After the annotation, Canary vocabulary and grammar rules were created to match the text fragments with maximum accuracy and generalizability.
6.3. Evaluation and Results
The evaluation follows the same scheme as described in section 4.3 above, with the exception that a held-out gold standard dataset of 242 provider notes were randomly selected and independently annotated by two trained pharmacy students. The results for the first stage of evaluation are listed in Table 2. We observe that at the note level, Canary achieved a sensitivity of 87.4% and a PPV of 99.4%. As expected, detection at the sentence level as more challenging, with slightly lower results.
Table 2.
Evaluation results on the test set of 242 manually annotated gold-standard notes. The 95% confidence interval is provided in parentheses.
| Sensitivity | Specificity | PPV | |
|---|---|---|---|
| Note-level | 87.4% (83.0–91.8) | 98.3% (96.5–100) | 99.4% (98.2–100 |
| Sentence-level | 80.6% (75.4–84.9 | N/A | 98.6% (96.9–100) |
Having validated the model and demonstrated that it can achieve high levels of accuracy, we proceeded to the second stage of our evaluation. Canary was used to process the full set of 4.7 million notes collected for the study. This resulted in the identification of 224,421 patients who were prescribed a statin during the study period, with 31,531 of these being flagged by our software as having had an adverse reaction to a statin side. However, only 9,020 (28.6%) of the patients had a statin reaction recorded in structured format (in PEAR).15 This result indicates that the great majority of care providers record drug reaction information only in unstructured data.
7. Bariatric Surgery Counselling
Bariatric surgery is the single most effective treatment for significant and sustained weight loss in obese patients and significantly improves numerous obesity-related comorbidities, including cardiovascular risk, hypertension, myocardial infarctions, strokes and cardiovascular deaths.
A critical step in a patient’s decision to undergo bariatric surgery is clinician discussion and recommendation of bariatric surgery to the patient. Little systematic data are available on the epidemiology of bariatric surgery recommendation. It is not known how commonly physicians discuss and recommend bariatric surgery to obese patients who are surgical candidates. One of the reasons for the paucity of research in this area is that information on physician recommendation of bariatric surgery is not easily available. This information is typically not reflected in either administrative or structured electronic clinical data, as no prescription or insurance claims data are generated. Instead, recommendation of bariatric surgery is primarily recorded in narrative notes, requiring labor-intensive chart review.
The methodology we have used thus far can be used to study this issue empirically. In attempting to quantify the recommendation of bariatric surgery within notes, any approach must be able to distinguish between discussion of the procedure and mentions of prior procedures. Consequently, the identification of notes containing these two categories of information the aim of our preliminary project.
7.1. Data Collection
Data for this study comes from the clinical records of all adult patients with body mass index (BMI) > 35 kg/m2 treated in primary care practices affiliated with Massachusetts General Hospital and Brigham and Women’s Hospital between 2000 and 2014.
7.2. Bariatric Surgery Counselling Language Model
A set of 300 notes were manually annotated by a trained pharmacy student and subsequently used by another researcher with no formal training in NLP or software development to create a Canary model to distinctly detect instances of prior surgery and surgery discussion. Canary vocabulary and grammar rules were created to match the text fragments with maximum accuracy and generalizability. This resulted in a model with 17 semantic word classes and 160 structures.
7.3. Evaluation and Results
A held-out test set of 300 notes were manually annotated by a trained pharmacy student and used for evaluation, which follows the same procedure as described in section 4.3. The results for both categories at the note- and sentence-level are listed in Table 3. This preliminary evaluation is very promising, showing that our models are able to achieve high accuracy and demonstrate the generalizability to new data.
Table 3.
Evaluation results for both categories on the test set of 300 manually annotated gold-standard notes.
| Sensitivity | PPV | |
|---|---|---|
| Bariatric Surgery Discussion (Note-level) | 90% | 90% |
| Bariatric Surgery Discussion (Sentence-level) | 85% | 69% |
| Prior Bariatric Surgery (Note-level) | 83% | 90% |
| Prior Bariatric Surgery (Sentence-level) | 44% | 96% |
8. Discussion and Conclusion
We described and demonstrated the application of information extraction for identifying actionable insights by mining clinical documents. We showed that this approach can assist with answering quantitative healthcare quality research questions whose answers are not easily derived from structured data sources.
As part of this approach we also presented Canary, an information extraction tool based on user-defined parameters and ontologies. The principal advantage of our tool is that it is a GUI-based software which does not require any technical background. In this study, this was underlined by the fact that it was used by several researchers without any such technical background to successfully create language models of important clinical phenomena. The feedback from these users was positive and the models were then tested on a large-scale set of provider notes.
This approach is also useful for outcomes research, which involves quantifying treatment patterns and measuring the associated patient outcomes, and is an important area of study for identifying potential areas for improving healthcare quality. To this end, the methodology described here could be used to quantitatively measure complex social and demographic issues in health services research.
References
- 1.Jick TD. Mixing qualitative and quantitative methods: Triangulation in action. Administrative science quarterly. 1979;24(4):602–611. [Google Scholar]
- 2.Pope C, van Royen P, Baker R. Qualitative methods in research on healthcare quality. Quality and Safety in Health Care. 2002;11(2):148–152. doi: 10.1136/qhc.11.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckmann U, Bohringer C, Carless R, et al. Evaluation of two methods for quality improvement in intensive care: facilitated incident monitoring and retrospective medical chart review. Critical care medicine. 2003;31(4):1006–1011. doi: 10.1097/01.CCM.0000060016.21525.3C. [DOI] [PubMed] [Google Scholar]
- 4.Allison JJ, Wall TC, Spettell CM, et al. The Art and Science of Chart Review. The Joint Commission Journal on Quality Improvement. 3// 2000;26(3):115–136. doi: 10.1016/s1070-3241(00)26009-4. [DOI] [PubMed] [Google Scholar]
- 5.Pivovarov R, Coppleson YJ, Gorman SL, Vawdrey DK, Elhadad N. Can Patient Record Summarization Support Quality Metric Abstraction? Paper presented at: AMIA Annual Symposium Proceedings. 2016 [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. Journal of the American Medical Informatics Association. 2010;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Chen ES, Pakhomov S, et al. Automated Extraction of Substance Use Information from Clinical Texts. Paper presented at: AMIA Annual Symposium Proceedings. 2015 [PMC free article] [PubMed] [Google Scholar]
- 8.D’avolio LW, Nguyen TM, Farwell WR, et al. Evaluation of a generalizable approach to clinical information retrieval using the automated retrieval console (ARC) Journal of the American Medical Informatics Association. 2010;17(4):375–382. doi: 10.1136/jamia.2009.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turchin A, Kolatkar NS, Grant RW, Makhni EC, Pendergrass ML, Einbinder JS. Using Regular Expressions to Abstract Blood Pressure and Treatment Intensification Information from the Text of Physician Notes. J Am Med Inform Assoc. 2006;13(6):691–695. doi: 10.1197/jamia.M2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraus S, Blake C, West SL. Information extraction from medical notes. Paper presented at: Medinfo 2007: Proceedings of the 12th World Congress on Health (Medical) Informatics; Building Sustainable Health Systems. 2007 [Google Scholar]
- 12.Malmasi S, Sandor N, Hosomura N, Goldberg M, Skentzos S, Turchin A. Canary: An NLP Platform for Clinicians and Researchers. Applied Clinical Informatics. 2017;8(2):447–453. doi: 10.4338/ACI-2017-01-IE-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubino A, McQuay L, Gough S, Kvasz M, Tennis P. Delayed initiation of subcutaneous insulin therapy after failure of oral glucose-lowering agents in patients with Type 2 diabetes: a population-based analysis in the UK. Diabetic Medicine. 2007;24(12):1412–1418. doi: 10.1111/j.1464-5491.2007.02279.x. [DOI] [PubMed] [Google Scholar]
- 14.Khan H, Lasker S, Chowdhury T. Prevalence and reasons for insulin refusal in Bangladeshi patients with poorly controlled Type 2 diabetes in East London. Diabetic Medicine. 2008;25(9):1108–1111. doi: 10.1111/j.1464-5491.2008.02538.x. [DOI] [PubMed] [Google Scholar]
- 15.Skentzos S, Shubina M, Plutzky J, Turchin A. Structured vs. unstructured: factors affecting adverse drug reaction documentation in an EMR repository. Paper presented at: AMIA Annual Symposium Proceedings. 2011 [PMC free article] [PubMed] [Google Scholar]


