Abstract
To help researchers better access clinical data, we developed a prototype query engine called DataSphere for exploring large-scale integrated clinical data repositories. DataSphere expedites data importing using a NoSQL data management system and dynamically renders its user interface for concept-based querying tasks. DataSphere provides an interactive query-building interface together with query translation and optimization strategies, which enable users to build and execute queries effectively and efficiently. We successfully loaded a dataset of one million patients for University of Kentucky (UK) Healthcare into DataSphere with more than 300 million clinical data records. We evaluated DataSphere by comparing it with an instance of i2b2 deployed at UK Healthcare, demonstrating that DataSphere provides enhanced user experience for both query building and execution.
Introduction
In the last decade, electronic health records (EHR) technologies have been increasingly adopted across the United States. According to a data brief from the Office of National Coordinator for Health Information Technology, the adoption of basic EHR technology between 2008 and 2015 rose from 9.4 to 83.8 percent1. This exponential growth directly resulted in a significant increase the rise of the secondary use of clinical data. Secondary use of clinical data aims to reuse the data captured from clinical care for purposes of research or quality improvement 2. A major source of valuable information for clinical and translational research resides in Enterprise Data Warehouses (EDWs) or integrated clinical data repositories (IDRs), as they are designed to integrate multiple facets of data generated from patient care. Surveys 3,4 show that many IDRs are built on the transaction-based clinical care systems in recent years. Previous research 5–8 also suggests that IDRs have facilitated researchers to conduct biomedical research in various areas such as monitoring infectious diseases, mining patterns between disease progression and management, and identifying eligible subjects for clinical trials.
A typical IDR query engine consists of 4 necessary components (see Figure 1): Ontology Concepts, Observational Facts, Query Interface, and Query Execution. The Ontology Concepts component manages terminology and knowledge information such as ICD-9-CM or ICD-10-CM diagnosis codes, RxNorm medication codes, and LONIC lab test codes. Observational Facts contain patient clinical information including but not limited to demographics, diagnoses, lab tests, procedures, and medications that are structured and coded by the concepts provided by the ontology concepts component. Query Interface is the component that directly interacts with end users providing them the interface to enter query terms and review query results. The Query Interface determines the effectiveness of an IDR directly. Behind the Query Interface is the Query Execution component, which translates queries composed in the Query Interface into query language of the backend database and retrieves query results.
Figure 1.
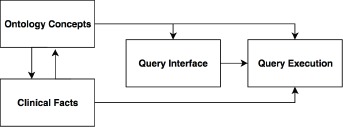
A typical IDR query engine structure.
Query exploration tools implemented as parts of query engines make IDRs easier and more practical to use. Existing query engines such as i2b29 and Harvest10 provide direct support for data query needs. These systems incorporate powerful interfaces to allow researchers to explore clinical data, build queries, and execute queries without the need for the users to understand the underlying data models. Previous research shows that properly designed query interfaces significantly lower decision maker’s mental workload11. However, the query interface is increasingly recognized as a bottleneck for the rate of return for investments and innovations in clinical research 12–14. Therefore, query interfaces need to continue to improve with the availability of advanced information technologies.
A challenge of current popular IDR query engines is that they use relational database management system (RDBMS) such as MySQL, PostgreSQL, Oracle and Microsoft SQL Server to manage IDR data. The relational database has been the foundation of enterprise applications for decades, but with the explosion in both the volume and variety of data in recent years, researchers have found that the traditional RDBMS has become a performance bottleneck and NoSQL database management systems such as MongoDB outperformed it in management of large complex biomedical data15,16.
Because of the rich variety of data sources integrated in an EDW, existing approaches to data integration leverages an amalgamation of controlled vocabularies (or ontologies) such as ICD, CPT, LOINC, and SNOMED CT. Too close of a coupling between the roles of these controlled vocabularies play, both for data coding/annotation, as well as for the interface for query construction, can limit the benefits of what informatics innovations can offer. The strategy of decoupling of the backend annotation and front-end query interface roles has been previously used successfully in three projects: VISAGE/PhysioMIMI17, x-search for the NHLBI-funded National Sleep Research Resource18, and the epilepsy-ontology-driven query interface for the NINDS-funded Center for SUDEP Research19. Since 2010, we have been applying an ontology-driven methodology to the interface design of clinical data management tools. A series of tools have been created under this philosophy including VISAGE (2010) 17, OPIC (2012) 20, FEMI (2013)21, MED- CIS (2014)22, and Neuro3D (2016)23. Using the role-decoupling strategy, we developed a prototype system called DataSphere, which incorporates ontologies in a streamlined interface with dynamic browsing, searching capabilities to facilitate the construction, management and sharing of queries for cohort discovery. We performed limited-scale comparative experiments both on usability and performance of DataSphere on UK Healthcare’s clinical data by porting our i2b2 data store to MongoDB to achieve enhanced flexibility and performance.
1. Background
1.1. Biomedical Ontologies
Biomedical ontologies play an important role in IDRs, which provide a standard and formalized way to specify entities (or concepts), their attributes and relationships among the entities. This enables integration, query, and exchange of heterogeneous biomedical data and interoperability between computer systems. The ontologies involved in this paper include International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) and International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) for diagnoses coding, Logical Observation Identifiers Names and Codes (LOINC) for lab tests coding, Current Procedural Terminology (CPT) for medical, surgical, and diagnostic procedures coding, and RxNorm for clinical drugs coding. DataSphere imports these ontology concepts into database and visualize them with its “Concept Finder.”
1.2. VISAGE
The VISual AGgregator and Explorer (VISAGE) is a domain ontology-driven interface for a federated approach to data integration. Interface design features in VISAGE include auto-generated slider bar, selection boxes, and built-in charting. DataSphere’s user interface is an extension of VISAGE, with new features (Section 2.2) developed such as: Concept Finder with browse and search modes to locate ontology concepts, Widget Canvas with single and disjunctive concept containers, logical annotation between different concept containers and widgets, and collapsable organization of different interface modules. All these new features work together to make query building more intuitive and efficient.
1.3. MongoDB
As an open-source, cross-platform document-oriented database, MongoDB24 provides us with advantages such as flexible data model, scalability, and improved performance. MongoDB uses a different set of terms for database storage and operations. The terms involved in this paper include “Collection,” “Document,” and “MQL” similar to the concepts “Table,” “Row,” and “SQL” in the traditional relational database. Unlike the traditional relational database that stores data in tables and uses structured query language (SQL) for data access, MongoDB stores data in JSON like documents that can vary in structure. Therefore, in MongoDB, related information can be stored together for fast access using MongoDB query language (MQL), which is the main motivation for choosing MongoDB as the backend database for DataSphere.
1.4. i2b2 and UK Healthcare IDR
i2b2 is an open source clinical data analytics platform originally developed with funding from the National Institutes of Health. i2b2 is widely used for querying patient data to address research questions. i2b2 aggregates a copy of the patient data from an EHR system and provide query services in parallel to the EHR system for research purposes. i2b2 provides a web client with which users can build customized queries and retrieve results. As an excellent query engine for IDRs, i2b2 has over 100 instances deployed internationally in various organizations including CTSA sites, academic health centers, HMOs, and companies.25
UK HealthCare is the hospitals and clinics of the University of Kentucky. It has captured clinical data for more than one million patients since 2006. The data include, but are not limited to, demographics, financial classification, provider level details, medical diagnoses, medical procedures, lab tests and results, medications, visit details, and vital signs. An i2b2 instance was deployed at UK Healthcare in 2016. It has more than 350 active users and has handled more than one thousand queries from UK Healthcare community. In this paper, we import UK Healthcare IDR data into DataSphere and evaluate it by comparing with UK Healthcare’s i2b2 instance.
2. Methods
2.1. Architecture Design
DataSphere follows the web-interface driven development methodology created in our Neuro3D work23 to involve the end users closely in the process of system design and implementation. As illustrated in Figure 2, DataSphere consists of four critical modules: Data Import, Query Interface, Query Translation and Optimization, and Query Management. First, DataSphere imports data from IDRs to its own MongoDB database; then based on the imported ontology concepts, it renders a query building interface with two critical components: Concept Finder and Widget Canvas. Following that, users can customize data queries using the building interface and the queries will be translated into MongoDB query language and optimized to run against the imported clinical data. At last, the queries can be saved for reuse or shared with other DataSphere users.
Figure 2.
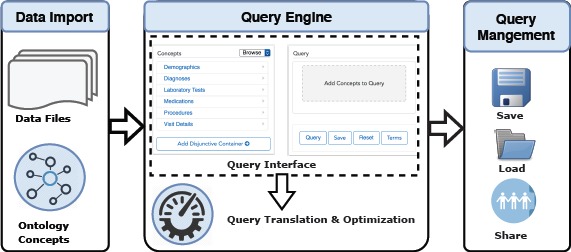
System Architecture of DataSphere.
Table 1 shows the architecture differences between DataSphere and the traditional IDR i2b2. DataSphere’s allows a flexible ontology library and the ontology library dynamically determines the rendering of DataSphere’s query interface. Instead of relying on specific extract, transform, load (ETL) process, DataSphere import patient data from files of common formats such as csv and json, which provides the general applicability of DataSphere to various IDRs. DataSphere adopts MongoDB as the backed database to replace the traditional RDBMS to avoid potential data processing bottlenecks. For query management tasks (save, load, share), DataSphere provides comprehensive supports to encourage the collaborations between researchers. In the following section, we describe in detail how each module works.
Table 1.
Architecture difference between DataSphere and i2b2.
| Function Module | i2b2 | DataSphere | |
|---|---|---|---|
| Ontology Library | Pre-defined | Plug | Play |
| Query Interface | Pre-defined | Dynamically Rendered | |
| Patient Data Import | via ETL | via Files of Common Formats | |
| Backend Database | RDBMS | MongoDB | |
| Query Management (Save | Load) | Limited | Comprehensive |
| Query Share | Not supported | Comprehensive |
2.2. IDR Data Import
The Data Import module handles the process of importing two kinds of data: data files and ontology concepts. Data files contain patient demographic (age, gender, race, etc) information and clinical (diagnoses, lab tests, procedures, medications, etc) information. Ontology concepts provide a standard way to describe patient data. Multiple ontologies are involved in IDRs. In our work, ICD-9-CM and ICD-10-CM are used for coding diagnoses, LOINC is used for coding lab tests, CPT is used for coding procedures, and RxNorm is used for coding clinical medications. DataSphere imports patient demographic information into a collection called “Patients,” clinical inforamtion into another collection called “Observational Facts” and ontology concepts into the third collection called “Concepts” with specific collection data schema. Figure 3 depicts the key fields of each collection and relationships among the collections.
Figure 3.
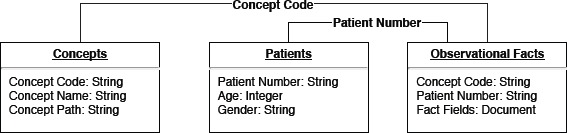
Three critical data collections of DataSphere.
2.3. Query Interface Design
DataSphere’s query interface consists of two critical components: Concept Finder and Widget Canvas.
Concept Finder – A Query of IDRs is built from the concepts that are used to code the patient data. Therefore, an appropriate way of locating or navigating concepts is a necessary component of the query interface. In DataSphere, we implement this component as “Concept Finder.” Concept Finder provides two modes: browsing and search. In browsing mode, concepts are organized in a hierarchical structure that users can navigate down level by level. Search mode performs keyword-based searches against concept names according to the user entry. These two modes are designed for users with different skill levels: more skilled users can search concepts directly by concept names within the searching mode, while less experienced users can navigate concepts level by level using the browsing mode. Designing the browsing mode for large-scale IDRs is a challenging task due to the large size of concepts. For example, UK Healthcare’s IDR contains about 200,000 concepts. Rendering the hierarchical structure of these concepts within a web page takes more than five minutes, which is not ideal for an interactive web application. Concept Finder overcomes this challenge by implementing a tailored concept hierarchy, which initially renders several top levels of concepts and displays the subsequent levels in real time with AJAX calls. In addition, the number of levels displayed initially is configurable. With appropriate configuration, the tailored hierarchy can tremendously reduce the size of concepts for initial display so that the rendering process is instant.
Widget Canvas – DataSphere users can add concepts to the Widget Canvas after they find their target concepts. Widget Canvas can render specific query widgets for concepts automatically according to their properties. For example, as illustrated in Figure 4(a), the concept “Age Today” is a numeric concept so that Widget Canvas renders a slider bar for it. Another example in Figure 4(a) is the concept “Gender” that is categorical with options “Male,” “Female,” and “Unknown.” Then Widget Canvas renders three checkboxes for it. Widget Canvas provides two kinds of concept containers: single concept container and disjunctive concept container. Intuitively, a single concept container only allows one concept in it while disjunctive concept container can consist of multiple concepts. The logical relationship among all concept containers is conjunctive while the relationship among all concepts within one disjunctive container is disjunctive. Semantically, the query displayed in Figure 4 is to find all adult (age ≥ 18) female patients with Parkinson’s disease or Dystonia.
Figure 4.
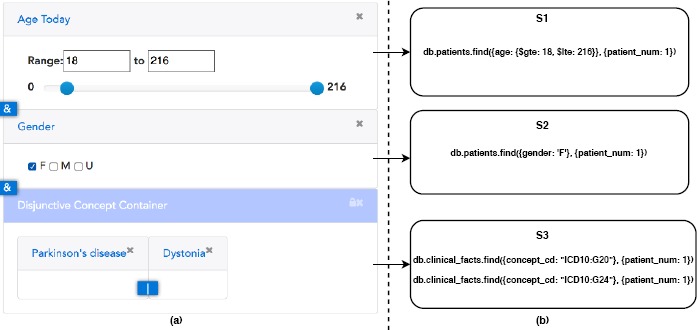
Widget Canvas of DataSphere and MongoDB query statements translated from query widgets.
2.4. Query Translation and Optimization
In DataSphere, all query widgets are eventually translated into MongoDB query statements and executed.
Query Translation – As illustrated in Figure 4(b), every concept container is translated to certain MongoDB query statements. The number of translated query statements is equal to the number of query widgets within each concept container since the translation from query widget to query statement is a “one to one” correspondence. The translation is directed by both concept properties and query widget details. In the first query container of Figure 4(a), the “Age Today” concept contains information that its target data collection is “Patients” and its matching column is “Age” And the query widget in the first concept container specifies that the age range to query is from 18 to 216. Therefore, the translated query (S1) is to scan patients collection with the condition as column age between 18 and 216. On the other hand, the last concept container in the figure is a disjunctive concept container with two diagnosis concepts in it: Parkinson’s Disease and Dystonia, with concept codes “ICD10:G20” and “ICD10:G24,” respectively. These two concepts state their target data collection is “Observational Facts” and matching column is “Concept Code” and there is no additional query widget information so that the translation results are two individual statements looking for documents in “Observational Facts” collection with column “Concept Code” as “ICD10:G20” and “ICD10:G24.” In summary, the query composed in Figure 4(a) is translated into query statements: S1, S2, and S3, within which S3 contains two individual MongoDB query statements. In DataSphere, we call S1, S2, and S3 together as a query statement workflow. Therefore, any semantic query in DataSphere is built into a set of query widgets and then translated into a query statement workflow.
Query Optimization x2013 Without any optimization, a query statement workflow is naturally executed by its creation order or translation order. For the example in Figure 4, the query statement workflow should be executed in the order: S1 → S2 → S3. However, the order matters. Queries in DataSphere look for the number of unique patients with characteristics specified in the query widgets. Each statement returns a list of patient ids. The latter executing statement can query within this patient list to reduce its searching space. As stated in the above section, every concept itself defines its target data collection and matching column. We can pre-compute the number of documents within the target data collection match to a concept without additional query widget level restriction. This number is the maximum number of document scanning needed when executing an individual MongoDB query statement translated from the concept. In DataSphere, we call this number “Concept Selectivity.” Concept selectivity intuitively tells us how long it takes to execute a statement because statement execution time is proportional to the number of documents to scan. In addition, a translated query statement’s selectivity is defined as the sum of concept selectivities of all involved concepts. To be clear, the selectivity of a query statement translated from a single concept container is equal to the concept’s selectivity within the container, whereas the selectivity of a query statement translated from a disjunctive concept container is equal to the sum of all concepts within that disjunctive concept container. We optimize the statement execution order by evaluating the selectivities of statements. We pre-compute all concepts’ selectivities so that the evaluation process is instant. We order all statements by selectivity in ascending order and execute them one by one. Using the above query as an example, it takes 24.66 seconds to complete the query in order: S1 → S2 → S3. After optimization, the execution order is: S3 → S2 → S1 and the execution time is 0.08 second.
2.5. Query Management
DataSphere supports collaboration by providing researchers with the ability to save and share queries.
Save a query – After building and executing a query, DataSphere users can choose to save the query and give it a name and description. Saved queries can be found in each user’s own query library: “My Queries.” Users can search by name or description in their own library and click query name to load the query details in the Widget Canvas. Saving queries in DataSphere is very straightforward and similar to taking “screenshots” for Widget Canvas. As mentioned above, a query is built with several concept containers and every concept container can have multiple concept widgets. When saving queries, every concept container is saved as a “Query Concept Group” and each concept widget is saved as a “Query Concept” within one specific “Query Concept Group.”
Share a query – DataSphere provides two ways to share queries: public sharing and private sharing. Public sharing means users can make their saved queries public so that all DataSphere users can view these queries. Private sharing means users can select specific users to share their queries with. Within “My Queries”, each user can see all public queries and shared queries. Users can choose to save public queries and shared queries into their own libraries, in which case the query will not be affected by the owner’s further changes.
2.6. Evaluation Method
We implemented one instance of DataSphere for the IDR of UK HealthCare and evaluated two aspects of DataSphere: usability and performance. UKHealthCare is currently using an i2b2 instance to support patient cohort identification for clinical research. We evaluated the DataSphere instance by comparing it with the i2b2 instance. We selected 8 representative queries from i2b2 query logs, and interpreted them into the human readable language.
Usability – We invited 4 data scientists as evaluators to build these 8 queries with both the i2b2 web client and DataSphere. We asked each evaluator to record the query building time in seconds. System usability is considered better if users require less time to build the same query.
Performance – To evaluate the performance, we performed the 8 selected queries independently in two systems. We executed each query in each system three times and calculate the average execution time. The system performance is better if the execution time is shorter for the same query.
3. Results
3.1. Data Import
We loaded a total of 24GB de-identified data from UK Healthcare, which contained over one million patients data with more than 300 million observational fact records. We imported the data into DataSphere’s data collections and Table 2 shows the details of the data import results. The data loaded to DataSphere followed the same ETL process of UK Healthcare’s i2b2 ETL pipeline so that the two systems eventually contain the same data set.
Table 2.
Data import results from UK Healthcare.
| Data File Name | Format | Size | MongoDB Collection Name | Number of Documents |
|---|---|---|---|---|
| Concept Dimension | CSV | 160MB | Concepts | 194,648 |
| Patient Dimension | CSV | 56MB | Patients | 978,144 |
| Visit Dimension | CSV | 854MB | Visits | 11,313,511 |
| Facts | CSV | 23.07G | Observational Facts | 309,053,442 |
3.2. Interactive Interface for Query Building
Figure 5(a) is a screenshot of the query interface of DataSphere consisting of four parts labeled 1 to 4. Label 1 shows Concept Finder’s browsing mode. Six top concepts are displayed in the figure. Hovering mouse over each concept will show the subsequent level of concepts. Each concept is clickable and clicking on the concept will add it to the right area labeled as 2. Label 2 denotes Widget Canvas. Widget Canvas contains a number of concept containers. These concept containers are connected with logical notation “&” indicating they are conjunctive. Concepts within a disjunctive container are connected with logical notation “|” indicating they are disjunctive within that container. Label 3 is the query control panel consisting of three buttons. The first query button triggers the query translation, optimization, and execution process. The second save button populates a form with fields name and description to save together with the query widgets. The last reset button removes all the query widgets in the current Widget Canvas. Saved queries can be found with the link “My Queries” labeled 4.
Figure 5.
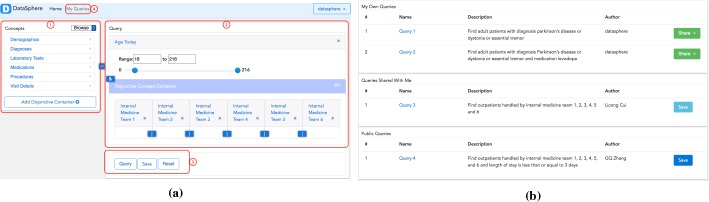
(a) Query Interface of DataSphere; (b) Query management of DataSphere.
3.3. Query Management
Figure 5(b) shows “My Queries.” Three different sections are displayed in my queries: “My Own Queries,” “Queries Shared With Me,” and “Public Queries.” “My Own Queries” are queries created by users themselves; “Queries Shared with Me” are queries that are shared privately by other users; and “Public Queries” are queries that are made public by other users. At the end of each row in my own queries, the button “Share” allows users to make their own queries public or visible to specific users. At the end of each row in “queries shared with me” and “public queries” the button “save” allows users to save these queries to their own queries.
4. Evaluation
The evaluation was designed to assess DataSphere’s usability and performance by comparing it with the i2b2 instance deployed at UK Healthcare. To perform the evaluation, we chose some representative queries from the i2b2’s query logs with three goals: (1) these queries should at least cover multiple concepts; (2) all queries should cover all the categories of concepts, and (3) every query should have a clear clinical meaning. With these goals in mind, 8 queries were filtered as shown in Table 3. These 8 queries covered all categories of concepts we imported: Demographics, Diagnoses, Procedures, Lab Tests, Medications, and Visit Details. The smallest number of involved concepts in the queries was 3 and each query had clear clinical meaning. The general purpose of each query was to find the number of patients that satisfied the query’s requirements.
Table 3.
8 evaluation queries picked from i2b2 query logs.
| Query Number | Query Description | Number of Concepts |
|---|---|---|
| No. 1 | Find adult patients with diagnosis of Parkinson's disease or dystonia or essential tremor. | 4 |
| No. 2 | Find adult patients who were diagnosed with Parkinson's disease, dystonia, or essential tremor and took levodopa as medicine. | 5 |
| No. 3 | Find outpatients handled by internal medicine team 1, 2, 3, 4, 5, and 6. | 7 |
| No. 4 | Find outpatients handled by internal medicine team 1, 2, 3, 4, 5, and 6, and length of stay was less than or equal to 3 days. | 10 |
| No. 5 | Find female patients that were of age greater than 35 years old, diagnosed with cerebral infarction unspecified and took medication estrogens conjugated. | 4 |
| No. 6 | Find adult patients that were diagnosed with malignant neoplasm of breast, and used emergency department services. | 3 |
| No. 7 | Find adult patients that were diagnosed with malignant neoplasm of breast, used emergency department services and stayed for one day. | 4 |
| No. 8 | Find male adult patients with BMI between 25 to 30 and lab test of transcutaneous measurement of total bilirubin | 4 |
4.1. Deployment Environment
UK Healthcare’s i2b2 environment is deployed on a dedicated server with 72 CPU cores and 378 GB of random-access memory (RAM). The i2b2’s deployed version is 1.7.08 and the backend database is PostgreSQL of version 9. On the other hand, the DataSphere instance is deployed on a high-end research server provided by the Institute for Biomedical Informatics of University of Kentucky with 80 processors (10 CPU cores for each processor) and 1 Terabyte RAM. Although the physical environments of deployment are different, neither of the two systems detect bottlenecks about physical configuration so that we assume these two systems are running on sufficient configurations of RAM and processing power.
4.2. Usability Evaluation
As we have stated in the methods Section 2.5, the usability measurement was the time cost to compose queries. We invited four non-technical users to complete the query building task and counted the time cost. These four users have no development experience with either i2b2 or DataSphere. i2b2 web client also provides browsing and search modes to find concepts. Therefore, we asked the evaluators to build the 8 queries in browsing and search modes separately in the two systems. Table 4 shows the average time cost in seconds to build the 8 selected queries. To avoid potential bias introduced by the order of building these queries in the two different systems, two of our evaluators were requested to build queries first in DataSphere then in i2b2 and the other two evaluators completed the query construction in the opposite order. The results show that DataSphere’s query building efficiency is consistently better than i2b2 in both browsing and search modes for all 8 queries. On average, evaluators built same queries with 36.99% less time using DataSphere’s browse mode than using i2b2’s browse mode; evaluators composed the same queries with 37.14% less time using DataSphere’s search mode than using i2b2’s search mode. Two paired t-tests were conducted to compare query building time in DataSphere and i2b2 within browsing and search mode respectively. Test results show that: in browsing mode, there is significant difference in the scores for DataSphere (Mean=46, SD=10) and i2b2 (Mean=73, SD=13) with a p value < 0.0001; in search mode, the difference between DataSphere (Mean=44, SD=9) and i2b2 (Mean=70, SD=17) scores is also very significant with a p value 0.0016.
Table 4.
Statistics for query building in usability evaluation.
| Query | i2b2 Browse | DataSphere Browse | i2b2 Search | DataSphere Search |
|---|---|---|---|---|
| 1 | 59 | 41 | 74 | 38 |
| 2 | 72 | 58 | 59 | 47 |
| 3 | 52 | 32 | 42 | 32 |
| 4 | 82 | 45 | 56 | 45 |
| 5 | 93 | 61 | 91 | 57 |
| 6 | 76 | 42 | 87 | 36 |
| 7 | 73 | 50 | 83 | 56 |
| 8 | 76 | 37 | 67 | 44 |
| Average | 73 seconds | 46 seconds | 70 seconds | 44 seconds |
4.3. Execution Time
We ran the 8 queries in i2b2 and DataSphere and recorded the running statistics in Table 5. We ran each query in the two systems for 10 times respectively and Table 5 shows the average execution time for each query. For each of the evaluated queries, i2b2 web client took more than five seconds while DataSphere took less than one second. The last row shows that the i2b2 web client took 81.4 seconds to run all 8 queries which were more than 30 times of DataSphere’s running time. DataSphere showed much better performance for the selected evaluation queries. A paired t-test was conducted to compare query execution times in i2b2 and DataSphere. There was a significant difference in the scores for i2b2 (Mean=10.2, SD=6.1) and DataSphere (Mean=0.3, SD=0.3) with a p value 0.0029. These results suggest that DataSphere has a better query execution performance than i2b2.
Table 5.
Performance statistics of query running of i2b2 web client and DataSphere.
| Query Number | i2b2 Running Time | DataSphere Running Time |
|---|---|---|
| No. 1 | 5.8 seconds | 0.2 seconds |
| No. 2 | 6.1 seconds | 0.1 seconds |
| No. 3 | 14.7 seconds | 0.1 seconds |
| No. 4 | 23.3 seconds | 0.2 seconds |
| No. 5 | 5.2 seconds | 0.3 seconds |
| No. 6 | 7.5 seconds | 0.7 seconds |
| No. 7 | 11.3 seconds | 0.9 seconds |
| No. 8 | 7.5 seconds | 0.1 seconds |
| Total | 81.4 seconds | 2.6 seconds |
5. Discussions
For a query engine for IDRs, effectiveness (or usability) and efficiency (or performance) are two important aspects to optimize for researchers to achieve better user experience.
Usability – For effectiveness of query engine DataSphere, we described two modes of concept finding in this paper: browse and search. We stated that search is for more skilled users and browse is for less experienced users. We expected search mode to be faster than browse mode in composing queries, but results suggested that also depended on specific queries. For example, in our comparative usability evaluation (Section 4.1), search mode outperformed browse mode for most queries except for query No. 7 when all evaluators reported that browse mode was faster. The reason for that is the evaluators spent a lot of time deciding on the proper search terms for concept “Malignant Neoplasms of Breast.” All combinations except the full name returned too many results, which made it difficult to find the correct concept. Therefore, an appropriate ranking algorithm is a key factor for a successful search function.
Performance – For the efficiency or performance of DataSphere, we observed significant improvement compared to i2b2’s web client. The query workflow optimization contributed substantially to the performance improvement. We verified that by running same queries with query workflow optimization disabled. Results showed that queries ran much slower. For example, query 6 took 19.5 seconds to complete in DataSphere without query workflow optimization, which is much slower than 0.7 seconds with optimization reported in Table 5.
General Applicability The simplicity of DataSphere’s data models makes it easy to integrate heterogenous clinical data from different data sources. Although we did not have a chance to import data from multiple IDRs into DataSphere, we expect it to do so in the future. One type of IDRs in mind are the ones following ETL process for OMOP Common Data Model26.
Limitation – First, we were only able to have four evaluators complete the usability testing. Second, the selection process of the evaluation queries is subjective although we stated 3 goals to direct the query selection (Section 4). DataSphere is in a prototyping stage with additional functionalities to complete such as temporal queries and federated search across different data sources.
6. Conclusion
In this paper, we introduced an ontology-driven, enhanced, general query framework DataSphere for large scale integrated data repositories. It uses document-oriented NoSQL MongoDB as the backend database, creates an interactive query interface that renders hierarchical Concept Finder automatically from concepts imported from IDRs, and applies query optimization strategies to improve query execution performance. The comparative evaluations with i2b2 demonstrated superior usability and performance of DataSphere, revealing the potential of DataSphere to support queries for large-scale IDRs.
7 Acknowledgements
This work is supported by University of Kentucky Center for Clinical and Translational Science (Clinical and Translational Science Award UL1TR001998) and National Science Foundation MRI Award No.1626364.
References
- [1].Henry J, Pylypchuk Y, Searcy T, Patel V. Adoption of Electronic Health Record Systems among US Non-Federal Acute Care Hospitals: 2008-2015. 2016 Mar 9; Retrieved from http://dashboard.healthit.gov/evaluations/data-briefs/non-federal-acute-care-hospital-ehr-adoption-2008-2015.php. [Google Scholar]
- [2].Weiner MG, Embi PJ. Toward reuse of clinical data for research and quality improvement: the end of the beginning? Annals of internal medicine. 2009 Sep 1;151(5):359–60. doi: 10.7326/0003-4819-151-5-200909010-00141. [DOI] [PubMed] [Google Scholar]
- [3].MacKenzie SL, Wyatt MC, Schuff R, Tenenbaum JD, Anderson N. Practices and perspectives on building integrated data repositories: results from a 2010 CTSA survey. JAMIA. 2012 Jun 1;19(e1):e119–24. doi: 10.1136/amiajnl-2011-000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Murphy SN, Dubey A, Embi PJ, Harris PA, Richter BG, Turisco F, Weber GM, Tcheng JE, Keogh D. Current state of information technologies for the clinical research enterprise across academic medical centers. Clinical and translational science. 2012 Jun 1;5(3):281–4. doi: 10.1111/j.1752-8062.2011.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Samore M, Lichtenberg D, Saubermann L, Kawachi C, Carmeli Y. A integrated data repository enhances hospital infection control. AMIA Annual Fall Symposium. 1997:56–60. [PMC free article] [PubMed] [Google Scholar]
- [6].Prather JC, Lobach DF, Goodwin LK, Hales JW, Hage ML, Hammond WE. Medical data mining: knowledge discovery in a clinical data warehouse. AMIA annual fall symposium. 1997:101–5. [PMC free article] [PubMed] [Google Scholar]
- [7].Breault JL, Goodall CR, Fos PJ. Data mining a diabetic data warehouse. Artificial intelligence in medicine. 2002 Oct 31;26(1):37–54. doi: 10.1016/s0933-3657(02)00051-9. [DOI] [PubMed] [Google Scholar]
- [8].Deshmukh VG, Meystre SM, Mitchell JA. Evaluating the informatics for integrating biology and the bedside system for clinical research. BMC medical research methodology. 2009 Oct 28;9(1):70. doi: 10.1186/1471-2288-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, Gainer V, Berkowicz D, Glaser JP, Kohane I, Chueh HC. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annual Symp Proc. 2007:548–52. [PMC free article] [PubMed] [Google Scholar]
- [10].Pennington JW, Ruth B, Italia MJ, Miller J, Wrazien S, Loutrel JG, Crenshaw EB, White PS. Harvest: an open platform for developing web-based biomedical data discovery and reporting applications. Journal of the American Medical Informatics Association. 2014 Mar 1;21(2):379–83. doi: 10.1136/amiajnl-2013-001825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Speier C, Morris MG. The influence of query interface design on decision-making performance. Mis Quarterly. 2003 Sep 1;:397–423. [Google Scholar]
- [12].Calinescu R, Harris S, Gibbons J, Davies J. Cross-trial query system for cancer clinical trials. Innovations and Advanced Techniques in Computer and Information Sciences and Engineering. 2007:385–90. [Google Scholar]
- [13].Murphy S, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, Gainer V, Berkowicz D, Glaser JP, Kohane I, Chueh HC. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annual Symp Proc. 2007:548–52. [PMC free article] [PubMed] [Google Scholar]
- [14].Weber GM, Murphy SN, McMurry AJ, MacFadden D, Nigrin DJ, Churchill S, Kohane IS. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. Journal of the American Medical Informatics Association. 2009 Sep 1;16(5):624–30. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schulz WL, Nelson BG, Felker DK, Durant TJ, Torres R. Evaluation of relational and NoSQL database architectures to manage genomic annotations. Journal of Biomedical Informatics. 2016 Dec 31;64:288–95. doi: 10.1016/j.jbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- [16].Chatterjee S, Stupp GS, Park SK, Ducom JC, Yates JR, Su AI, Wolan DW. A comprehensive and scalable database search system for metaproteomics. BMC genomics. 2016 Aug 16;17(1):642. doi: 10.1186/s12864-016-2855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang GQ, Siegler T, Saxman P, Sandberg N, Mueller R, Johnson N, Hunscher D, Arabandi S. VISAGE: a query interface for clinical research. AMIA Jt Summits Transl Sci Proc. 2010 Mar 1;2010:76–80. [PMC free article] [PubMed] [Google Scholar]
- [18].National Sleep Research Resource. 2016. Mar 9, Retrieved from https://sleepdata.org/
- [19].The Center for SUDEP Research. 2016. Mar 9, Retrieved from http://csr.case.edu/index.php/MaimPage.
- [20].Sahoo SS, Zhao M, Luo L, Bozorgi A, Gupta D, Lhatoo SD, Zhang GQ. OPIC: ontology-driven patient information capturing system for epilepsy. AMIA Annual Symp Proc. 2012:799–808. [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang GQ, Cui L, Teagno J, Kaebler D, Koroukian S, Xu R. Merging ontology navigation with query construction for web-based Medicare data exploration. AMIA Summit on Clinical Research Informatics (CRI) 2013 Mar 18;:285–9. [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang GQ, Cui L, Lhatoo S, Schuele S, Sahoo S. MEDCIS: Multi-Modality Epilepsy Data Capture and Integration System. AMIA Annual Symp Proc. 2014:1248–57. [PMC free article] [PubMed] [Google Scholar]
- [23].Tao S, Walter BL, Gu S, Zhang GQ. Web-Interface-Driven Development for Neuro3D, a Clinical Data Capture and Decision Support System for Deep Brain Stimulation. International Conference on Health Information Science; 2016. Nov 5, pp. 31–42. [Google Scholar]
- [24].MongoDB Documents. 2016. Mar 9, Retrieved from https://www.mongodb.com.
- [25].i2b2 Installations. 2016. Mar 9, Retrieved from https://www.i2b2.org/work/i2b2_installations.html.
- [26].Observational Medical Outcomes Partnership. 2016. Mar 9, Retrieved from http://omop.org/


