1. Introduction
A disease registry is an organized system that uses observational study methods to collect uniform data to evaluate specified outcomes for a population defined by a particular disease, condition, or exposure, and that serves one or more predetermined scientific, clinical, or policy purposes[1]. Different from Electronic Health Record (EHR), whereas the research of understanding a particular disease belongs to the secondary use of data, the disease registries serve as a primary source to study the target disease. As a primary source, the registry data usually involves data generated from known and comprehensive clinical assessments for the target disease, and therefore a disease registry can be a powerful tool to observe the course of disease, to understand variations in treatment and outcomes, to examine factors that influence prognosis and quality of life, and to assess the clinical cost and quality of care.
The natural course of a disease can be characterized by the onset and progression of symptoms. Measures from clinical assessments are collected in a disease registry for tracking various symptoms of a target disease. However, such measures can be biased since they not only can be affected by the progression of the target disease, but also can be influenced by non-disease-related factors, e.g. selection bias, clinical instrument sensitivity and participant compliance. Moreover, for a symptom of interest, its onset and progression indicator are decided by threshold values of recorded clinical measures. Such threshold values are currently determined based on evaluation of measures recorded in comparable historical studies or by domain expert, and are applied to all participants in a registry. Consequently, they do not take individual variations into account. In order to address the aforementioned issues, data-driven methods are needed to generate robust (less biased) and personalized symptom onset indicators, and as a result, in return for better understanding the natural course of a target disease.
A disease registry may include only people with disease of interest, or may also include one or more comparison groups for which data are collected using the same methods during the same period. Hereinafter, we refer to patients with disease of interest as case participants, while patients in the comparison group who are not at risk as control participants. Control participants may share some similar traits or are exposed to similar environmental factors as case participants. In this article, we propose a data-driven method to generate robust symptom onset indicators based on assessments collected in disease registry data with the requirement that this disease registry should have included control participants. According to our knowledge, the proposed method is the first of its kind that uses control participants in a disease registry to adjust the biases inherited in the raw clinical measurements among case participants. The biases are caused by non-disease-related factors such as natural aging process, education level and marital status. In the remainder of this paper, we will exemplify the application of this novel method to integrated data from observational Huntingtons Disease studies.
Huntington’s Disease (HD) is an autosomal dominant fully-penetrant neurodegenerative disorder, which is caused by an abnormal expanded trinucleotide (CAG) repeat in the Huntingtin (HTT) gene[2]. Owing to its monogenic nature, predictive genetic testing is able to determine whether the disease will manifest in an individual. Among genetically confirmed HD patients, a clinical diagnosis of HD is typically made when an individual exhibits overt, otherwise unexplained extrapyramidal movement disorder. While motor impairment is currently the primary indicator of clinical onset, cognitive[3] and certain behavioral disorders[4] are also known to surface years before motor onset. As such, clinical measurements along these dimensions are also important for understanding the progression of the disease. Functional assessments in particular are important in measuring overall quality of life of individuals with HD and prove useful for a descriptive characterization of HD progression.
In recent years, several large-scale observational studies have been conducted in HD gene expansion carriers (HDGECs) with the hope to understand the natural history and pathophysiology of the disease. A diverse range of clinical assessments have been designed in HD observational studies to record the triad of motor, cognitive/behavior, and functional symptoms of HD. While accessibility to a wide range of clinical assessments from these domains has helped gain insightful information about the natural history of HD, these clinical assessments are often influenced by factors other than HD disease status and progression. For instance, natural aging processes are especially known to affect participants cognitive and functional abilities. Therefore, absolute changes in most cognitive and functional assessment scores can be attributed to multiple factors including both HD disease progression and the natural aging process, rendering assessment scores less robust for tracking disease progression.
Onset of new symptoms is useful in characterizing the course of HD. Certain clinical measures collected have predefined thresholds to indicate reliability of clinical diagnosis. For example, a Diagnostic Confidence Level (DCL) of 4 is used to indicate motor symptom onset, thereby leading to a confirmed diagnosis of HD onset. However, other measures (e.g. the SDMT score, which is used to assess cognitive abilities) do not have clearly defined thresholds for disease onset. To address this inconsistency in a systematic manner, a data-driven method is needed for generating robust symptom onset indicators for better understanding of the progression of the disease.
In this article, we propose a data-driven procedure for adjusting the values of clinical assessments and generating robust symptom onset indicators. Its worth pointing out that although this article and methodology described here relies on HD observational data sets, the methodology can be generalized to other disease registry data if they have also recruited control participants.
For readers benefit, the contents of this paper are organized as follows. In Section 2, we describe the HD observational data used in this work. In Section 3, we describe a novel method for generating robust clinical assessments from the outcome measures recorded in the observational datasets. In Section 4, we use two cognitive assessments to demonstrate the properties of the generated assessment score. Lastly, Section 5 provides a summary and brief discussion of the new method.
2. Data Sources
In this study, we integrated data from four large prospective observational studies of HD, namely Enroll-HD[5], REGISTRY[6], TRACK-HD/TRACK-ON[7, 8], and PREDICT-HD[9], respectively.
Enroll-HD is a worldwide observational study of Huntington’s Disease families. The study aims at providing a platform to support the design and conduct of future clinical trials, improving the understanding of the phenotypic spectrum and the disease mechanisms of HD and improving health outcomes for the participant/family unit. The study monitors how HD appears and changes over time in different subjects. It recruits confirmed HD patients, HD at-risk patients, HD genotype negative participants as well as control participants from HD family. Study participants are required to visit study sites annually, and undergo a comprehensive battery of clinical assessments. In this work, we used the ENROLL-IDS-2015-10-R1 version of the Enroll-HD periodic data, which contains un-monitored data from 7614 subjects who made their baseline visits prior to October 2015. Among the participants, 5475 are Huntington’s disease gene expansion carriers (HDGECs) with CAG length greater than 35, 1613 participants are control subjects with CAG length less than or equal to 35, and the other 527 have unknown CAG length. Subjects have up to four annual visits, with an average number of visits being 1.44.
REGISTRY is a multi-center, multi-national observational study, managed by the European Huntington’s Disease Network (EHDN), with no experimental intervention. REGISTRY aims at obtaining natural history data on many HD mutation carriers and individuals who are part of an HD family, relating phenotypical characteristics of HD, expediting the identification and recruitment of participants for clinical trials, developing and validating sensitive and reliable outcome measure for detecting onset and change over the natural course of pre-manifest and manifest HD. The REGISTRY cohort used in this study consists of 12108 participants, among which 7988 are HDGECs (i.e. CAG > 35), 758 are control participants (i.e. CAG length ≤ 35), and the other 3894 participants do not have CAG length information. Participants have up to 15 annual visits, with the average number of visits equals to 2.9.
TRACK-HD is a multinational study of HD that examines clinical and biological findings of disease progression in individuals with pre-manifest HD and early-stage HD. Participants in the study underwent annual clinical assessments for 36 months. At the baseline visit, 402 participants were enrolled. Among the participants, 127 participants were control subjects, 144 participants were pre-manifest subjects who had not reached HD clinical onset, and 130 were post-manifest subjects who had already reached HD clinical onset. 298 participants completed the 36-month follow-up, among which 97 were controls, 104 were pre-manifest subjects at their baseline visits, and 97 were post-manifest subjects at their baseline visits.
TRACK-ON is a follow-up study of TRACK-HD with the aim of testing for the compensatory brain networks after structural brain changes in TRACK-HD pre-manifest participants. Participants in the study underwent annual clinical assessment for 24 months. At the baseline visit, 245 participants were enrolled, among them 181 were participants of TRACK-HD who have not reached HD clinical onset at the end of TRACK-HD, and 64 participants were newly recruited in the study. 112 participants in TRACK-ON were control subjects, and others are HDGECs.
PREDICT-HD is another longitudinal observational study of subjects who chose to undergo predictive testing for the CAG expansion in the HD gene but did not meet criteria for a diagnosis of HD (Diagnostic Confidence Level = 4). Participants were recruited from 32 sites worldwide beginning in October 2002. The goal of PREDICT-HD is to define the neurobiology of Huntington’s disease (HD) and to develop tools to allow clinical trials of potential disease- modifying therapies before at-risk individuals have diagnosable symptoms of the disease. It collected a variety of biosamples including MRI, blood and urine samples, and comprehensive assessments of cognitive, motor, functional and psychiatric outcomes to characterize the pre-manifest syndrome in HD, to document the rate of change of these variables during the years leading up to and following a clinical diagnosis of HD, and to investigate the relationship among neurobiologic factors, clinical diagnosis and CAG repeat length. The PREDICT-HD data used in this study consists of 1481 participants. Among them 316 were control subjects. Participants have up to 14 annual study visits, with the average number of visits equals to 5.2.
3. Materials and Methods
3.1. Integration of Multiple HD Data Sets
The four studies introduced in Section 2 contain a diverse set of clinical assessments that span a spectrum of clinical symptoms expressed by HD patients. In this section, we briefly describe the process of integrating data from these four studies.
We began by matching subjects across studies using a unique Recoded HD participant ID. This unique identifier also allows us to recognize the subjects who participated in multiple studies. In the four HD observational studies, participants visited study sites approximately annually and were evaluated by a diverse range of clinical assessments. In the rest of this paper, we refer to the data generated from one visit of one participant as an observation. In each of the four studies, the date of a participant’s first study visit in the study, referred to as the baseline visit, was used as the reference date for the participant and was set to 0. The visit dates of all his follow-up visits in the same study were aligned with the reference date and measured in days. In addition, for a subject who participated in multiple studies, the time gaps between the multiple reference dates from different studies were also available. Therefore, subjects’ records from multiple studies could be stitched together when they were available.
The second step of combining the multiple data sets was matching and merging variables. Not all variables were named consistently across studies. We analyzed data dictionaries, study protocols and guidelines from the four studies and manually matched variables across studies. We also corrected coding inconsistencies across studies.
We categorized variables into two groups, namely, assessment score and demographical information. The assessment score group consists of measurements from clinical assessments performed at annual study visits to capture wide range of clinical symptoms among HDGECs, such as motor impairment, cognitive deficits, functional decline, and behavioral disorder. The demographic information group includes participants’ demographics (e.g. age, sex, education level, etc.), CAG length, medical history (e.g. drug abuse history, alcohol abuse history, etc.), and other information related to study designs (e.g. region, study site). The integrated data set contains 106 variables from the participants demographic information group and 2079 variables from the assessment scores group.
Finally, we performed cross-study distributional check to filter out obvious erroneous measurements in the integrated data set. After all these steps, we ended up with a data set containing 55782 observations from 16553 HDGECs and 2716 control participants. The average number of observations per participant of the integrated data is 2.9.
3.2. Flow of the proposed new method
The aim of this work is to develop a data-driven method to generate robust symptom onset indicators based on the clinical assessment scores collected in HD registry data. In the rest of this paper, we will refer to the original assessment scores as the ‘raw assessment scores’, and the newly generated scores as the ‘robust scores’. A robust score is generated from its raw assessment score. As proof of concept and application, we will showcase the proposed method using two cognitive assessments in the integrated data, which are Symbol Digit Modalities Test (SDMT) total correct score, and the Stroop Word Reading Test (SWRT) total score.
As discussed in Section 1, raw assessment scores can be influenced by both disease-related and non-disease-related factors. While influence of the disease-related factors on these assessment scores is desirable, influence of nondisease-related factors on these scores can result in misleading follow-up analysis and conclusions. Figure 1 illustrates an example from the raw SDMT total correct scores. The left panel (1a) depicts the distributions of raw SDMT scores vs. the levels from International Standard Classification of Education (ISCED). The right panel (1b) depicts the distributions of raw SDMT scores vs. HD stages of participants in the aggregated data. HD clinical stage has been discretized here into early pre-manifest, late pre-manifest, HD1, HD2, HD3 and HD4. It is used here as a surrogate for HD disease progression. From the figures, it is clear that SDMT scores not only depend on HD disease status and progression, but also depend on other factors such as education levels.
Figure 1.
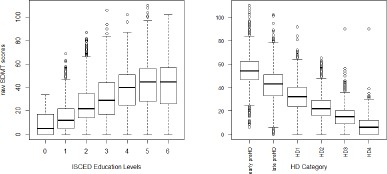
(a) Boxplot of raw SDMT score versus ISCED education levels; (b) Boxplot of raw SDMT total correct scores versus canonical HD clinical stages
The integrated HD observational data includes control participants who do not carry HD gene expansions. Control participants by definition are not affected by HD disease progression. Therefore, raw assessment scores of control participants are expected to be influenced by non-disease-related factors. Throughout this study, we assume that the underlying effects of a non-disease-related factors on an assessment score are the same for both control participants and HDGECs. Any differences in the effects between the two groups can be attributed to the disease. The basic idea of the proposed procedure is to utilize the control cohort to evaluate the effects of non-disease-related factors on an assessment score of interest. Then for a case participant, the predicted control value of the assessment score can be produced from the control-based model. The predicted control value gives an estimate of what the expected assessment score look like for a hypothetical control participant with similar characteristics as the case participant. Subtracting the predicted control value from the observed value can remove the effects of non-disease-related factors. The remaining part of the assessment score is less subject to changes of non-disease-related factors, and is more robust in terms of reflecting the effects of HD related factors. The onset of a new symptom (e.g. cognitive impairment) is defined to be the critical point at which a case participant exhibits significant difference from control participants. Comparing the observed assessment scores with the distributions of the predicted control values could lead to personalized symptom onset indicators.
Next we describe the work flow of generating the robust measure of a target assessment score. For each target assessment score, the framework consists of a sequence of steps: (1) check missing values. If there are missing values in the data, perform imputation to generate multiple sets of complete data sets. (2) For each imputed dataset, build a model for the target assessment with participants’ characteristics, using control subjects only. The model is referred to as the control model for the imputed dataset. (3) For each imputed dataset, get predicted control values and the prediction confidence interval for HDGEC based on the control model from step (2). (4) For each imputed dataset, generate robust assessment scores for target assessment scores. (5) Aggregate the robust assessment scores from multiple imputed datasets. If there is no missing values in the data, steps (2)-(4) will be performed on the observed data set, and step (5) will no longer be needed. We discuss each step in detail below.
- Step 1. Check missing values and perform multiple imputation.
-
All four HD observational studies have missing values. Therefore, the aggregated data contain missing values. To cope with this problem, we performed Multiple Imputation (MI) with the Fully Conditional Specification method [10] and Predictive Mean Matching [11] to impute the missing values and generated multiple sets of complete data sets.
Multiple Imputation[12] is a statistical technique for analyzing incomplete data sets. Instead of filling in a single value for each missing value, MI procedure replaces each missing value with a set of plausible values. Uncertainty about the value to impute can be represented by the multiple imputed values. These multiple imputed data sets are analyzed individually. Results from the multiple sets of complete data sets are then aggregated to generate the final results. In this paper, we applied the MI procedure using the MICE package in R [13] and generated ten sets of complete data sets.
- Step 2. Build control models.
With each imputed data set, we build a model for the target assessment score using available patient characteristics as the predictors. The goal of this step is to build a model with high predictive power. For each target assessment score of interest on each imputed dataset, we compared multiple candidate predictive models, and choose the one with the highest predictive power (measured by R-squared) as the model of choice for the target assessment score on the imputed dataset. In this study, we used three types of models as candidate control models in the experiments: the generalized linear regression model, Support Vector Machines (SVM) with RBF kernel, and Multivariate Adaptive Regression Splines(MARS). The proposed method is not limited to the three types of models. Other types of predictive models can be included in this step.
- Step 3. Get prediction confidence interval of a target assessment score on HDGECs from control models.
Once a candidate model is selected as the control model for a target assessment score on an imputed data set, we obtain predicted control values of the target assessment scores for HDGECs on the imputed data set. We also obtain the lower and upper bounds of the 95% confidence interval of the predicted control value (PCI) for each case observation. In this paper, we used bootstrap method to obtain the PCI for case observations on each imputed dataset.
- Step 4. Generate robust assessment scores.
The PCI obtained from Step 3 gives an interval estimate of what an assessment score would be for a hypothetical control participant with similar characteristics as a case participant. We define the symptom onset as the event when a case participant can be distinguished from the control participants. The PCI obtained from the previous step can be used to mark the boundary to determine whether a case participant presents significant difference from the controls. Therefore, the time of symptom onset is defined to be the first time that an assessment score of a case participant falls out of the PCI. Following the above definition, the robust assessment score is defined as the distance between the observed assessment score and the boundary of the PCI. Figure 2 summarizes the method. If the raw assessment score decreases with time (left panel of Figure 2), the difference between the observed value and the lower bound of the PCI is used as the robust assessment score. If the raw assessment score increases with time (right panel of Figure 2), the difference between the upper bound of PCI and the observed value is used as the robust assessment score. The sign of a robust assessment score from an observation can indicate whether the participant show significant difference from controls in the symptom assessed by the corresponding raw assessment score. A positive sign indicates that the participant cannot be distinguished from controls. A negative sign indicates that the participant can be distinguished from controls. The value 0 serves as a natural threshold for deciding the onset of a new symptom. Therefore, the robust assessment score can be used as a symptom onset indicator. Note that a robust assessment score can be generated from each raw assessment score. A robust assessment score assesses the same symptom as the symptom targeted by its corresponding raw assessment score. Multiple robust assessment scores together could be used to provide a comprehensive view of the progression pathway of the target disease.
- Step 5. Aggregate results from multiple imputed data.
Up to Step 4, for each HDGEC observation, we generate ten robust assessment scores, each from one imputed data set. The final step is to aggregate the ten sets of robust assessment scores. In this study, we used the average across the ten sets as the final robust assessment scores. When there is no missing values in the observed data set, steps 2-4 will be performed on the observed data set and step 5 will be skipped.
Figure 2.
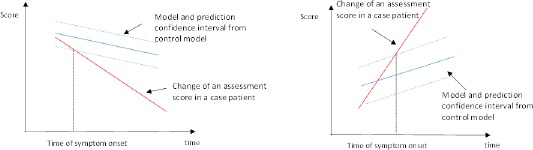
Description of the method in Step 4
4. Results
We applied the proposed procedure to the integrated HD observational data. In this section, we demonstrate the characteristics of the robust assessment scores using two cognitive assessments: the Stroop Word Reading Test (SWRT) score and the Symbol Digit Modalities Test (SDMT) total correct score.
Both the two assessments have missing values in the data. Step 1 of the proposed method generated multiple complete data sets using the Multiple Imputation method. We first check the quality of the imputed data. The imputed values should 1) have the same support as the observed data; 2) have similar distributions as the observed data. We adopted the Predictive Mean Matching method in the MI step, therefore the imputed values were guaranteed to have the same support as the observed data. In this section, we compare the distributions of the imputed values with the observed values by visual inspection. One example of the inspection is showed in Figure 3. The left panel of Figure 3 shows the distribution of observed SDMT scores in each age group, and the right panel shows the distribution of imputed SDMT scores in each age group. The plots demonstrate that the distributions of the imputed value are similar to that of the observed values. Similar inspections were performed for observed and imputed values versus other factors. Due to lack of space, we do not show the details in this paper.
Figure 3.
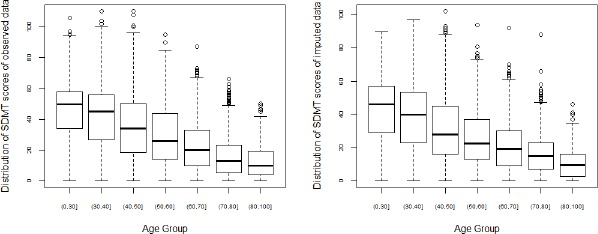
Boxplots of imputed and observed SDMT scores in each age group
After obtaining the ten sets of imputed data, we build control models for SDMT and SWRT separately on each complete data set. Patient characteristics, such as study ID, age, gender, and education levels, were used as predictors in the control models. For each assessment score on each imputed dataset, three types of candidate models (Generalized Linear Regression (GLM), Support Vector Machine (SVM) and Multivariate adaptive regression splines (MARS)) were compared. The type of model with the highest R-squared value was selected to build the control model on the imputed data set. A summary of the R-squared values from the selected models are listed in Table 1. After building the control models, bootstrap method was used to obtain the 95% PCI for each case observation. The robust assessment scores for each complete dataset were calculated following the descriptions in Section 3. In the final step, we aggregated the robust scores from the ten sets of complete data sets by calculating the mean values across the ten sets of complete data sets.
Table 1:
Summary of selected model types and R-squares of the control models
| Mean of R-squared | std. of R-squared | |
|---|---|---|
| SDMT | 0:3994 | 3:46 × 10–5 |
| SWRT | 0:2519 | 1:68 × 10–5 |
Next we discuss the properties of the robust assessment scores. Values of raw assessment scores were not only influenced by HD disease status and progression, but also by other non-disease-related factors. The proposed method utilizes the control cohort to model and adjust the effect of non-disease-related factors. The robust assessment scores are expected to be less subject to the non-disease-related factors. Figure 4 and 5 show two examples comparing the distributions of raw and robust SDMT scores among the HDGECs versus two patient characteristics, which are age groups and ISCED education levels. The left panels show the distributions of the raw SDMT scores vs. age groups and ISCED education levels, respectively. The right panels show the distributions of the robust SDMT scores vs. age groups and ISCED education levels, respectively. The raw SDMT scores demonstrate strong correlation with age and education levels, while the robust SDMT scores demonstrate decreased correlation with the two factors.
Figure 4.
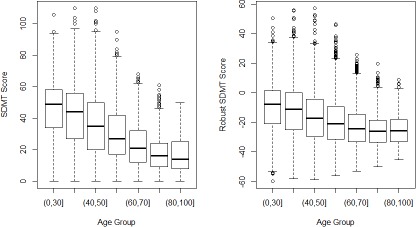
Boxplots of the Original and robust SDMT scores vs age groups
Table 2 summarizes the influences of non-disease-related factors in raw and robust SDMT/ SWRT scores. For categorical factors, we calculate the average Cohen’s d effective sizes of the scores between pairs of levels of the factor. For continuous factors, we report the Spearman’s correlation coefficients. A smaller average effective size or smaller absolute value of Spearman’s correlation coefficient indicates decreased influence of the non-disease-related factor. Most non-disease-related factors show decreased influence in the robust scores. The few exceptions can be attribute to the imbalance of factors’ effects in the control model. In general, the robust scores are less subject to changes in non-disease-related factors.
Table 2:
Correlations and average effective sizes of raw and robust SDMT/SWRT scores with patient characteristics
| Study | Age | Marital Status | Education Level | Gender | Region | Tobacco abuse history | Drug abuse history | |
|---|---|---|---|---|---|---|---|---|
| SMDT | 0.798 | –0.378 | 0.493 | 0.918 | 0.216 | 0.715 | 0.574 | 0.674 |
| Robust SDMT | 0.679 | –0.269 | 0.277 | 0.287 | 0.309 | 0.622 | 0.596 | 0.514 |
| SWRT | 0.793 | –0.320 | 0.438 | 0.902 | 0.169 | 0.653 | 0.660 | 0.636 |
| Robust SWRT | 0.587 | –0.280 | 0.254 | 0.250 | 0.129 | 0.795 | 0.511 | 0.407 |
The values of raw SDMT scores range from 0 to 120. A few previous literature [14, 15] reported some threshold values to distinguish normal vs. impaired cognitive abilities. However, whether these threshold values can be applied to HDGECs has not been systematically tested. There is no clear threshold to decide when a HDGEC participant starts to show the symptom of cognitive impairment. A robust SDMT score comes with a natural threshold (i.e. 0) for deciding whether a HDGEC starts to show cognitive impairment. Since influences of the non-disease-related factors for the observation have been adjusted in the robust assessment score, the threshold is personalized and specific to HD. Similarly, a value of 0 serves as a natural personalized threshold for other robust assessment scores to determine the onset of other symptoms in HDGECs. It is worth mentioning here that ‘onset’, for a case participant, is being identified as a deviation from the model learned based on control participants. This data driven ‘onset’ is a hypothesis that needs further clinical validation.
5. Discussion
In this paper, we proposed a method for generating robust assessment scores and symptom onset indicators from patient registry data set which have recruited both case and control participants. The generated scores are more robust in the sense that they are less subject to changes due to non-disease-related factors. Therefore are more relevant when evaluating subjects’ disease status and tracking disease progression. The signs of the robust assessment scores indicate whether an observation can be distinguished from the control cohort. The value 0 serves as a natural indicator to indicate the onset of the symptom assessed by the score.
We applied the proposed procedure to an integrated HD observational data set and discussed the properties of the robust assessment scores using two cognitive assessments. The proposed procedure is not limited to these two assessment scores. It could be applied to other assessment scores in HD observational data.
The proposed method may also be applied to other disease registry data that have recruited both case and control participants. However, the application of the proposed method to other disease registry data should be conducted with caution. Owing to the monogenic nature of HD, the identification of the control participants in HD observational data is relatively clearer compared to other types of diseases. Participants in the control cohort of HD data sets by definition are not expected be affected by any direct (or known) HD disease-related factors. The identification of a control cohort in other disease registry may require more efforts such as matching and stratification.
One limitation of the proposed method comes from the nature of observational studies. The proposed method can only adjust the effects of non-disease-related factors that are available among both case and control participants. If a factor is not collected in the patient registry data or is only available in one of the groups, its influences cannot be adjusted. In other words, the proposed procedure only mitigates the effects of non-disease-related factors, but does not guarantee elimination of their effects entirely.
Despite the aforementioned limitations, the proposed procedure is useful in improving the quality of assessments scores in patient registry data. The value of zero serves as an indicator for detecting symptom onsets. Better understanding of the course of the disease could be obtained by comparing the times and order of multiple symptoms. We will explore the clinical insights of the generated symptom onset indicators in future work.
Figure 5.
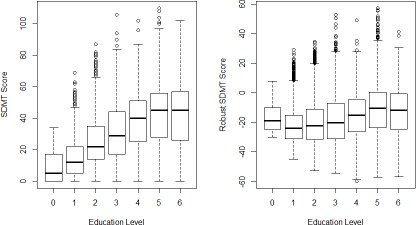
Boxplots of the Original and adjusted SDMT scores vs ISCED education levels
Acknowledgement
Enroll-HD: Data used in this work was generously provided by the research participants in ENROLL-HD and CHDI Foundation, Inc that financially supports the study.
TRACK (TRACK-HD & TRACK-ON): Data used in this work was generously provided by the research participants from the TRACK-HD and TRACK-ON studies and made available by the TRACK Investigators.
REGISTRY-HD: Data used in this work was generously provided by the the participants of the European Huntington’s Disease Network (EHDN) REGISTRY study and made available by the EHDN REGISTRY Investigators.
PREDICT-HD: Data used in this work was generously provided by the participants in PREDICT-HD and made available by the PREDICT-HD Investigators and Coordinators of the Huntington Study Group, Jane Paulsen, Principal Investigator.
References
- [1].Gliklich R, Dreyer N, Leavy M. Registries for Evaluating Patient Outcomes: A User’s Guide. (Prepared by the Outcome DEcIDE Center [Outcome Sciences, Inc., a Quintiles company] under Contract No. 290 2005 00351 TO7.) AHRQ Publication No. 13(14)-EHC111. Rockville, MD: Agency for Healthcare Research and Quality. [Google Scholar]
- [2].Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nature reviews Neurology. 2014;10(4):204–216. [Google Scholar]
- [3].Stout JC, Paulsen JS, Queller S, Solomon AC, Whitlock KB, Campbell JC, et al. Neurocognitive signs in prodromal huntington disease. Neuropsychology. 2011;25(1):1. doi: 10.1037/a0020937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tabrizi SJ, Scahill RI, Owen G, Durr A, Leavitt BR, Roos RA, et al. Predictors of phenotypic progression and disease onset in premanifest and early-stage huntingtons disease in the track-hd study: analysis of 36-month observational data. The Lancet Neurology. 2013;12(7):637–649. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- [5].Mestre T, Fitzer-Attas C, Giuliano J, Landwehrmeyer B, Sampaio C. Enroll-hd: A global clinical research platform for huntingtons disease (s25. 005) Neurology. 2016;86(16 Supplement):S25–005. [Google Scholar]
- [6].Orth M, Handley OJ, Schwenke C, et al. Observing Huntingtons disease: the European Huntingtons disease networks REGISTRY. PLoS Currents. 2011;2:RRN1184. doi: 10.1371/currents.RRN1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tabrizi SJ, Scahill RI, Owen G, Durr A, Leavitt BR, Roos RA, et al. Predictors of phenotypic progression and disease onset in premanifest and early-stage huntingtons disease in the track-hd study: analysis of 36-month observational data. The Lancet Neurology. 2013;12(7):637649. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- [8].Papoutsi M, Labuschagne I, Tabrizi SJ, Stout JC, et al. The cognitive burden in huntingtons disease: pathology, phenotype, and mechanisms of compensation. EBioMedicine. 2015;29(5):673–683. doi: 10.1002/mds.25864. [DOI] [PubMed] [Google Scholar]
- [9].Paulsen J, Langbehn D, Stout J, Aylward E, Ross C, Nance M, et al. Detection of huntingtons disease decades before diagnosis: the predict-hd study. Journal of Neurology, Neurosurgery & Psychiatry. 2008;79(8):874880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stef van Buuren. Multiple imputation of discrete and continuous data by fully conditional specification. Statistical Methods in Medical Research. 2007;16(3):219–242. doi: 10.1177/0962280206074463. [DOI] [PubMed] [Google Scholar]
- [11].Roderick J. A. Little. Missing-data adjustments in large surveys. Journal of Business & Economic Statistics. 1988;6(3):287–296. [Google Scholar]
- [12].Rubin D.B. Multiple Imputation for Nonresponse in Surveys. Wiley Classics Library: Wiley; 1987. [Google Scholar]
- [13].Stef van Buuren, Karin Groothuis-Oudshoorn. mice: Multivariate imputation by chained equations in r. Journal of Statistical Software. 2011;45(3) Open Access. [Google Scholar]
- [14].Mariana Lpez-Gngora, Luis Querol, Antonio Escartn. A one-year follow-up study of the Symbol Digit Modalities Test (SDMT) and the Paced Auditory Serial Addition Test (PASAT) in relapsing-remitting multiple sclerosis: an appraisal of comparative longitudinal sensitivity. BMC Neurology. 2015;15(40) doi: 10.1186/s12883-015-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ugo Nocentini, Angela Giordano, Sarah Di Vincenzo, Marta Panella, Patrizio Pasqualetti. The Symbol Digit Modalities Test - Oral version: Italian normative data. Functional Neurology. 2006;21(2):93–96. [PubMed] [Google Scholar]


