Abstract
Bar charts are crucial to summarize and present multi-faceted data sets in biomedical publications. Quantitative information carried by bar charts is of great interest to scientists and practitioners, which make it valuable to parse bar charts. This fact together with the abundance of bar chart images and their shared common patterns gives us a good candidates for automated image mining and parsing. We demonstrate a workflow to analyze bar charts and give a few feasible solutions to apply it. We are able to detect bar segments and panels with a promising performance in terms of both accuracy and recall, and we also perform extensive experiments to identify the entities of bar charts in the images of biomedical literature collected from PubMed Central. While we cannot provide a complete instance of the application using our method, we present evidence that this kind of image mining is feasible.
Introduction
Bar charts are necessary resources for scientists and practitioners to describe experimental results such as comparisons among categories or grouped data in published biomedical literature. Recently, there has an enormous increase in the amount of open-access heterogeneous biomedical image production and publication, and a trend in the area of literature mining is allowing users to query the figures of biomedical articles which are otherwise not readily accessible.1, 2, 3 These researches focus mostly on image retrieval, image classification and making text within image available. However, biomedical images contain much structured information and quantitative information, which is not yet accessible through search. Below, we present our approach to detect and access bar charts in biomedical publication. For the purpose of further study, we also illustrate some possible situations which take advantage of our bar charts mining method.
Image types are of great importance for image mining. Kuhn et al. manually annotate the segment sub-graphs in the constructed corpus of 3000 images and classify them into five basic categories: Experimental/Microscopy, Graph, Diagram, Clinical, and Picture. The results show that bar chart is the most common types of subfigure accounting for 12.4% of entire set of images,4 which reinforces our determination to study bar charts. Furthermore, the bar charts share the common patterns that have at least two straight and visible axes that one represents specific categories being compared, and the other represents a discrete value, and uses rectangular bars with lengths proportional to the values that they represent.
A closer look at bar chart reveals that they follow regular patterns to encode their semantic relations. Since the priority target of our approach is to automatically extract the relation of the corresponding quantitative proportion to the categorical data, we focus on the text information (the axes labels and the legends) as well as the length proportion of the bars. Figure 1 shows four typical bar chart images. The slight differences about the text and the bars should be noticed. Panel (a) shows the most common bar chart, which has a horizontal x-label, y-label and legends.Panel (b) shows a figure with a horizontal grid and a slanted x-label. Panel (c) shows bars on both sides of the x-axis, and the x-label is vertical. Panel (d) shows the condition in which the bars are filled with slash. These kinds of bar charts represent the most typical bar chart images.
Figure 1.
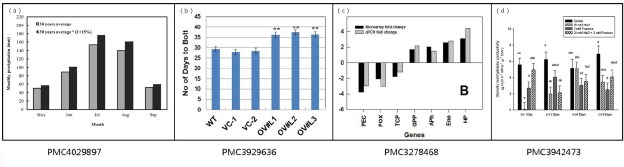
Four typical examples of bar charts.
Figure 2 shows two common bar charts together with a table representation of the extracted information. The first one shows the expression patterns of the GmIFR genes in various tissues. The x-label represent the category of the tissues while the y-label represent the expression level of the gene. A bar chart can be considered a kind of matrix with pictures of experimental artifacts as content. The tables to the right illustrate the semantic relations encoded in the bar charts. Each relation instance consists of a condition, a measurement and a result. Gene is the entity being measured under the conditions of the different tissues. The result is a certain level of expression indicated by the lengths proportion to the bars. Second example is a slightly more complex one. The mRNA transcript level of the GmPAD4, GmEDSl and GmPRl gene on AtPAD4-overexpressing and control roots. More than one gene are tested against each other in a way that involves more than two dimensions. In this case, legend is the major technique to denote the different possible combinations of a number of conditions.
Figure 2.
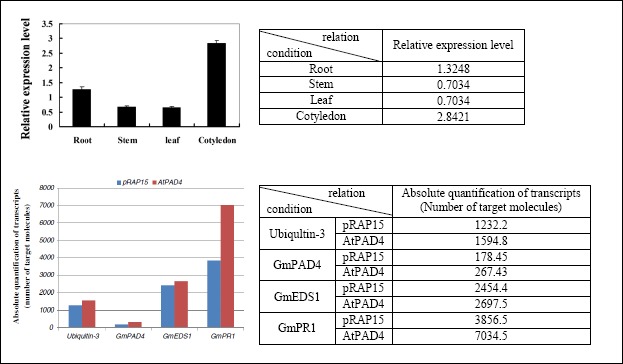
Two examples of bar charts from biomedical publications (PMC4655237 and PMC3648381) with tables showing the relations that could be extracted from them.
Background and related work
Image classification and retrieval are two of the most active areas in biological image mining research. Rafkind et al.5 proposed a classification system using coherence and frequency features to divide figures into five sets. Then, a retrieval method,6 which had better flexibility than the system of Rafkind et al.5, was proposed to retrieve figure types defined conceptually by taking advantage of principles in image understanding, text mining, and optical character recognition (OCR). For searching information in text embedded in figures, YIF (Yale Image Finder)7 has been proposed to retrieve biological figures and associated papers in which the retrieved images allow users to find related papers by linking to their source papers. Hearst et al.8 developed the BioText Search Engine, which is a freely available web-based application for searching and browsing figures in articles as well as their captions. The study of image processing also includes image segmentation, optical character recognition (OCR) and interpretation of figure captions. Li et al.9 proposed an algorithm to segment images that consisted of multiple subfigure into single images and then extract the title and tag information from the graph. Lopez et al.10 developed a robust image segmentation algorithm in order to perform text retrieval based on images. Kim et al.11 developed an image and text extraction tool (figtext) through the combination of image preprocessing, character recognition, and text correction to improve the performance of OCR tools.
Research involving biological images is currently subject to some specific limitations. Only a small among of research has been conducted into the mining of information from image data in biological research or into the construction of a structured biological knowledge base. SLIF (Structured Literature Image Finder) has been developed for automated information extraction from fluorescence microscopy image information, and a series of follow-up studies have been reported for promotion. Their project included image classification1,12, figure title understanding13, figure segmentation14, and the relationships between sub-images and sub-captions.15,16 Also, there is a large amount of existing work on how to process bar charts. Zhou et al.17 proposed a modified probabilistic Hough transform algorithm to detect and recognize bar charts, which is, however, just the first step in locating them and analyzing their labels and their structure. Al-Zaidy et al.18 attempts to extract information from bar charts automatically. However, the method take plain bar chart as input, which are not readily accessible from biomedical papers, because they make up just parts of the figures. Furthermore, the method mentioned above is designed for researchers who want to analyze their bar charts and not to read bar charts that have already been analyzed and annotated by a researcher. Therefore, these approaches do not tackle the problem of recognizing and analyzing bar charts. Some attempt to classify and redesign biomedical images include bar chart, but they are only tested on small data sets that do not satisfy the diversity of images in biological documents.19To the best of our knowledge, there is little research into the mining of bar charts, which is a frequently used means of demonstrating the differences among data.
Method
Our approach to image mining from bar charts consists of 6 components: figure extraction, image preprocessing, bar segment detection, in-image text recognition, panel segmentation, and quantitative information extraction (Figure 3). Figure extraction module that extract images accessed in PubMed. Image preprocessing module that removes non-informative figures to reduce computing cost. For bar segment detection, we use a detection procedure based on hand-coded rules and convolutional neural network (CNN) method to detect bar segments. In-Image Text Processing module identifies the corresponding text region in the image by performing a text location method. Additionally, the Panel Segmentation module combines the results obtained from the previous two modules to first estimate. Finally, we extract the quantitative information among the entities.
Figure 3.
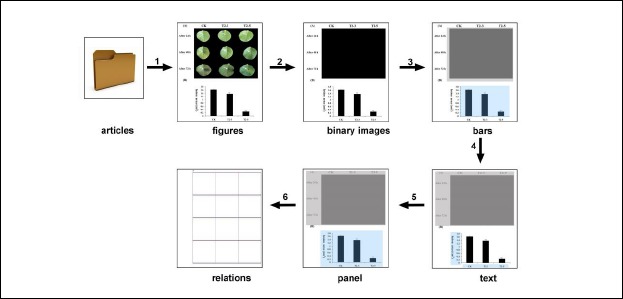
The procedure of our bar charts mining method. (from PMC4482714)
Figure extraction. The data interface (OA web service) are manually accessed in PubMed, which makes it convenient to download article folds consist of documents with different formats (PDF/NXML format articles, GIF/JPEG format images, etc.). We obtain a subset for our future use case by searching the keyword combination of “soybean”, “gene” and “expression.” We only use this subset so far, which makes it easy for us to extract figure. Although it would be definitely feasible, it would also be more trivial for automatic extraction of figures from biomedical articles in PDF format.
Image preprocessing. The download figures from PubMed Central typically incorporate a significant number of non-informative figures (e.g., conference and journal logos, formula), which lead to additional computational processing cost. We separate the objects from the background image by using pixel level features. When converting an image into binary image, we give a fixed threshold value of 0.9 instead of automatic threshold value because we want to get a more complete bar segment. Three factors are considered here to filter out figures that are definitely not bar charts: (1) The number of connected domains is less than 5; (2) the absolute width or height is more than 100 pixels, and (3) .the 0 value rate is more than 95% of binary image.
Bar Segment Detection. We first extract connected components from the filtered image by grouping adjacent pixels of similar color, (for binary image, the adjacent pixels of 1 belong to the same component), and then represent the objects in the figure as a bounding box. The small components (e.g., characters) are filtered according to the area ratio of the connected domains firstly.
As a baseline, we first propose a relatively simple hand-coded bar segment detection method. Such bar segments typically have several bars with the same width distributed uniformly on the x-axis, which is the most distinct feature of the bar chart. For this reason, a projection method is used to detect such bar segments. All the columns are subsequently vertically projected to the x-axis when the area value of the connected domain reached the threshold. The bar charts are detected through defining rules for the ratio of columns that are completely blank and the ratio of the sum of equal columns.
There is an alternative method on using machine learning approaches for image classification. In this paper, we choose a simple CNN model to be the training model for bar charts detecting. With the analysis of the intermediate results (bar segments) generated by the hand-coded method, we can easy get the dataset of CNN. The training set consists of 12000 positive samples and 4000 negative samples, while the test set consists of 3000 positive samples and 1500 negative samples. Experiment of different parameters is conducted and the best model is chosen with the learning rate 1 and iteration times 800.
In-image text recognition. Because the text in figures is of great significance, a text extraction method is used to extract the important text in the figure. Not only should we extract the text from the image, but also the position of the text. Text localization detects different text regions in images. In this part, we focus on x-label, y-label, and legends. All above information are obtained respectively for the necessity of constructing complete corresponding relation. To do this, we use a detection procedure with simple rules. The coordinate axes are used to partition the region. Then the direction of the character is distinguished through combination of the character spacing and character size, and the character region, which is vertical and slanted, is rotated. For optical character recognition (OCR), the open-source tool OCROPY (https://github.com/MissCrastal/ocropy) is used. Sometimes the deletions, insertions, and substitutions of letter or number tokens appeared in the extracted text information from bar chart can be found from the related figure caption text. Spell correction of the extracted in-image text is realized by computing Levenshtein Distance between the extracted in-image text and the corresponding figure caption text.
Panel Segmentation. The last step consists of segmenting the figure into panels using the information extracted from the previous two modules. The sub bar charts detected in the bar segment detection step are found in most case that it is a part of a whole figure in which some parts are not bar charts. Although in our study we just extract information from the image, the information from the article text can be potential additional information to the image. Hence, the order of the sub charts is an important information for further study.
A rectangle segmentation algorithm is used to extract the potential sub bar graphs contained within the figures. After bar segment detection and in-image text recognition processes, the detected bar segments are extended to produce a larger rectangle containing important information such as x-label, y-label, and legends of a sub bar chart. If the area cover the whole figure, the segmentation process is not able to continue because it have only one sub chart. Otherwise, the multiple sub bar charts are generated by reducing the enlarged rectangle based on the in-image text detected. The sub charts that are not sub bar graph were filtered out. On the basis of our observation, sub charts are usually arranged from left to right and from top to bottom. Therefore, we name the sub charts according to the panel position.
Quantitative Information Extraction. The quantitative information in bar charts is obtained through recognition of the x-axis and the height of each bar. Twenty uniformly distributed rows per graph are traversed. If the row crossed the bar, it is traversed up and down to find the critical point and the row index is recorded. The x-axis is here defined as the row at which the row index is recorded the greatest number of times. If the x-axis is in the bottom of the figure, then all of the bars are on top of the x-axis. All the graduated lines including the x-axis are removed to separate all the bars. Then the bars are filled in to make them all solid. Then all of the columns are traversed up to down. The first row index for which the pixel is 0; otherwise, 1 was set as the value, and it is denoted as vector U. Then vector U is traversed the height of a bar is defined if there are no fewer than 5 consecutive identical values. If the x-axis is in the center of the figure, and the bars are located on both sides of the x-axis. All the columns are traversed from both sides, and steps similar to those outlined above are used.
Results
To test our approach, we created a gold standard corpus of images. We randomly selected a sample of 300 open-access articles folds from PubMed Central that consist of jpg image. Altogether, these 300 articles contain 1769 figures, an average of about 6 figures per article. For these figures, we manually annotated the number of image containing bar charts and the number of bar charts, resulting in 534 (32.2%) images containing at least one bar chart and in total 1659 sub bar chart. The analysis demonstrated that bar charts are indeed a very important image type in the biomedical literature. Among the figures contain at least one bar chart, 87 (16.4%) thereof contain more than 5 bar charts. This corpus was selected and annotated after we finished implementing the algorithm presented here. While we develop the method, another corpus is downloaded from PubMed Central. This two corpus have no overlap.
As shown in the top part of Table 1, the hand-coded algorithm is able to detect the bar charts with a high precision of 95.47% and a recall 59.08%. The CNN classifiers based on image features alone have lower precision than the hand-coded algorithm, but higher recall. Overall, the CNN classifier combined with hand-coded algorithm performs better than the hand-coded classifier and CNN classifier alone with F-measures of 88.66%. To clarify the cause of the result, we analyze the result of the combined method. Most of the false positive cases are axis diagram (e.g., line chart), and most of the true negative cases are bar chart with no distinct bars (e.g., very low bars).
Table 1.
Evaluation of our approach
| Task | Method | Precision | Recall | F-measure |
|---|---|---|---|---|
| Bar segment detection | Hand-coded | 0.9547 | 0.5908 | 0.7299 |
| CNN | 0.8945 | 0.8545 | 0.8691 | |
| Hand-coded + CNN | 0.9770 | 0.8115 | 0.8866 | |
| Panel segmentation | Hand-coded | 0.9811 | 0.5273 | 0.6859 |
| Information extraction | Hand-coded | 0.8240 | 0.3981 | 0.5368 |
The results of the bar panel segmentation algorithm are shown in the middle part of Table 1. We define the complete panel segmentation as to segment the panel with the x-label, y-label and the legends (if the bar chart has legends). Our method correctly produce 98.11% of the panel segmentation at a recall of 52.73%, which leads to an F-measure of 68.59%. In order to determine the different reasons of the low recall, we analyze the figure which is detected as the bar segments but not obtained the complete bar chart panel. (10.31%) Figures identified by our system share at least one data (e.g., x-label), which leads to incomplete panel. Other factors such as the sub charts are very close in a big image, the legends of the bar chart are not below the x-axis or is described in the caption could also make the panel incomplete.
Results of the extracted information are shown in the bottom part of Table 1. If information extracted from the bar charts can fill all the columns of the corresponding table, it is defined as a correct extraction. Almost 39.81% of the bar panels are extracted. 82.40% thereof are correct. Low quality of images is the most important reason of incorrect information extraction. The incorrect cases could be split into two classes of roughly the same size: the wrong number of categories in the group data, the wrong number of bars. The primary cause for the first class of incorrect cases was the fact that different forms of x-label (such as slanted x-label) make it hard to segment the labels. The causes for the second class are the facts that some images contain large numbers of very thin bars that are difficult to detect and the interferences such as the graduated lines, the slash bars of some bar chart lead to more bar numbers than exactly being contained. Usually, the number of detected bars was greater than the number of real bars, owing to the margin of error occurring in lines.
Table 2 shows the results of running the pipeline on the subset of PubMed Central. We start with about 14596 articles folds, which is accessed from the data interface (OA web service) in PubMed and searched by the keyword combination of “soybean”, “gene” and “expression.” About 18% article folds are discarded by the reason of containing no article with XML format or no JPG image. Remained articles contain around 80378 figures. In order to reduce additional computational cost, no-information figures are filtered in the image preprocessing steps. We ended up with more than 61238 figures, in which about 44537 bar charts are detected.
Table 2.
The results of running the pipeline on the open access subset of PubMed Central
| Total articles | 14596 |
|---|---|
| Processed articles | 11973 |
| Total figures from processed articles | 80378 |
| Processed figures | 61238 |
| Detected bar charts | 44537 |
Discussion
In this paper, we have developed a comprehensive system for automatically extracting information from bar chats. Our result confirm that our bar chart detection method can achieve a high accuracy, allowing us to segment the bar panel with a high precision. While the bar charts are so frequent, the low recall of the panel segmentation is not a severe problem at this point. To extract the relation of the categorical data in perspective, the conditions (x-labels and legends) should match to the bars exactly. Although the recall is low, about 40% is still in a reasonable range.
Since we invested effort in extracting information from bar charts, we give our opinion on the use of such information. Our method can help to construct dataset provided direct reference and evidence for the researchers. For example, the statistical analysis of PCR and phenotypic data are often expressed by means of bar chart. PCR is the gold standard, used to validate the results of RNA-seq.20 PCR data are more accurate and repeatable than high-throughput RNA-seq data. However, PCR data are always reported in academic papers, which have seldom been collected from the specialized databases. A combination of text mining method and our bar chart mining method is able to construct the database not only containing information from articles, but also the quantitative information of the experimental results. Illustration of phenotypic data also account for a large proportion in the bar charts, which is currently a bottleneck of genome-wide association study (GWAS) and molecular breeding. Currently, most public genotype-phenotype databases did not include the corresponding data regarding soybean organisms.21 For this reason, one of the practical values of the current dataset is that it contain correlation information for genotype and phenotype, which could be extracted from bar charts in publications.
It seems reasonable to assume that these results can provide necessary information to biomedical application. We plan to investigate this in future work. The results obtained from bar chart processing pipeline indicate that it is feasible to extract relations from bar charts, but it is clear that this procedure is far from perfect. Aiming to the problem of low image resolution, the automatic analysis of vector diagram seems to be an efficient way to extract such relations from existing publications in the future.
Conclusion
In this article, we present the most common image — bar charts — to be automatically assessed in a reliable way. Our results show that the hand-coded algorithm and the CNN method we proposed can detect the bar segment at a high accuracy. We also depict that relation and quantitative information can be extracted from the bar charts with satisfactory precision. In order to demonstrate and exemplify the algorithm’s ability and advantage, the literatures related to soybean gene expression are selected and tested. Based on these results, we believe that our proposed bar chart mining method is a viable and promising approach to provide more power to gather relations such as gene regulation. All in all, we have demonstrated a novel methodology that detects bar charts for our use case. Further research will be to apply the methodology to a larger set of citations with more diverse content.
References
- 1.Ahmed A, Arnold A, Coelho LP, Kangas J, Sheikh AS, Xing E, Cohen W, Murphy RF. Structured literature image finder: Parsing text and figures in biomedical literature. Web Semantics: Science, Services and Agents on the World Wide Web. 2010;8(2):151–4. doi: 10.1016/j.websem.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed Z, Zeeshan S, Dandekar T. Mining biomedical images towards valuable information retrieval in biomedical and life sciences. Database 2016. 2016:baw118. doi: 10.1093/database/baw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coelho LP, Ahmed A, Arnold A, Kangas J, Sheikh AS, Xing EP, Cohen WW, Murphy RF. Structured literature image finder: extracting information from text and images in biomedical literature. Linking Literature, Information, and Knowledge for Biology. 2010:23–32. [Google Scholar]
- 4.Kuhn T, Nagy ML, Luong T, Krauthammer M. Mining images in biomedical publications: Detection and analysis of gel diagrams. Journal of biomedical semantics. 2014;5(1):10. doi: 10.1186/2041-1480-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rafkind B, Lee M, Chang SF, Yu H. Proceedings of the Workshop on Linking Natural Language Processing and Biology: Towards Deeper Biological Literature Analysis. Association for Computational Linguistics; 2006. Exploring text and image features to classify images in bioscience literature; pp. 73–80. [Google Scholar]
- 6.Rodriguez-Esteban R, Iossifov I. Figure mining for biomedical research. Bioinformatics. 2009;25(16):2082–2084. doi: 10.1093/bioinformatics/btp318. [DOI] [PubMed] [Google Scholar]
- 7.Xu S, McCusker J, Krauthammer M. Yale Image Finder (YIF): a new search engine for retrieving biomedical images. Bioinformatics. 2008;24(17):1968–1970. doi: 10.1093/bioinformatics/btn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hearst MA, Divoli A, Guturu H, Ksikes A, Nakov P, Wooldridge MA, Ye J. BioText Search Engine: beyond abstract search. Bioinformatics. 2007;23(16):2196–2197. doi: 10.1093/bioinformatics/btm301. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Shijian L, Tan CL. A figure image processing system. In: Proceedings of the Workshop on Graphics Recognition. Recent Advances and New Opportunities. 2008:191–201. [Google Scholar]
- 10.Lopez LD, Yu J, Tudor CO, Arighi CN, Huang H, Vijay-Shanker K, Wu CH. Robust segmentation of biomedical figures for image-based document retrieval. In Bioinformatics and Biomedicine 2012. IEEE International Conference on; 2012; pp. 1–6. [Google Scholar]
- 11.Kim D, Yu H. Figure text extraction in biomedical literature. PloS one. 2011;6(1):e15338. doi: 10.1371/journal.pone.0015338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii N, Koike A, Yamamoto Y, Takagi T. Figure classification in biomedical literature towards figure mining. Bioinformatics and Biomedicine, 2008. IEEE International Conference on; 2008; pp. 263–269. [Google Scholar]
- 13.Cohen WW, Wang R, Murphy RF. Understanding captions in biomedical publications. In Proceedings of the ninth ACM SIGKDD international conference on Knowledge discovery and data mining; 2003; pp. 499–504. [Google Scholar]
- 14.Kou Z, Cohen WW, Murphy RF. A stacked graphical model for associating information from text and images in figures. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. NIH Public Access. 2007:257. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou YP, Tan CL. Hough technique for bar charts detection and recognition in document images. Image Processing, 2000. International Conference on IEEE; 2000. pp. 605–608. [Google Scholar]
- 16.Lopez LD, Yu J, Arighi CN, Huang H, Shatkay H, Wu C. An automatic system for extracting figures and captions in biomedical pdf documents. In Bioinformatics and Biomedicine, 2011. IEEE International Conference on; 2011. pp. 578–581. [Google Scholar]
- 17.Murphy RF, Kou Z, Hua J, Joffe M, Cohen WW. Extracting and structuring subcellular location information from on-line journal articles: The subcellular location image finder. In Proceedings of the IASTED International Conference on Knowledge Sharing and Collaborative Engineering. 2004:109–114. [Google Scholar]
- 18.Al-Zaidy RA, Giles CL. Proceedings of the 8th International Conference on Knowledge Capture. ACM; 2015. Automatic extraction of data from bar charts; p. 30. [Google Scholar]
- 19.Savva M, Kong N, Chhajta A, Fei-Fei L, Agrawala M, Heer J. Proceedings of the 24th annual ACM symposium on User interface software and technology. ACM; 2011. Revision: Automated classification, analysis and redesign of chart images; pp. 393–402. [Google Scholar]
- 20.Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, Mason CE, Socci ND, Betel D. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome biology. 2013;14(9):3158. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groth P, Pavlova N, Kalev I, Tonov S, Georgiev G, Pohlenz HD, Weiss B. Phenomic DB: a new cross-species genotype/phenotype resource. Nucleic acids research. 2007;35(suppl 1):D696–D699. doi: 10.1093/nar/gkl662. [DOI] [PMC free article] [PubMed] [Google Scholar]


