Abstract
De-identification of protected health information is an essential method for protecting patient privacy. Most institutes require de-identification of patient data prior to conducting scientific studies; therefore, it is important for clinical scientists to be cognizant of all modes of de-identification and all services provided by their de-identification tools. In this article, we discuss eight different modes of de-identification that yield de-identified data at different levels of quality. Most of these modes can be used in combination to achieve the best performance.
Introduction
De-identification is a process of detecting identifiers (e.g., personal names and social security numbers) that directly or indirectly point to a person (or entity) and deleting those identifiers from the data. Health information containing personal identifiers that are linked to the subject of the information is defined as protected health information1 and is protected under the federal law.2 The deleted personal identifier (e.g., “James Smith”) can be replaced with information describing the type of identifier (e.g., [Personal Name]) or with fake identifiers called pseudonyms (e.g., “John Doe”). Replacing personal identifiers in the clinical data with non-identifying terms minimizes the chance of re-identification of the patient during the use of the data for scientific purposes;* hence, it is one of the most essential tools for protecting patient privacy.
According to the Privacy Rule of the Health Insurance Portability and Accountability Act (HIPAA), the de-identification process can be accomplished using two separate pathways: Safe Harbor method and Expert Determination approach (see Figure 1). The Safe Harbor method requires all 18 personal identifiers to be eliminated. The latter approach uses the preservation of certain personal identifiers (usually dates and demographics) combined with an expert’s assurance that these identifiers could not be used to re-identify the patient. Note that the Privacy Rule neither defines the qualifications of the expert or the expertise nor sanctions any set of methods that would yield such a determination.
Figure 1.
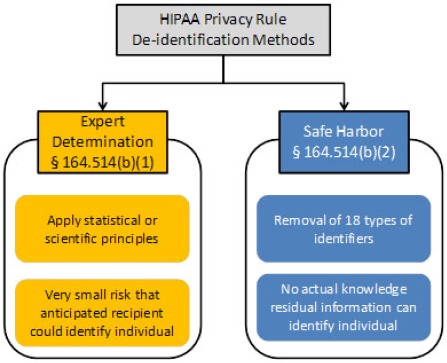
Two pathways for de-identification of protected health information as sanctioned by HIPAA Privacy Rule as depicted in the guidelines9 prepared by the Office of Civil Rights, Department of Health and Human Services
If dates, ages over 89 years, and/or detailed geographic information (five-digit ZIP code or at the town level of details) are required in the study, Safe Harbor method can be used along with another HIPAA Privacy Rule provision called Limited Data Set. Under this provision, researchers can access those pieces of information if they agree to sign a data use agreement established by the data-providing institute. Compared to the Safer Harbor, the expert determination approach is less definitive, so we do not consider it here. For this article, the term de-identification implies the Safe Harbor method only.
Anonymization is closely related to de-identification. It is sometimes used interchangeably and confused with de-identification. Although both anonymization and de-identification aim to protect the privacy of the subject of information, they are semantically different concepts. As a federal regulation, HIPAA Privacy Rule defines 18 types of personal identifiers. State laws and institutional review board (IRB) regulations that are more stringent than the Privacy Rule may require the removal of an additional set of identifiers such as provider identifiers and specimen slide numbers.4 However, in all cases, the process of de-identification is defined explicitly; that is, finding specific identifiers and deleting them. Thus, de-identification is a well-circumscribed process with explicit specifications of what needs to be done. The same cannot be said for anonymization. Anonymization is not a method per se but a goal, which may be achieved using different methods and strategies. Anonymization does not imply a standard nor does it specify what needs to be done to attain that goal.
Another crucial difference between anonymization and de-identification pertains to the claim of the outcome. The Department of Health and Human Services (DHHS) “is cognizant of the increasing capabilities and sophistication of electronic data matching used to link data elements from various sources and from which, therefore, individuals may be identified” (see Federal Register p. 53232).5 The de-identification process minimizes the risk of re-identification but has no claim to make it impossible. On the other hand, methods for anonymization found in the literature such as aggregations of microdata6 7 are almost always applicable to tabular data only and attempt to guarantee a certain level of anonymity usually as a function of aggregation.8
Automatic De-identification
We can de-identify tabular datasets manually if we know which fields contain personal identifiers. Unfortunately, de-identifying narrative text is significantly more challenging than de-identifying tabular data. Narrative text has neither a schema nor enumerated set of fields to guide the process. Narrative report structures tend to differ randomly in location and labeling among different providers. Moreover, the ambiguous nature of natural languages makes the problem of narrative text de-identification especially difficult.
Automatic text de-identification tools can de-identify clinical text with high accuracy comparable to the performance of human annotators.10 Furthermore, automatic de-identification is fast and inexpensive. Unlike human annotators, computers do not get tired but instead, consistently produce the same level of quality for every document as long as these documents do not differ from each other in style and context.
Modes of Operations
A software application may be used in different configurations, forms or modes, e.g. batch vs. interactive mode, text vs. graphic mode, beginner vs. expert mode. In the literature, there are plenty of de-identification papers about automatic clinical text de-identification systems, but there is little information on how to use them to get the best de-identification results. In this paper, we discuss different modes of de-identification, each of which may yield results at different levels of quality.
Most IRBs require de-identification as a prerequisite for clinical research using retrospective patient data; therefore, it is important for every clinical scientist to be cognizant of all options and services provided by de-identification systems.
Institutional Responsibilities
De-identification plays a crucial part in protecting patient privacy by enabling institutions to share a large volume of clinical information for clinical research while maintaining patient privacy. However, de-identification alone is not sufficient for protecting patient privacy. This is a difficult task that can only be achieved through a close collaboration of all parties in the healthcare system chain who create, handle, store, and transmit health information. Furthermore, institutions have overarching responsibilities to establish the right policies and monitor their compliance through IRBs. Institutions have to ensure that the data leaving their control has been properly de-identified.
As part of this responsibility, data managers in clinical institutions should make the necessary efforts to provide requisite patient and provider information to their de-identification system for achieving the highest de-identification rate possible. For example, an automatic de-identification system capable of accepting patient and provider identifiers as inputs should be provided to the system, if possible, so that institutions are not left with subpar de-identification results. Similarly, a de-identification system that requires annotated clinical reports for training should be provided with the requisite training dataset. To ensure that the training dataset is large enough, the machine learning system should be continuously fed with new sets of annotated reports until the de-identification performance increases become negligible. Furthermore, this process should be repeated in scheduled cycles as clinical practice, use of terminology, reporting styles and reporting physicians tend to change over time.
Different Modes of De-identifying Reports Using Automatic De-identification Systems
In this article, we consider any automatic de-identification application as a black box and remain indifferent to the methods used inside the box. The focus is on the process of de-identification from the perspective of the user. The user needs to provide a set of input to the system, so we are interested in the variety of information that the system is capable of taking as input and how the user can operate a given de-identification system to achieve the best patient privacy protection while preserving the integrity of de-identified data. We discuss eight modes of de-identification (see Table 1).
Table 1.
Modes of de-identification
| A. | Repository-wide batch de-identification |
|---|---|
| B. | On-demand cohort-specific de-identification |
| C. | On-demand de-identification of query results |
| D. | De-identification withpatient and provider identifiers |
| E. | Scientist involved de-identification |
| F. | Patient involved de-identification |
| G. | Physician involved de-identification |
| H. | Online de-identification by honest brokers |
Repository-wide batch de-identification. This is the simplest among all the alternative processing methods; thus, perhaps the most commonly used among institutions. When institutions obtain a de-identification system, some may immediately create a de-identified version of their entire repository, so that researchers with proper credentials can quickly access what they need in a de-identified format.
On-demand cohort-specific de-identification. An alternative to the repository-wide batch de-identification method is providing on-demand de-identification for a defined cohort of patients at the request of the scientist. Unlike the repository-wide batch de-identification approach, the de-identified data is not ready to be provided to the research group, but given the speed of modern systems, automatic de-identification is almost instantaneous. The time difference between these two approaches would be negligible compared to the selection of the cohort reports.
On-demand de-identification of query results. If the de-identification system is attached to a clinical database querying system, the de-identification can also be obtained on the fly. This approach would probably be much faster than the cohort-specific approach, since there would be no need to wait for the availability of the data manager who, in the previous two modes, de-identifies the data and provides the de-identified data to the scientist. In this mode, the data manager is out of the loop.
De-identification with patient and provider identifiers. Research results indicate that providing patient and provider identifiers likely to be in the report yields significantly better de-identification results.11 Personal identifiers can be provided to a de-identification system in four different ways: report-specific identifiers, cohort-specific identifiers, repository-wide identifiers, or a combination of the above. For provider identifiers, repository-wide mode, purging all provider identifiers listed in the institution’s file is an attractive approach,† because each patient can be seen by many of the providers who may be mentioned in the patient’s reports.
Patient identifiers are the most important identifier that need to be detected and eliminated from reports. Each report in an electronic health record system (EHR) is typically tied to many patient identifiers. The de-identification system can take advantage of that information and can detect those identifiers in the text at the highest sensitivity level. For example, if the last name of the patient is Cushing, the de-identification system would not overlook the word by erroneously presuming that it refers to Cushing’s disease.
In certain record types, specifically in HL7 v.2 records, additional personal identifiers, such as the patient’s address, telephone number, medical record number, and next of kin identifiers are also available as part of the PID and NK1 segments.
In some circumstances, providing patient identifiers for each report may not be feasible but the identifiers of the patients retrieved for a specific research cohort could be provided as a single set of input to the de-identification system.
Identifiers of providers who dictate and sign reports are also found in EHRs. Inputting both repository-wide and report- specific provider information may improve de-identification results.
Scientist involved de-identification. Clinical text de-identification systems are not perfect. Because their primary goal is to remove all possible words and other alphanumeric strings that might identify the patient, they tend to favor sensitivity (for identifiers) over specificity, and may inadvertently remove non-identifying health information in the process. Users of the de-identification systems (i.e., scientists, directly or data managers, indirectly) can compensate for the problem by providing clinical terms that they would like to preserve from excessive de-identification. A capable system may automatically increase its sensitivity level when it receives such clinical metadata.
Patient involved de-identification. Many clinical institutions allow patients to access their health information stored in EHRs or personal health record systems. As today’s consumers become more and more suspicious of how their data have been shared by Internet companies without their knowledge, the transparency in the healthcare sector could alleviate such concerns. Institutions can recruit patients who volunteer to help de-identify their health records and/or verify that their personal identifiers have been properly purged from their data. Although it is not in current practice, we suspect some patients would demand this level of transparency and the option of self-verification from their providers in the near future.
Note that some identifying information can be inferred from context of the narrative or through circumstantial information instead of just personal identifiers. For example, “the examination of his injury that he endured during his US championship match today…” We call such information personally identifying context,12 which is hard for any automatic de-identification system to detect. The patient’s assistance would be very valuable for de-identifying such information.
Physician involved de-identification. Patients’ names and chart numbers get into narrative reports because physicians use patient identifiers when they dictate. In a few special cases, e.g. pathology reporting, a requirement exists to dictate both the patient identifying information and the specimen number into the report to tie that report securely to the specimen and the patient. For most dictated reports, however, recording the patient’s identifiers in the body of the report is not needed because the patient is usually identified to the transcriber prior to the dictated note. But from their habits in conversation with patients or other care providers, many providers often say “Mr. Jones,” or “Fred Jones” instead of simply “the patient.” Medical schools and professional societies should discourage the recording of patient names, or identifiers in the body of the dictation. De-identification systems can highlight names and identifiers in transcribed dictation pre signature, which may help providers to break this habit.
Online de-identification by honest brokers. With the advances on big data, scientists start accessing larger patient cohorts than what is available in their own institutes. As this trend continues, we can expect some of the big data processes, including de-identification, are going to be centralized.13–15 At that point, de-identification of clinical data can be provided as an online service by these centers. Small research institutes with less funding may also find this option attractive as they can rely on external expertise for proper de-identification. These centers can act as honest brokers providing expertise and assistance to scientists and clinical institutions for their de-identified data needs.
Discussions
We have introduced eight distinct modes of de-identification. Each of these modes comes with advantages and disadvantages, which we discuss in this section.
Institutes with smaller budgets for de-identification may find repository-wide batch de-identification attractive because of its simplicity, but they may not repeat the de-identification process on the same repository data. Since automatic clinical text de-identification systems improve continuously, the quality of the de-identified data improves over time; therefore, the repository data de-identified a number of years ago may not be on par with the current standards. Relying solely on this mode has the danger of stale data, which contain too many intact personal identifiers and too many false positives in the “de-identified” data according to state-of-the-art. If an institution would like to use this mode alone, the IRB should impose a policy for daily or weekly de-identification of new incoming data and cyclical re-de-identification of the repository data using the latest version of the automatic de-identification software.
On-demand de-identification practices solve the stale data problem since the institution could use the latest technology available to produce up-to-date de-identification. Furthermore, the de-identification output can be tailored according to the scientist’s needs. For example, if certain demographic information is necessary, it can be preserved in the output, producing a limited data set. If the de-identified data is produced via batch mode, the output would be either a fully de-identified set (hence the scientist’s needs could not be addressed) or a particular type of limited data set. In other words, the batch mode would be a generic one-size-fits-all approach.
On the other hand, producing a liberal limited dataset may be a viable, middle ground solution for those institutions, if their IRBs allow it. Due to the complexity of natural languages, no de-identified clinical report should be considered safe to share openly except for trivial cases; thus, even fully de-identified clinical reports should not be shared without a data use agreement, which is imposed by HIPAA Privacy Rule for all limited data sets. Because scientists, including those who would receive fully de-identified reports, should sign into a data use agreement as if they were receiving limited data sets, the institution may develop a policy to produce limited data sets only.16 Since limited data sets comprise PHI, this approach would not be in accord with the Privacy Rule for entities covered by HIPAA. Privacy Rule establishes the standard called the “minimum necessary” indicating limiting the content of PHI to the necessary minimum. 45 CFR 164.502(b): “When using or disclosing protected health information or when requesting protected health information from another covered entity or business associate, a covered entity or business associate must make reasonable efforts to limit protected health information to the minimum necessary to accomplish the intended purpose of the use, disclosure, or request.”17
Institutes that use an on-demand de-identification mode can employ a separate batch mode as well. Before submitting a research protocol to their IRBs, scientists need to analyze existing datasets to understand whether available datasets are suitable to their research needs. A de-identified copy of the entire repository would be beneficial for scientists at this stage of their research. A policy that would require such a research repository would better protect patient privacy.
On-demand de-identification of query results can be done quite easily due to the fast de-identification performance of modern automatic de-identification systems such as NLM-Scrubber,11 18 19 which de-identifies a typical report in a fraction of a second, much faster than the time required for a person to read the same text. The downside of this mode is that it is more difficult to integrate a standalone application to an EHR. On the other hand, if the de-identification system is integrated to the EHR, not only this particular mode but also the other two modes in which physicians and patients are involved, can be put in practice. When all of these modes are used in parallel, the performance of de-identification can be maximized.
Cohort-specific on-demand de-identification requires more time to access the data than the query results de-identification, but some institutions may prefer this approach, since the data manager could provide oversight to the de-identification process and monitor the level of privacy protection.
Properly providing patient and provider information to the de-identification system is necessary for better de-identification if the de-identification system is capable of accepting them. This mode requires extra steps from the institutions and data managers. Those steps might not be taken unless the institution establishes a policy to require these actions and the IRB monitors the compliance.
De-identification may inadvertently eliminate some informative clinical terms (e.g., newly introduced gene names) that mimic personal identifiers. A modern de-identification system should be capable of accepting new lists of words and concepts and set the bar of their removal higher to preserve the integrity of the content. If a repository-wide batch de-identification is adopted, it would be more difficult to introduce such functions.
If a de-identification system is used with the repository-wide batch mode, it would have to employ a single set of sensitivity and specificity levels to maximize the utility of the system. When the designer of a de-identification system (and in capable systems, the user of the system) adjusts the de-identification sensitivity of the system to eliminate all personal identifiers, the number of false positives could increase in tandem. If an on-demand de-identification mode is adopted and the scientists provide their metadata, the system can perform at a higher sensitivity level to eliminate all personal identifiers.
Online de-identification mode has a promising future. Large non-profit research organizations such as NIH or State Cancer Registries can employ the requisite expertise and human capital and provide these services to other research institutes free of charge. The benefits of this mode are that the de-identification can be improved and stringent policies for patient privacy can be employed. Furthermore, the data from a number of institutions can be hosted at these sites or in the cloud; thus, scientists can access much larger cohorts and conduct research on big data. Since these datasets for cancer have already been collected at the state level, cancer registries can take this responsibility if necessary funding can be provided. At some point in the future, this could be achieved at the federal level as well, but governance of the data at the federal level could be more complicated and may require new laws and regulations as well as a lot of convincing before allowing data to be shared across states and institutions.
Note that a number of these modes can be used in parallel. Consider the following as a possible scenario. An institute may use repository-wide batch de-identification so that the scientists at the institution can study the repository data for preliminary research. Since the preliminary study was done on de-identified data, some clinical terms could be deleted by the de-identification process and the scientist might not have retrieved all patient cases. During the study, the scientist can query the original data containing personal identifiers and receive the fully de-identified results. After studying the data and observing the eliminated terms, the scientist can provide a list of terms to be preserved to the data manager and may request certain date and age information be preserved. The data manager can repeat the de-identification by using patient and provider information as well as the list of clinical terms as inputs to the system and adjust the system to preserve date and age information. After the data manager’s review of the results, the scientist would receive the resulting limited data set with all pertinent clinical information preserved. If available, the scientist can repeat this process by accessing a clinical data warehouse in the cloud to enhance the patient cohort using big data. This process can be improved further by involving patients and physicians in the de-identification process.
Conclusions
There are many different ways to perform de-identification. Most of these modes of de-identification operations can be combined in order to reach ideal results. The success of the de-identification process depends not only on the competency of the automatic de-identification systems, but also on the competency, dedication, and discipline of the institution that is responsible for de-identification of protected health information. Users of de-identification systems need to learn how to operate the different modes properly, recognize and obtain all available patient and provider identifiers, gather the terminology needs of the scientist, and tailor the de-identification to achieve the highest quality of research data and patient privacy.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Competing Interests
The author is the principal investigator in NLM-Scrubber project at NIH. He receives royalties from University of Pittsburgh for his contribution to a de-identification project; the resulting product was acquired by a third party, which today is known as De-ID Data Corp. NLM’s Ethics Office reviewed and approved his appointment.
Footnotes
Although the secondary use of clinical information may go beyond scientific purposes, such as financial analysis, fraud detection and marketing,3. American Medical Informatics Association. A Taxonomy of Secondary Uses and Re-Uses of Healthcare Data. Policy Meeting, 2007. our scope is limited to non-commercial scientific use only.
Provider identifiers are not necessarily part of the protected information, at least according to HIPAA Privacy Rule, but other laws, regulations and policies may require their elimination through de-identification.
References
- 1.U.S. Department of Health and Human Services. The public welfare; administrative data standards and related requirements; general administrative requirements; general provisions; definitions. 45 CFR § 160.103.: U.S. Department of Health and Human Services. 2002.
- 2.The Public Health and Welfare, Health Information Technology, Privacy. 42 U.S. C. § 17921. 2009.
- 3.American Medical Informatics Association. A taxonomy of secondary uses and re-uses of healthcare data. Policy Meeting. 2007.
- 4.Office of Civil Rights. Does the HIPAA Privacy Rule preempt state laws? Health Information Privacy. 2013.
- 5.Federal Register (Part V) 45 CFR Parts 160 and 164 Standards for privacy of individually identifiable health information; final rule: National Archives and Records Administration. 2002. pp. 53182–273. [PubMed]
- 6.Feige EL, Watts HW. An investigation of the consequences of partial aggregation of micro-economic data. Econometrica: Journal of the Econometric Society. 1972:343–60. [Google Scholar]
- 7.Anonymization techniques for knowledge discovery in databases. Knowledge Discovery & Data Mining; Proc. 1st Int. Conf.1995. [Google Scholar]
- 8.Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. Proc AMIA Annu Fall Symp; 1996. pp. 333–37. [PMC free article] [PubMed] [Google Scholar]
- 9.Office of Civil Rights. Guidance regarding methods for de-identification of protected health information in accordance with Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. U.S. Department of Health and Human Services, editor. 2012.
- 10.Deleger L, Molnar K, Savova G, Xia F, Lingren T, Li Q, et al. Large-scale evaluation of automated clinical note de-identification and its impact on information extraction. J Am Med Inform Assoc. 2013;20(1):84–94. doi: 10.1136/amiajnl-2012-001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayaalp M, Browne AC, Callaghan FM, Dodd ZA, Divita G, Ozturk S, et al. The pattern of name tokens in narrative clinical text and a comparison of five systems for redacting them. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2013-001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kayaalp M, Sagan P, Jones JK, Browne AC, McDonald CJ. Guidelines for annotating personal identifiers in the clinical text repository of the National Institutes of Health. 2016 [Google Scholar]
- 13.Department of Health and Human Services. Big data to knowledge. 2015. https://www.hhs.gov/open/plan/big-data-to- knowledge.html.
- 14.Penberthy L. SEER registries: population-based infrastructure to support cancer research: National Cancer Institute, National Institutes of Health. 2016 [Google Scholar]
- 15.Devers K, Gray B, Roamos C, Shah A, Blavin F, Waidmann T. The feasibility of using electronic health records (EHRs) and other electronic health data for research on small populations: Urban Institute. 2013 [Google Scholar]
- 16.Cimino JJ, Ayres EJ, Beri A, Freedman R, Oberholtzer E, Rath S. Developing a self-service query interface for re-using de-identified electronic health record data. Studies in health technology and informatics. 2013;192:632–6. [PMC free article] [PubMed] [Google Scholar]
- 17.Sebelius K. 45 CFR Parts 160 and 164. Modifications to the HIPAA Privacy, Security, Enforcement, and Breach Notification Rules Under the Health Information Technology for Economic and Clinical Health Act and the Genetic Information Nondiscrimination Act; Other Modifications to the HIPAA Rules; Final Rule In: Office of the Secretary of the Deparment of Health and Human Services. 2013;78(17):5566–702. Federal Register. [PubMed] [Google Scholar]
- 18.Kayaalp M, Browne AC, Dodd ZA, Sagan P, McDonald CJ. Clinical text de-identification research. A report to the Board of Scientific Counselors: U.S. National Library of Medicine, National Institutes of Health. 2013 [Google Scholar]
- 19.Kayaalp M, Browne AC, Dodd ZA, Sagan P, McDonald CJ. De-identification of address, date, and alphanumeric identifiers in narrative clinical reports. Proc AMIA Annual Fall Symp; 2014. [PMC free article] [PubMed] [Google Scholar]


