Abstract
Monitoring the future health status of patients from the historical Electronic Health Record (EHR) is a core research topic in predictive healthcare. The most important challenges are to model the temporality of sequential EHR data and to interpret the prediction results. In order to reduce the future risk of diseases, we propose a multi-task framework that can monitor the multiple status ofdiagnoses. Patients’ historical records are directly fed into a Recurrent Neural Network (RNN) which memorizes all the past visit information, and then a task-specific layer is trained to predict multiple diagnoses. Moreover, three attention mechanisms for RNNs are introduced to measure the relationships between past visits and current status. Experimental results show that the proposed attention-based RNNs can significantly improve the prediction accuracy compared to widely used approaches. With the attention mechanisms, the proposed framework is able to identify the visit information which is important to the final prediction.
Introduction
Disease monitoring is often limited by physician experience, test time, economic barriers and so on. The Electronic Health Record (EHR), which consists of longitudinal health information of patients, is a valuable source for exploratory analysis to monitor diseases and assist clinical decision making. However, due to the complexity of EHR data, the efficient mining of EHRs is not trivial. Firstly, EHR data is heterogeneous which contains various types of features. For example, type of visit is a categorical feature while body mass index is continuous. In addition, some features are static through the lifetime while some change dynamically. Models should be able to capture the essence of heterogeneous features. Secondly, the data is inherently sparse and noisy, due to patient’s irregular visits, absence of tests, and incomplete recording, etc. Thirdly, result interpretation in healthcare applications is essential, and the lacking of interpretability often hinders the adaption of models in clinical settings. Thus, how to correctly model heterogeneous and sparse EHR data and reasonably interpret the prediction results is a challenging problem for disease prediction.
Recent work has made rapid progress in utilizing EHRs for predictive modeling tasks in healthcare, including predicting unplanned readmission1, early prediction of chronic disease2, adverse event detection3 and monitoring disease progression4. In these settings, the EHRs are typically represented as temporal sequences of medical visits, and each visit contains a set of objects (such as diagnosis and procedure codes). The main idea is to learn a good representation of a patient’s historical health information, in order to improve the performance of the prediction for future risks. To capture the progression of the patient’s health status, much effort has been made on regression models5 and Markov models6. However, these models cannot take into account the long-term dependencies of diagnoses, which may miss several severe symptoms in the past and reduce the performance of disease monitoring.
In order to model the dependencies of diagnoses, deep leaning techniques, such as recurrent neural networks, can be employed. Recent work1,2,7–9 shows that deep learning can significantly improve the prediction performance. To handle the temporality of multivariate sequences, dynamically modeling the sequential data is necessary. Recurrent neural networks (RNNs), in particular Long Short-Term Memory (LSTM) and Gated Recurrent Units (GRU), have achieved stat-of-the-art performance in handling long-term dependencies and nonlinear dynamics. Taking advantage of the capability of RNN in memorizing historical records, multiple recent models based on RNNs are employed for deriving accurate and robust representations of patient visits. The work by Lipton et al.10 applies LSTM to a multilabel classification task for diagnosing multiple diseases in the future, and the contemporary work by Choi et al.11 applies GRU to predict codes for subsequent visits. Both of them show the efficacy of basic RNN models in modeling longitudinal healthcare data. Among the state-of-the-art models, RETAIN12 adopts a temporal attention generation mechanism to learn both visit level and code level weights; GRAM13 is a graph-based attention model, which uses medical ontologies to handle data insufficiency and combines with an RNN to learn robust representations; and Diploe14 uses bidirectional recurrent neural networks (BRNNs) to further improve the prediction accuracy.
The aforementioned models are RNN-based frameworks which use medical codes as inputs to predict whether the diagnosis or treatment will appear in the future visit, i.e., binary prediction. However, for some diseases, the doctor may care about the transition and severity level of the clinical event, i.e., multiple prediction. For example, if a person is likely to have osteopenia, the doctor may suggest more exercises and supplements, while if osteoporosis occurs, medications will be necessary. To measure the severity of diagnoses, the diagnosis values may be discretized into multiple status: normal range and abnormal range of different severity (i.e. low/high abnormal range), following doctor’s advice or medical references. As a disease may be characterized by multiple important observations or diagnoses, we need to monitor these variables simultaneously.
In this paper, our goal is to predict the status of multiple diagnoses (or observations), with each diagnosis having multiple severity levels. We form our problem as multi-task learning, which first learns a shared representation from all the features, and then performs task-specific predictions. We propose an attention-based RNN model to monitor patient’s longitudinal health information. First, we use an RNN to memorize all the information from historical visits, and then attention mechanisms to measure visit importance. Based on the latent representation, we train multiple classifiers and each focuses on the prediction of a specific task. We perform our model on two applications: predicting chronic states for bone health, and monitoring BloodTest values for cardiovascular disease. Our main contributions can be summarized as follows:
We propose a multi-task framework to monitor the future status of different clinical diagnoses. We process the monitored diagnoses to multiple severity status following medical references, which can help doctors to make more precise decisions on controlling risks.
We employ three attention mechanisms to evaluate the importance of previous visits to prediction tasks. This gives the explanation of visit importance, which can provide suggestions for doctors to pay more attention on the information from a specific timestamp.
Our experiments show promising results of using RNNs to handle historical health information from longitudinal records. We empirically show that the proposed attention-based RNNs outperform widely used methods in multi-diagnoses prediction on real world EHR datasets.
This work will result in an effective tool for the physicians to monitor disease progression for early treatment in a more efficient way. This framework can be used real-time on a regularly scheduled basis which highlights patients whose disease state is more likely to worsen.
Method
In this section, we first introduce the format of our healthcare datasets and some basic notations. Then we describe the details of the proposed framework, including the preliminary of RNN structure, proposed attention mechanisms, and the multi-task model. Finally, we describe the interpretation for analyzing the importance of different visits.
Basic Notations
The EHR data contains heterogeneous variables such as diagnosis results, lab data, and physical functions. Among them, diagnosis results are what we care about most, and we want to monitor their progression. Other variables are risk factors that may potentially influence patient’s health status. Assume that there are N patients and M diagnoses to be monitored, and the total number of visit records for the n-th patient is T(n). The health record of a patient can be represented by a sequence of visits V1, V2,…, VTn. Each visit Vt is denoted by a vector of feature variables xt. To monitor patient’s health status progression, the diagnosis results are discretized into several classes, indicating the severity level of the disease, following doctor’s opinions or medical references. For example, in a patient’s visit for bone health test, bone mineral density (BMD) in different areas such as femoral neck and intertrochanteric is measured, and the X-ray scan results can be diagnosed as normal, osteopenia and osteoporosis. We want to predict the severity range of BMD value in each bone area in this patient’s next visit. For simplicity, we describe the proposed method for a single patient and drop the superscript (n) in the following notations when it is unambiguous. Figure 1 illustrates the health records of one patient in our data. The patient has multiple visits, and each visit contains multiple variables. Each monitored variable falls into a severity range. Suppose that we are currently at time t and want to know the diagnoses at time (t + 1), this patient’s historical records from Vj to Vt can be utilized for the training of the model.
Figure 1.
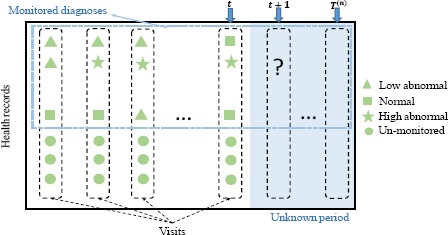
An example of health records of the n-th patient.
Model
The basic component of our framework is gated recurrent unit, which is a state-of-the-art deep learning architecture for modeling long range sequences. To further improve its performance, we apply attention mechanisms to measure the importance of historical sequences. To predict the status of multiple diagnoses, we add a multi-task classification layer on top of the learned representations.
Recurrent Neural Network
Recurrent neural network (RNN) captures the characteristics of the input sequence by recursively updating its internal hidden states. Figure 2(a) shows the unfolded RNN structure for a general classification task. For the first visit Vj (i.e., t = 1), RNN learns a hidden state h j to represent the input feature vector x j; as time moves to t = 2, feature vector x2 together with hj are fed into the RNN to update parameters in the network, and the learned hidden state h2 contains information from both x2 and x j. Through updating the network parameters recursively, the hidden state ht learns all the previous information from xj to xt_ j. Then a softmax classifier is applied on ht to perform classification. As the parameters of the network are shared by each visit, RNN can handle patients with different visit lengths.
Figure 2.

Illustration of RNN models.
We implement our RNN with Gated Recurrent Units (GRU)15, which has been shown to have comparable performance as Long-Short Term Memory (LSTM), while employing a simpler architecture. The structure of GRU is shown in Figure 2(b). A GRU has two gates, a reset gate r and an update gate z. Intuitively, the reset gate determines the combination of the new input and the previous memory, which allows the hidden layer to drop irrelevant information that is not useful to the prediction, and the update gate controls how much information from the previous hidden layer should keep around. The mathematical formulation of GRU can be described as follows:
| (1) |
where о denotes the entry-wise product, σ is the activation function, rt and zt represent the reset gate and update gate at time t respectively, is the intermediate memory unit, and ht is the hidden unit. Matrices Wr, Wz, W, Ur, Uz, U and vectors br, bz, b are model parameters to be learned. At time t, we take the hidden state ht to predict the labels of time (t + 1).
Attention Mechanism
As mentioned in the above section, RNN can remember the past information for future prediction. However, it is limited to only a few latest steps, with more impact from later ones, and may not be able to discover major influences from earlier timestamps. Therefore, we apply attention mechanisms to memorize the effect from long-time dependencies, which have gained success in many tasks. In neural machine translation18, the attention mechanism can be intuitively described as follows: given a sentence of length S in the original language, RNN is adopted to generate the word representations h1,…, h|S|. To find the t-th word in the target language, we assign each word in the original language an attention score ati, and then calculate a context vector to perform prediction. Through attention mechanism, RNN can focus on specific words when generating each target word. Similarly, in diagnoses prediction, we use a temporal attention mechanism to predict medical results in the (t + 1)-th visit, according to visit records from x1 to xt. The hidden state ht from the t-th visit can be estimated as a representation for the (t + 1)-th visit. However, it may not contain enough long-term visit information. Therefore, we need to derive a context vector ct which captures relevant information to help prediction. We propose three methods to compute attention score at in order to obtain the context vector ct: location-based attention, general attention and concatenation-based attention.
The attention mechanisms are illustrated in Figure 3. The general procedure goes as follows: we first obtain a set of hidden states through the GRU layer, and then calculate the attention score αt for each of them, in order to obtain the context vector ct; an attentional hidden state ht is then calculated by combining ct and ht. Thus ht contains both current and historical information. Location-based attention, as in Figure 3(a), calculates the attention score solely from each individual hidden state hi(1 ≤ i ≤ t –1) using formula: , where Wα and bα are parameters to be learned. Since location-based attention mechanism only considers each hidden state individually, it does not capture the relationships between the current hidden state and all the previous hidden states. The other two attention mechanisms, as shown in Figure 3(b), calculate attention weight αti by considering the relationship between ht and hi. General attention uses a weight matrix Wα to connect ht and hi through formula: . For concatenation-based attention, we first concatenate the current hidden state ht and the previous state hi, and then calculate a latent vector by multiplying a weight matrix Wα. Thus the attention weight vector is generated as: .
Figure 3.

Attention mechanisms.
After obtaining αt, we can obtain the context vector ct through formula which contains the weighted hidden representations of the past visits from x1 to xt−1. To combine the information from context vector ct and the current hidden state ht, we employ a simple concatenation layer to generate an attentional hidden state using , where Wc is the weight matrix to be learned. contains all the information from x1 to xt, such that the prediction task can be performed on top of .
Multi-task Diagnosis Prediction
Our task is to predict the status of multiple measurement results at the time (t +1) given the historical records from x1 to xt Figure 4 shows a high-level overview of the proposed model. Given the information from time 1 to t, the i-th visit’s health record xi is fed into an RNN network, which outputs a hidden state hi as the representation of the i-th visit. Along with the set of hidden states , we compute their relative importance αt, and then obtain a context state ct. From the context state ct and the current hidden state ht, we can obtain an attentional hidden state , which is used to predict diagnoses in the (t + 1)-th visit. For the prediction, we use M softmax classifiers, which correspond to the M different diagnoses, to predict the severity level for each diagnosis. The representation ht contains the visit information of all the input features, and the task-specific classifier focuses on the prediction of each diagnosis.
Figure 4.

Overview of the proposed model.
To perform the multi-task classification, we feed the hidden state into each task through a classification layer. Thus the information of the k-th diagnosis in the (i + 1)-th visit can be produced by , where and are parameters to be learned. We use cross-entropy between the ground truth yt and the predicted to calculate the classification loss. The total loss is the sum of cross-entropy among all the diagnoses categories in predicted visits of patients. The loss function L can be described as:
| 2 |
where N is the number of patients, M is the number of monitored diagnoses, and T(n) is the number of visits of the n-th patient. In the training procedure, we estimate parameters in the proposed models by minimizing the loss function (2).
Interpretation
In healthcare applications, giving interpretation of the learned representations is important. Here we evaluate the contribution of the past visits to the prediction of future status in the process of learning latent representations. Since we adopt attention mechanisms, the importance of each visit can be found by analyzing its attention score. For example, for the t-th prediction, if the attention score ati is large, then the probability of the (i + 1) -th visit information related to the current prediction is high. In most cases, the last visit is usually important for chronic diseases, as patient’s health status usually does not change much during two visits. However, since disease progression is complex and affected by many factors, the disease can get better or worse. Thus the health information of specific earlier visits may be more important for some patients. Therefore, the attention mechanism can help doctors to pay attention to specific important visits in the past.
Experiments
We conduct experiments on two real-word datasets, and evaluate the performance of the proposed attention-based RNN models compared to other prediction methods. Moreover, we use case studies to understand the behavior of the proposed models.
Datasets
Study of Osteoporotic Fractures Dataset. The study of osteoporotic fracture (SOF)20 is the largest and most comprehensive study focused on bone diseases. It includes 20 years longitudinal data about osteoporosis of 9,704 Caucasian women aged 65 years and older. Potential risk factors and confounders belong to several groups such as demographics, family history, and lifestyle. We process people’s bone health diagnoses of different areas using the bone mineral density (BMD) values by comparison with young healthy references17, resulting in three BMD levels: normal, osteopenia and osteoporosis.
BloodTest Dataset. This dataset21 contains multivariate blood tests of 3,000 patients affected by cardiovascular disease from the University Hospital of Catanzaro, Italy. For each patient, there are several blood tests during their in-hospital stay, such as hemoglobin, triglycerides, glucose, and calcium. As suggested by doctors, we pick 12 blood analytes variables which are important to cardiovascular. Each variable has a normal range provided by doctors. Knowing variable transitions in advance can alarm doctors to take actions before the abnormal occurs, in order to reduce the risk of diseases.
As a common issue of EHR, these datasets are irregular sampled and sparse, so that data preprocessing is needed. For each person, we remove those visits without any monitored variables recorded, and remove patients with less than three visits. We use simple imputation to fill missing variables. For the SOF data, we fill the missing variables with the values in the previous visit. For the BloodTest data, we impute missing sequences (where a single variable is missing entirely) with a clinical normal value. This is based on an assumption that clinicians believed it to be normal so that they did not measure it. Other missing variables are filled with the median value of other patients. After data preprocessing and extraction, we obtain the datasets with statistics shown in Table 1.
Table 1:
Statistics of datasets.
| Dataset | SOF | BloodTests |
|---|---|---|
| Number of patients | 5,318 | 2,055 |
| Number of visits | 22,313 | 18,758 |
| Average number of visits per patient | 4.19 | 9.13 |
| Number of normal claims | 25,145 | 221,642 |
| Number of low abnormal claims | 55,399 | 17,407 |
| Number of high abnormal claims | 31,021 | 79,837 |
| Total number of features | 42 | 17 |
| Number of monitored diagnoses | 5 | 17 |
Experiment Setup
For each patient, we want to predict the diagnosis results of each visit based on his/her previous records. To validate the performance of the proposed models in this diagnosis prediction task, we conduct experiments on two categories of methods: baselines and RNN-based models.
We set up two kinds of baselines. The first baseline is to use the median value of each monitored variable from V1 to Vt to predict Vt+1 for continuous variables. This is based on a heuristic assumption that the most frequent state is more likely to occur. For each patient, we use his/her most popular health status as the current status, regardless of time variations. The second baseline is a multi-task logistic regression (LR). To predict information at Vt+1, we feed the health records at Vt to a logistic regression model with multiple softmax classifiers. This can be viewed as a simplified model of Figure 4 without using RNNs and attention mechanism to learn latent states. This model only considers the effect from previous one time step, rather than long time history.
For the proposed methods, we have several variants, including a plain RNN or attention-based RNNs. For RNN, the architecture is similar to the proposed model, but without the attention mechanism. RNN;, RNNg and RNNc are three attention-based RNN models, whose architecture is shown in Figure 4. RNN;, RNNg and RNNc stands for location-based, general and concatenation-based attention respectively. The attention mechanism of RNN; can be seen in Figure 3(a), and attention mechanism of RNNg and RNNc can be seen in Figure 3(b).
The proposed approaches are implemented with Theano 0.7.022. Adadelta16 with a mini-batch of 50 patients is used to optimize Eq. (2). To evaluate prediction performance, we define the accuracy as the ratio between correctly predicted severity status and the total number of variables to be predicted.
Results of Diagnosis Prediction
Table 2 shows the accuracy of the proposed approaches in comparison with baselines on the two datasets. For each patient in the testing set, we predict the health conditions for the subsequent visits using his/her historical health records. For the SOF dataset, we predict the probability of BMD states of normal, osteopenia and osteoporosis for different measurements such as hip and femoral neck. For the BloodTest dataset, we predict the probability of each blood analyte falling into normal, low abnormal and high abnormal. The results are averaged over 5 random trials of 5-fold cross validation. Avg.# Correct represents the average number of correctly predicted claims of 5 random trials. Accuracy represents the ratio between correctly predicted claims and total number of claims to be predicted.
Table 2:
Prediction performance on two datasets.
| Method | SOF | BloodTest | ||
|---|---|---|---|---|
| Avg.# Correct | Accuracy | Avg.# Correct | Accuracy | |
| Median | 10,509 | 0.8209±0.0057 | 32,253 | 0.7616±0.0013 |
| LR | 10,125 | 0.7909±0.0069 | 34,836 | 0.8225±0.772 |
| RNN | 10,769 | 0.8412±0.0042 | 36,167 | 0.8540±0.0051 |
| RNNl | 10,822 | 0.8454±0.0031 | 36,443 | 0.8605±0.0056 |
| RNNg | 10,805 | 0.8440±0.0027 | 36,423 | 0.8600±0.0059 |
| RNNc | 10,816 | 0.8449±0.0023 | 36,560 | 0.8632±0.0051 |
We can observe that RNN based methods outperform other baselines. The method of predicting with median values considers each variable separately, without taking into account the time trend and feature relationships. This intuitive method is very sensitive to noise, and cannot capture the correlation between variables. It performs the worst in the BloodTest dataset, possibly due to the reason that variables in that dataset are not independent but have strong correlations (e.g. MYO-CKM-TRHS-GPT-GL-GOT-LDH). Logistic regression (LR) takes the whole inputs into a classifier with multiple softmax functions, in order to classify each monitored variable. The structure of LR can be viewed as the framework of Figure 4 without latent representations to memorize historical information. The inputs of LR include all the features, such that features can impact on each target task. However, there is no way for logistic regression to memorize historical information, as it only takes information from the nearest one visit. Attention-based RNNs outperform the above baselines. This owes to the capability of RNN in memorizing long-term dependencies of patient’s longitudinal health records, and the attention-based mechanisms can further enhance this capability. For the two datasets, RNN;, RNNg and RNNc can clearly outperform plain RNN. Since the prediction of RNN mostly depends on recent visits, it may not memorize all the past information. Through attention-mechanism, RNN;, RNNg and RNNc can fully take all the previous visit information into consideration, assign different attention scores for past visits, and achieve better performance compared to RNN.
Visit Interpretation
The attention mechanism can be used to understand the importance of historical visits to the current visit. As an example, here we analyze the concatenation-based attention mechanism on the SOF dataset. Figure 5 shows a case study for predicting the diagnoses in the sixth visit through the previous five visits. The concatenation-based attention weights are calculated for the visits from V2 to V5 according to the hidden states hi, h2, h3 and h4. Thus, we have four attention scores corresponding to the visits from V2 to V5. In Figure 5, we select five patients for visualization. The X-axis represents patients, and Y-axis is the attention score calculated for each visit. We can observe that for different patients, the attention scores learned by this attention mechanism are different.
Figure 5.
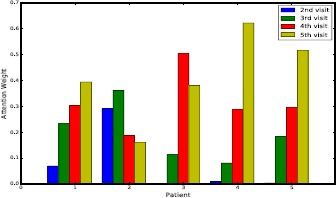
Attention weights of five persons, each with four visits.
For chronic diseases, the last visit is often the most important since patients’ health conditions change slowly. As in the figure, for the first, fourth and fifth patients, the importance of visit increases with time going on. However, this is not always the case due to the complexity of disease progression and impact from risk factors. Table 3 shows the variation of bone mineral density (BMD) diagnoses and attention scores of different visits of the second patient. In each visit, there are five different BMD diagnoses, and the values in the table indicate the severity of bone density loss. Although V4 and V5 are closer to V6 in terms of time, V2 and V3 have the same condition as V6. Thus health records of V2 and V3 are more important to V6. We can see that the attention mechanism correctly assigns larger weights to V2 and V3. As for the BloodTest dataset, using attention mechanism to memorize all the past information is also important. An abnormal blood analyte can temporarily turn into normality via medicine, but it may fall back after some time. Therefore, interpreting visit importance through the attention mechanism can help to better monitor disease progression.
Table 3:
BMD diagnoses and attention scores of one patient with six visits on SOF dataset. 0 is normal, 1 is osteopenia, and 2 (osteoporosis) does not occur for this patient.
| Diagnosesn\Visits | V2 | V3 | V4 | V5 | V6 |
|---|---|---|---|---|---|
| Total hip | 0 | 0 | 0 | 0 | 0 |
| Femoral neck | 1 | 1 | 0 | 0 | 1 |
| Intertrochanteric | 0 | 0 | 0 | 0 | 0 |
| Trochanteric | 0 | 0 | 0 | 0 | 0 |
| Wards | 1 | 1 | 1 | 1 | 1 |
| Attention weights | 0.290 | 0.361 | 0.187 | 0.162 | – |
In diagnosis prediction, making decisions using very recent record is usually not enough, and it is important to lookup long term health information. To understand the relationship between the length of patient medical history and the prediction performance, we select 1,000 patients from the BloodTest dataset with more than seven visits. Table 4 shows the accuracy of RNNl in predicting the diagnoses from V2 to V7. We can see that with the number of visit increasing, the performance can often improve. We believe that it is due to the fact that RNN is able to learn better estimates of patient information as it memorizes longer health records.
Table 4:
Prediction accuracy for V2 to V7 on BloodTest dataset.
| Visit | Accuracy |
|---|---|
| V2 | 0.8579 |
| V3 | 0.8624 |
| V4 | 0.8706 |
| V5 | 0.8792 |
| V6 | 0.8780 |
| V7 | 0.8735 |
Conclusions
In this paper, we introduce attention-based RNN architectures to predict patients’ disease progression. In particular, we monitor multiple diagnoses status simultaneously, based on patients’ historical health records. By employing recurrent neural network, our model can remember hidden knowledge learned from previous visits. Three attention mechanisms allow us to interpret the prediction results reasonably. Experimental results on two real world EHR datasets show the effectiveness of the proposed attention-based RNN models for simultaneously predicting multiple diagnoses. Analysis shows that the attention mechanisms can assign meaningful weights to previous visits when predicting the future visit information. The proposed approach can be widely used for the prediction of a variety of different diseases.
Acknowledgement
The authors would like to thank Dr. Giovanni Cuda (MD, PhD) for his help and advice. This work was supported in part by the US National Science Foundation under grants NSF IIS-1218393 andIIS-1514204. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
References
- 1.Nguyen P, Tran T, Wickramasinghe N, Venkatesh S. Deepr: A convolutional net for medical records. IEEE Journal of Biomedical and Health Informatics. 2016 Dec 1; doi: 10.1109/JBHI.2016.2633963. [DOI] [PubMed] [Google Scholar]
- 2.Cheng Y, Wang F, Zhang P, Hu J. Risk prediction with electronic health records: A deep learning approach. In Proceedings of the 2016 SIAM International Conference on Data Mining; 2016 Jun 30; Society for Industrial and Applied Mathematics. pp. 432–440. [Google Scholar]
- 3.Ma F, Meng C, Xiao H, et al. Unsupervised Discovery of Drug Side-Effects from Heterogeneous Data Sources. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. [Google Scholar]
- 4.Wang X, Sontag D, Wang F. Unsupervised learning of disease progression models. In Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining; 2014 Aug 24; ACM. pp. 85–94. [Google Scholar]
- 5.Zhou J, Liu J, Narayan VA, Ye J. (and Alzheimer’s Disease Neuroimaging Initiative). Modeling disease progression via multi-task learning. NeuroImage. 2013 Sep 30;78:233–48. doi: 10.1016/j.neuroimage.2013.03.073. [DOI] [PubMed] [Google Scholar]
- 6.Henriques R, Antunes C, Madeira SC. Generative modeling of repositories of health records for predictive tasks. Data Mining and Knowledge Discovery. 2015 Jul 1;29(4):999–1032. [Google Scholar]
- 7.Li H, Li X, Ramanathan M, Zhang A. Prediction and informative risk factor selection of bone diseases. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB) 2015 Jan 1;12(1):79–91. doi: 10.1109/TCBB.2014.2330579. [DOI] [PubMed] [Google Scholar]
- 8.Suo Q, Xue H, Gao J, Zhang A. Risk Factor Analysis Based on Deep Learning Models. In Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics; 2016 Oct 2; ACM. pp. 394–403. [Google Scholar]
- 9.Che Z, Kale D, Li W, Bahadori MT, Liu Y. Deep computational phenotyping. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2015 Aug 10; ACM. pp. 507–516. [Google Scholar]
- 10.Lipton ZC, Kale DC, Elkan C, Wetzell R. Learning to diagnose with LSTM recurrent neural networks. 2015 Nov 11; arXiv preprint arXiv:1511.03677. [Google Scholar]
- 11.Choi E, Bahadori MT, Sun J. Doctor ai: Predicting clinical events via recurrent neural networks. 2015 Nov 18; arXiv preprint arXiv:1511.05942. [PMC free article] [PubMed] [Google Scholar]
- 12.Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W. RETAIN: An Interpretable Predictive Model for Healthcare using Reverse Time Attention Mechanism. In Advances in Neural Information Processing Systems. 2016:3504–3512. [Google Scholar]
- 13.Choi E, Bahadori MT, Song L, Stewart WF, Sun J. GRAM: Graph-based Attention Model for Healthcare Representation Learning. 2016 Nov 21; doi: 10.1145/3097983.3098126. arXiv preprint arXiv:1611.07012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma F, Chitta R, Zhou J, et al. Dipole: Diagnosis Prediction in Healthcare via Attention-based Bidirectional Recurrent Neural Networks. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. [Google Scholar]
- 15.Chung J, Gulcehre C, Cho K, Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. 2014 Dec 11; arXiv preprint arXiv:1412.3555. [Google Scholar]
- 16.Zeiler MD. ADADELTA: an adaptive learning rate method. arXiv preprint arXiv:1212.5701. 2012 Dec 22; [Google Scholar]
- 17.Bonnick SL. Totowa, NJ: Humana Press; 1998. Jun 24, Bone densitometry in clinical practice. [Google Scholar]
- 18.Luong MT, Pham H, Manning CD. Effective Approaches to Attention-based Neural Machine Translation. In Empirical Methods in Natural Language Processing. 2015 Aug [Google Scholar]
- 19.Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. 2014 Sep 1; arXiv preprint arXiv:1409.0473. [Google Scholar]
- 20. https://www.sof.ucsf.edu.
- 21.Canino G, Guzzi PH, Tradigo G, Zhang A, Veltri P. On the analysis of diseases and their related geographical data. IEEE journal of biomedical and health informatics. 2015 Oct 30; doi: 10.1109/JBHI.2015.2496424. [DOI] [PubMed] [Google Scholar]
- 22.Bergstra J, Breuleux O, Bastien F, et al. Theano: A CPU and GPU math compiler in Python. In Proc. 9th Python in Science Conf; 2010 Jun; pp. 1–7. [Google Scholar]


