Abstract
The use of automatic speech recognition (ASR) to create clinical notes has the potential to reduce costs associated with note creation for electronic medical records, but at current system accuracy levels, post-editing by practitioners is needed to ensure note quality. Aiming to reduce the time required to edit ASR transcripts, this paper investigates novel methods for automatic detection of edit regions within the transcripts, including both putative ASR errors but also regions that are targets for cleanup or rephrasing. We create detection models using logistic regression and conditional random field models, exploring a variety of text-based features that consider the structure of clinical notes and exploit the medical context. Different medical text resources are used to improve feature extraction. Experimental results on a large corpus of practitioner-edited clinical notes show that 67% of sentence-level edits and 45% of word-level edits can be detected with a false detection rate of 15%.
Introduction
The use of ASR has increased within the clinical setting as speech recognition technology has matured and the availability of computational resources has increased1. The creation of clinical notes using ASR offers system-level benefits, like short document turnaround time; however, note quality is negatively impacted by speech recognition errors, including clinically significant errors, and higher document creation times for practitioners associated with editing1. Automatic detection and flagging of likely edits in ASR transcripts through a correction tool could reduce the time required to edit ASR transcripts and improve note quality by reducing the prevalence of uncaught errors. In this work, we investigated the automatic detection of sentences and words that are likely to be edited in the clinical note ASR transcripts by applying data-driven, machine learning detection strategies.
Practitioners may edit ASR transcripts to correct errors and disfluencies, but also to rephrase portions of the transcript. ASR errors are portions of the dictation that are incorrectly transcribed. Disfluencies include dictation that is repeated (e.g. “the patient the patient”), repaired (e.g. “hypertension I mean hypotension”), and restarted (e.g. “heart has abdomen is”). Rephrasing is associated with changes to the transcript that are not ASR errors or disfluencies (e.g. changing “patient got up” to “patient awoke” or changing “nothing by mouth” to “NPO”). Practitioners may also edit the transcripts as a continuation of the note creation process, deleting information that is no longer relevant or correct and inserting additional information such as test results and new plans for patient care. Word sequences and sentences in the ASR transcript that are edited by practitioners during editing are collectively referred to in this paper as transcript edits.
We identified ASR transcript edits within a corpus of clinical notes created through the voice-generated enhanced electronic note system (VGEENS) Project2. We applied ASR error detection techniques to the detection of transcript edits within the VGEENS Corpus, utilizing medical domain knowledge, including clinical note structure and medical terminology. ASR error detection identifies discrepancies between what is dictated and what is transcribed, while our investigation of transcript edits focused on identifying differences between what is transcribed and what the practitioner wants. Table 1 contains an example transcript edit, with the ASR transcript text and the corresponding text from the final note. The ASR transcript includes an incorrect categorization of the patient’s cognitive status and a disfluency with the repair word “correction.” In the final note, the cognitive status is corrected and the disfluency is deleted. In this example, our edit detection model correctly identified all of the words in the ASR transcript that should be replaced or deleted (indicated by bold font).
Table 1.
Transcript edit example (bold font indicates words flagged as likely edits by detection model)
| Source | Text |
|---|---|
| transcript | Alert and oriented 4, pleasant mood, blunted affect correction for affect, thought process is clear |
| final note | Alert and oriented x4, pleasant mood, full affect, thought process is clear |
Within the VGEENS corpus, practitioners deleted words and entire sentences from the ASR transcripts. We hypothesized that sentence-level deletions and word level deletions within the ASR transcripts have different characteristics and split the edit detection task into two tasks: sentence-level edit detection and word-level edit detection. The primary goals of this investigation were to evaluate the performance of ASR error detection techniques on the broader category of transcript edits at the sentence-level and word-level and explore methods for leveraging medical context and text resources to improve detection. We find that a substantial fraction of the errors can be detected with simple lexical context features but further gains are possible by leveraging medical context.
The rest of this paper is organized as follows. The Related Work section presents relevant ASR error detection work. The Methods section describes the data used in experimentation and the modeling approaches used to automatically detect transcript edits. The Results section presents the performance of the detection models, and the Conclusions section summarizes this investigation and discusses future work.
Related Work
There is a significant body of ASR error detection work (also known as ASR confidence estimation). ASR error detection has been approached using a range of discrete models, including the hidden Markov model (HMM), maximum entropy (MaxEnt) model, and conditional random fields (CRF) model, as well as continuous sequence models, including recurrent neural networks (RNN)3-11.
Many studies have explored ASR error detection using the linear chain CRF model, which is a discriminative sequence modeling variant of the general CRF framework12. Bechet and Favre created a CRF error detection model using ASR posterior probabilities, lexical features (word n-grams and word length), and syntactic features (part of speech (POS) and dependency labels)5. In their work, the inclusion of both lexical and syntactic features improved error detection performance. Ghannay, Esteve, and Camelin explored the use of the Multi Layer Perceptron (MLP) neural network architecture, as well as a CRF model as a baseline6. The input features included ASR posterior probabilities, word representations (orthographic for CRF and embeddings for MLP), lexical features (word length and trigram indicator function), and syntactic features (POS tags and dependency labels). The best results were achieved using the MLP.
RNNs are currently a popular neural sequence model for ASR error detection, and several RNN variants have been used. Kalgaonkar, Liu, Yifan, and Yao compared the performance of a MLP model with a standard RNN and an RNN with an output decoder consisting of a two-state (no error/error) bigram language model9. The input features consisted of acoustic, linguistic, and confidence scores from the speech recognizer. The recurrent approaches outperformed the MLP, and the output decoder provided a small benefit. Ogawa and Hori created ASR error detection models based on CRF and bidirectional RNN frameworks, using acoustic and linguistic features and speech recognizer states and scores10. The bidirectional RNN outperformed the CRF. Ángel del-Agua, Piqueras, Giménez, Sanchis, and Civera explored speaker-adapted ASR confidence estimation using NaÏve Bayes, CRF, and long short-term memory (LSTM) RNN models11. The models used pre-trained word vectors and speech recognizer-derived features, and the LSTM RNN achieved the best performance.
Much of the work on ASR error detection has focused on constrained domain human-computer interaction or human-directed broadcast news. The creation of clinical notes using ASR differs from other ASR transcription tasks in that the goal is note creation, not faithful transcription of what was dictated. The goal of note creation is to create a medically accurate document that articulates exam findings and plans for patient care and that meets formatting requirements/norms. Edit detection is motivated by this note creation goal and attempts to find a range of edit types (ASR errors, disfluencies, rephrasing, and note continuation) that impact the quality of the clinical notes. Edits within clinical ASR transcripts include sentence-level and word-level edits, where sentence-level edits tend to be a continuation of the note authoring process and word-level edits tend to be associated with disfluencies, speech recognition, and rephrasing. Another important difference with respect to ASR error detection work is that widely used medical dictation systems do not provide the detailed acoustic scores available in research systems, which makes error detection more challenging. On the other hand, in medical dictation, there are more context constraints that can be used to identify errors. Domain contextual constraints includes clinical note structure (e.g. topical sectioning), structured patient data in the Electronic Health Record (EHR), and ranges of numerical values (e.g. drug dosages and vital signs) and motivates the exploration of different features.
There is a relatively small body of work related to ASR error detection within the medical domain. Voll explored the automatic detection of ASR errors in radiology notes using language models, point-wise mutual information, and hand-crafted rules13,14. Schreitter and Trost investigated the correction of medication dosages within ASR transcripts by extracting medications and the associated dosages and then evaluating dosages based on medication databases15.
Methods
Medical transcript edits comprise more than ASR errors and impact a larger portion of the note. Edit regions within ASR transcripts can be identified by aligning pairs of ASR transcripts and final notes. In clinical settings where practitioners dictate to a recording device without viewing ASR output in real time (noninteractive setting), ASR transcript-final note pairs are created as part of the existing workflow, providing efficient and low cost access to training data for detection models.
Data
The automatic detection of transcript edits was explored through the corpus of free-text clinical notes created through the VGEENS Project, which was conducted at the University of Washington Medical Center and Harborview Medical Center. As part of the VGEENS Project, inpatient progress notes were created by resident and attending internal medicine physicians using ASR through a multi-step process. First, a doctor dictated the note to a recording device during rounds, verbalizing punctuation and topical section headings (e.g. Chief Complaint). Then the dictation was transcribed using a commercial ASR system (Dragon Medical by Nuance Inc.) and automatically post-processed to format section headings. Lastly, the ASR transcript was reviewed and edited by the doctor, and the final note was entered into the EHR. The VGEENS Corpus of clinical notes includes 669 records created by 15 practitioners, where each record consists of an ASR transcript-final note pair.
Transcript edits were identified through the alignment of each ASR transcript-final note pair using Gestalt Pattern Matching16. Gestalt Pattern Matching finds the longest sequence of matching tokens and then finds the next longest sequence of matching tokens to the left and right of the longest matching sequence. This process is applied recursively, until all of the matching sequences are identified. Based on the alignment, each word within the ASR transcripts was labeled as keep or delete. The capitalization of tokens was ignored during the alignment of the note pairs.
Sentence boundaries were determined based on the location of colons, periods, and line breaks, rather than using an off-the-shelf sentence boundary detector, because of the structure of the VGEENS notes (verbalized punctuation; section headings, numbered lists, etc. explicitly indicated). Approximately 10% of the sentences within the ASR transcripts were deleted during editing. In the subset of sentences that are not deleted, approximately 9% of the words were deleted during editing. Given the difference between the characteristics of sentence-level and word-level deletions, the detection of transcript edits was split into two tasks: sentence-level edit detection and word-level edit detection. Word-level gold standard labels were determined based on the keep or delete labels from the note alignments. Sentence-level gold standard labels were determined as follows: sentences were labeled as delete when all word-level labels were delete and sentences were labeled as keep when at least one word-level label in the sentence was keep.
The labeled ASR transcripts associated with the VGEENS Corpus were used in model training and testing (80% training/20% testing). Since this data set was relatively small, we explored use of the MedTrans17 and the i2b218 corpora of clinical notes (referred to as the External Corpora) for learning word classes and embeddings and a language model (LM). Table 2 contains a summary of the corpora used. In addition to these corpora, feature extraction utilized a list of medical terms derived from SNOMED CT, RxNorm, and the UMLS SPECIALIST Lexicon19-21.
Table 2.
Corpora summary
| Corpus | Description | Note count | Word count | Sentence count |
|---|---|---|---|---|
| VGEENS | Clinical notescreated using ASR | 669 | ASR transcripts: 483 k final notes: 695 k | ASR transcripts: 46 kfinal notes: 57 k |
| MedTrans17 | Example clinical notes created by human transcriptionists | 2.37 k | 1.51 M | 135 k |
| i2b218 | De-identified clinical notes, including onlyunique notes from 2006-2012 competition data sets. | 4.32 k | 4.75 M | 461 k |
Detection Models
The goal of the sentence-level edit detection task was to label sentences within the ASR transcripts as keep or delete. Logistic regression, which is a binary discriminative classifier, was selected for the sentence-level edit detection model because it is known to work well for text classification with relatively small amounts of training data when combined with regularization. The goal of the word-level edit detection task was to label words within the ASR transcripts as keep or delete. Word-level edit detection models were only trained and evaluated on sentences from the ASR transcripts that had sentence-level keep labels from the note alignment. Because of the importance of sequential context at the word level, word-level edit detection was explored using the linear-chain CRF modeling framework, which estimates the highest probability sequence of labels given a sequence of observations12. We also experimented with an LSTM RNN in the word-level edit detection task; however, the LSTM RNN did not outperform the CRF, and only the CRF modeling results are presented in this paper.
Feature extraction
The VGEENS Corpus only includes the text output of the ASR system and does not include acoustic information or speech recognizer internal states (confidence scores, alternative word sequences, etc.), which are often used in ASR confidence estimation tasks. For both detection tasks, we utilized domain knowledge and unlabeled training data, in order to compensate for the limited information and relatively small size of the VGEENS Corpus. We explored interpretable features, like topical coherence, that may be useful to physicians.
A fixed vocabulary was selected based on the words that occur in the VGEENS Corpus training subset and External Corpora at least four times, resulting in a vocabulary size of 20.7 k words. Out of vocabulary (OOV) tokens were mapped to one of seven OOV tokens, depending on whether the token was a medical term, numerical, lower case, upper case, title case, alphanumeric, or other.
Word-based Features
In both detection tasks, text-based features were created using discrete and continuous word representations. In the sentence-level edit detection task, word-based features were intended to automatically learn relevant sentence attributes, like: numbered lists (e.g. “1. Liver cirrhosis…”), additional information required (e.g. “…waiting for LFT results…” or “Continue to monitor”), or topical headings not conforming to EHR format (e.g. “Cardiovascular:”). In the word-level edit detection task, word-based features were intended to automatically identify frequent ASR errors (e.g. phonetically similar words like “he” and “she”), disfluencies (e.g. repair words like “I mean” and “correction”), and rephrasing (e.g. abbreviating “daily” to “qd”).
Discrete word representations included orthographic and word class forms. Word classes were used to reduce data sparsity by grouping words based on syntactic/semantic similarity. Because of the small amount of VGEENS data, we leveraged external data to learn more reliable classes7,22. Two types of classes were used: manually-defined (e.g. “hypertension” ⟶ “<med_term>”) and automatically learned (e.g. “patient” ⟶ “class00101”). Manual classes (rule-based classes) were created indicating punctuation, capitalization, numbers, and medical terms. Automatically learned word classes were created using unsupervised clustering approaches where words that appear in similar context are merged into the same class so as to maximize the mutual information between consecutive words, referred to as Brown clustering22. 500 Brown classes were learned from a merged corpus of the final notes in the VGEENS Corpus training set and the External Corpora.
Words were also represented as continuous word embeddings (vectors of real numbers), in which a sparse (one-hot) representation of words is mapped to a low-dimensional continuous space capturing syntactic, semantic, and topical information. Word embeddings were created using two unsupervised learning approaches. The first method starts with a term frequency-inverse document frequency (TF-IDF) representation of 4 k VGEENS note sections and learns a linear transformation of words (and documents) into a 200-dimensional space using non-negative matrix factorization (NMF). (Performance was similar with 100 dimensions.) Word embeddings were also created using the neural word2vec skip-gram model, which is a single-layer neural network that predicts context words given the current word23. The skip-gram embeddings were created using the final notes in the VGEENS Corpus training set and the External Corpora (embedding size 200, context width 10). K-mean clustering was used to create 500 discrete skip-gram classes from the skip-gram embeddings.
Language Modeling
We hypothesized that atypical (infrequent) word sequences were more likely to reflect ASR errors or disfluencies (and require editing) than frequent sequences and therefore used word sequence probability as an input feature in both detection tasks. In order to create a LM that did not include the ASR transcript edit regions and did not overfit to the final notes in the VGEENS Corpus, a LM was trained on a subset of the External Corpora that best matched the VGEENS Corpus. The Moore-Lewis data selection approach was used to select the subset of the External Corpora24. An LM was created using the final notes in the VGEENS Corpus training set, and a second LM was created from a random sampling of the sentences within the External Corpora of similar size to the final notes in the VGEENS Corpus training set. The cross entropy of each sentence in the External Corpora was calculated using each LM, and the difference between the cross entropy scores was used to select the best matching sentences in the External Corpora (approximately half of the corpora selected, 3.3 M words). A LM based on the selected subset of the External Corpora was used in subsequent experimentation. All LMs were trigrams models with Kneser-Ney smoothing25.
Topical Coherence
The topical coherence between the target word or sentence and the local context was scored using word embeddings. In the sentence-level edit detection task, topical coherence features were intended to identify sentences that were out of place within the section, potentially belonging in a different section of the note. A sentence-level vector representation was created by averaging the embeddings of each word in the target sentence, and a section-level vector representation was created by averaging the embeddings of each of the remaining words in the note section. The cosine similarity and the vector difference between the representations was then calculated. In the word-level edit detection task, topical coherence features were intended to identify words that did not fit the surrounding context, due to an ASR error. The target word embedding was compared to the averaged vector representation of words in the local context (current sentence without this word and +/- two sentences) using cosine similarity and vector difference.
Task-Specific Feature Sets
Table 3 and Table 4 contain a list of the features for the sentence-level and word-level edit detection tasks, respectively.
Table 3.
Sentence-level edit detection features
| Feature category | Feature set | Description |
|---|---|---|
| Structure | length/position | target sentence section, positionin line (first, middle, or last), and token count |
| Words | words | word occurrence (indicator) vector for target sentence |
| Brown classes | Brown classoccurrence vector fortarget sentence | |
| skip-gram classes | skip-gram class occurrence vector for target sentence | |
| adjacent words | word occurrence vector forthe last three words in the previous sentence and the first three words in the next sentence | |
| LM | perplexity | average per-word perplexity of the target sentence |
| Topical | VGEENS section | cosine similarity and vector difference of averaged TF-IDF/NMFembedding representations of the target sentence and note section |
| skip-gram section | cosine similarity and vector difference of averaged skip-gram embedding representations of the target sentence and note section |
Table 4.
Word-level edit detection features
| Feature category | Feature set | Description |
|---|---|---|
| Words | words | word n-grams (n=1-3) in window size of5 |
| manual classes | Manual class unigrams in window size of 5 | |
| Brownclasses | Brown class n-grams (n=1-3) in window size 5 | |
| skip-gramclasses | skip-gram class n-grams (n=1-3) in window size of5 | |
| LM | probability | probability of word sequences inwindowsizes 3 and 5 |
| Topical | VGEENS context | cosine similarity and vector difference of averaged TF-IDF/NMFembeddingre presentations of the target word and local context |
| skip-gram context | cosine similarity and vector difference of averaged skip-gram embedding representations of the target word and local context |
Training and Evaluation
The edit detection models (logistic regression and CRF) were trained using the VGEENS Corpus training set. Cross validation (three folds) was used to determine the best regularization type (L1-norm, L2-norm) and regularization weight for each feature set. For each feature set, the optimum L1-norm weight was determined with the L2-norm weight set to zero, and the optimum L2-norm weight was determined with the L1-norm weight set to zero. During cross validation, model performance was assessed using the Receiver Operating Characteristic (ROC) area under the curve (AUC). The final model for each feature set was trained on the entire training set using the selected regularization type and weight from cross validation.
The performance of the edit detection models was evaluated using the VGEENS Corpus testing set. Model performance was evaluated through ROC AUC and a performance analysis. In the edit detection tasks, the false detection rate (Pf) is the frequency of labeling a target as delete when the true label is keep, and the missed detection rate (Pm) is the frequency of labeling a target as keep when the true label is delete. In the performance analysis, Pf was fixed at 15%, and Pm was calculated. Conversely, Pm was fixed at 15%, and Pf was calculated. These Pf and Pm values were selected to understand model performance at different precision-recall operating points.
Results
Sentence-level Edit Detection
The sentence-level edit detection ROC AUC test results are presented in Table 5. The best performing single feature set was the words feature set, followed closely by the Brown classes. This suggests that the Brown classes capture the salient aspects of word context and syntax. The Brown classes, which are based on bigram occurrences, outperformed the skip-gram classes, which are based on word cooccurrence within a context window of length 10. The length/position feature set, which leverages the section structure of the clinical note, achieved high performance, despite a relatively small number of features (16 features). The TF-IDF/NMF topic modeling (VGEENS section feature set) achieved higher performance than the skip-gram topic modeling (skip-gram section feature set), even though the skip-gram embedding training set was approximately 13 times larger than the TF-IDF/NMF embedding training set. The TF-IDF/NMF approach utilized the structure of the note during topic learning, which may account for the higher performance. Two combined feature sets were tested: combined VGEENS using features based only on the VGEENS data and combined external, which added features based on external data. The combined feature sets outperformed the highest performing single feature, but the features based on external data had no added benefit. Figure 1 presents the error tradeoff curves for a subset of the feature sets evaluated.
Table 5.
Sentence-level edit detection ROC AUC test results. Features that leverage external text resources are indicated with (*).
| Feature categories | Feature set | AUC |
|---|---|---|
| Structure | length/position | 0.75 |
| Words | words | 0.81 |
| (*) Brownclasses | 0.80 | |
| (*) skip-gram classes | 0.77 | |
| adjacentwords | 0.65 | |
| LM | (*) perplexity | 0.65 |
| Topic | VGEENS section | 0.73 |
| (*) skip-gram section | 0.68 | |
| combined VGEENS | length/position + words+adjacentwords+ VGEENS section | 0.83 |
| combined external | (*) length/position + words +Brownclasses+ adjacentwords+ perplexity + VGEENS section | 0.83 |
Figure 1.
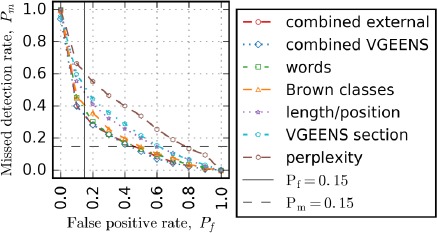
Sentence-level edit detection error tradeoff curves
Table 6 presents more detailed performance analysis for three systems at three different operating points of the sentence-level edit detector. First, we compare Pm for the different systems at Pf=15 %. At this operating point, the Pm of the combined VGEENS feature set was 67% lower than the status quo where all targets are labeled keep (no edit detection used). Next, we compare Pf for different systems at Pm=15%. At this operating point, Pf of the combined VGEEN feature set is 57% lower than the status quo. Since many studies assess performance using F-score, we include the results for the operating point with the best F-score for each system. The best performing feature set for all metrics was the combined VGEENS feature set, and the word indicator feature set performed only slightly worse than the combined VGEENS feature set.
Table 6.
Sentence-level performance analysis
| Feature categories | Feature set | Fixed Pf | Fixed Pm | Optimized F1 | ||||
|---|---|---|---|---|---|---|---|---|
| Pf | Pm | Pf | Pm | Precision | Recall | F1 | ||
| Structure | length/position | 15% | 46% | 59% | 15% | 0.35 | 0.44 | 0.39 |
| Words | words | 15% | 39% | 45% | 15% | 0.37 | 0.55 | 0.44 |
| combined VGEENS | length/position + words + adjacent words + VGEENS section | 15% | 33% | 43% | 15% | 0.40 | 0.59 | 0.48 |
Table 7 presents section-level edit examples based on predictions from the detection model trained on the combined VGEENS feature set at Pf = 15%. The first example, which was correctly labeled as delete, illustrates a case where the sentence indicates that additional information is required. We hypothesized that during the time between dictation and editing, new findings or conclusions were available, resulting in the sentence being deleted. The second example, which was incorrectly labeled as delete, is similar in that it includes the word “continue;” however, it does not imply additional follow-up. The third example was correctly labeled as delete and has a similar format (short phrase followed by a colon) to the topical headings within the notes. This heading does not conform to the section headings defined by the EHR, and the practitioner appears to have deleted this section heading and the associated note content. In example 4, the sentence was incorrectly labeled as keep. Similar to the first example, the example 4 implies additional information is needed, but there were insufficient cues for the system to predict a delete label. The sentence may have been deleted because the required consultation was performed.
Table 7.
Sentence-level edit detection examples
| No. | Label | Examplefrom ASR transcript | |
|---|---|---|---|
| Truth | Predicted | ||
| 1 | delete | Delete | Continue to monitor |
| 2 | keep | Delete | Continuerifaximin |
| 3 | delete | Delete | Ins and outs : |
| 4 | delete | Keep | Discuss with hepatology regarding further management |
Word-level Edit Detection
The word-level edit detection test ROC AUC results are presented in Table 8. For the CRF approach, the single feature sets with the highest performance were Brown classes and words. Similar to the sentence-level edit detection task, the discrete features defined by Brown classes outperformed those based on the skip-gram classes. While both word class approaches reduced the size of the feature space significantly, the Brown classes appeared to better capture the salient syntactic aspects of the words. In contrast to the sentence-level edit detection task, the skip-gram topic modeling approach outperformed the VGEENS topic approach for characterizing the word-level context match. This suggests that the external text is useful for characterizing words but not sentences, as is not surprising because of the more controlled format of the VGEENS notes.
Table 8.
Word-level edit detection test results. Features that leverage external text resources are indicated with (*).
| Feature categories | Feature set | AUC |
|---|---|---|
| Words | words | 0.72 |
| (*) Manual classes | 0.62 | |
| (*) Brown classes | 0.72 | |
| (*) skip-gram classes | 0.68 | |
| LM | (*) probability | 0.64 |
| Topic | VGEENS context | 0.60 |
| (*) skip-gramcontext | 0.65 | |
| combined VGEENS | words+ VGEENS context | 0.72 |
| combined external | (*) words + Manual classes+ Brownclasses+ probability + skip-gram context | 0.74 |
The combined VGEENS feature set was created using the words and VGEENS section feature sets. The combined external feature set was created using the best performing word class (Brown classes) and topic model (skip-gram context) feature sets and the remaining feature sets. The combined VGEENS feature set did not outperform the words feature set, but the combined external feature set outperformed the highest performing single feature set and the combined VGEENS feature set by approximately 3%. Figure 2 presents the error tradeoff curves for a subset of the feature sets evaluated.
Figure 2.
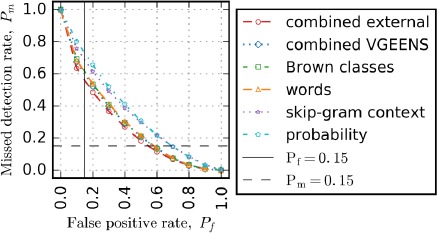
Word-level edit detection error tradeoff curves
Table 9 has more detailed performance analysis again at three different operating points for two word-level edit detectors. Comparing Pm for the different systems at Pf = 15%, the Pm of the combined external feature set was 45% lower than the status quo (assuming all targets are keep). With Pm fixed at 15%, the combined external feature set Pf was 46% lower than the status quo. Again, the best performing feature set for all metrics was the combined external feature set, and the words feature set performed only slightly worse than this combined feature set.
Table 9.
Word-level edit detection performance analysis
| Feature categories | Feature set | Fixed Pf | Fixed Pm | Optimized F1 | ||||
|---|---|---|---|---|---|---|---|---|
| Pf | Pm | Pf | Pm | Precision | Recall | F1 | ||
| Words | words | 15% | 60% | 58% | 15% | 0.23 | 0.36 | 0.28 |
| combined external | words + Manual classes + Brownclasses+ probability + skip-gram context | 15% | 55% | 54% | 15% | 0.25 | 0.42 | 0.31 |
Table 10 presents word-level edit detection examples based on predictions from the model trained on the combined external feature set with Pf=15%. Example 1 is from the Laboratory results section of the note and includes the word “restaurant,” which is a low probability word in this context and is not topically relevant. At Pf = 15%, the model misses the deletion associated with the word “restaurant;” however the model correctly identifies this deletion at higher Pf. Example 2 includes the disfluency repair word “correction,” and the detection model correctly identified the deletion but incorrectly labeled “Temperature” as delete. Example 3 includes a common phrase that is abbreviated, which the model correctly labeled as delete; however, the model incorrectly labels additional words in the sentence as delete. Example 4 appears to include an ASR error (transcription of “technetium” instead of “magnesium”), which the model correctly labels as delete. Similar to examples 2 and 3, a false delete label is also applied within the sentence.
Table 10.
Word-level edit detection examples (true delete are bold strikethrough, false delete are bold underline, false keep are italics strikethrough, and true keep are unformatted)
| No. | Example |
|---|---|
| 1 | ASR: …heart rate 79 restaurant rate 16 blood pressure 121 / 76… Final: …heart rate 79, blood pressure 121 / 76… |
| 2 | ASR: Temperature31 correction 37.1, heart rate 90… Final: Temperature 37.1, heart rate 90… |
| 3 | ASR: Macrocytic anemia, present on admission, chronic and related… Final: Macrocytic anemia, POA, chronic and related… |
| 4 | ASR: …Glucose 90, calcium 8.4, technetium 2.1… Final: …Glucose 90, calcium 8.4, magnesium 2.1… |
Conclusions
In this paper, we applied ASR error detection techniques to the automatic detection of sentence-level and word-level edits within clinical ASR transcripts. The results demonstrate that a substantial number of sentence- and word-level edits can be automatically detected with a small false detection rate. In both tasks, the word and word class feature sets were the highest performing single feature sets, indicating that word classes learned from external medical text resources using unsupervised Brown clustering are effective prediction features. Although the language model and topic-based features achieved lower performance than the word-based features, the results show that these features are relevant for edit detection in that the best performance in both detection tasks was achieved through a combination of features. The high performance achieved in the sentence-level tasks suggest a strong relationship between sentence editing habits and topical coherence within note sections. The best performance in the word-level task was achieved through the incorporation of external data.
This work is limited by the size of the corpus of clinical notes created using ASR and the number of practitioners involved in the creation of this corpus. A larger corpus, created by a larger sample of practitioners, would likely improve detection performance and improve the generalizability of the detection models to notes created with different dictation protocols. The methods used were also constrained by the ASR system configuration; access to alternative recognizer hypotheses or word confidence estimates would lead to further improvements in performance.
This work was motivated by the hypothesis that a correction tool that automatically detects and flags likely edits within ASR transcripts could improve note quality and accuracy. While the performance achieved in this investigation is likely not adequate to create a viable correction tool at this point, this work produced promising results that warrant further exploration, including the procurement of additional training data. Future work to improve performance would likely include incorporating additional, unlabeled data through semi-supervised learning and the inclusion of biomedical knowledge sources. A user study with practitioners is required to assess the required level of performance, determine the appropriate performance metrics and thresholds (precision, recall, etc.), and design the correction tool interface.
Acknowledgements
We thank Eve Riskin, Thomas Payne, and Andrew White for their assistance. The data used was provided through the support of grant number R21HS023631 from the Agency for Healthcare Research and Quality (AHRQ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the AHRQ.
Table and Figures
References
- 1.Hodgson T, Coiera E. Risks and Benefits of Speech Recognition for Clinical Documentation: A Systematic Review. J Am Med Informatics Assoc. 2015;23(1):69–179. doi: 10.1093/jamia/ocv152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne T, Zhou L. Webinar: Improving Health IT Safety through the Use of Natural Language Processing [Internet] Agency for Healthcare Research and Quality. [cited 2017 Jan 1]. Available from: http://www.ahrq.gov/news/webinar.html.
- 3.Liu Y, Shriberg E, Stolcke A, Member S, Hillard D, Ostendorf M, et al. Enriching Speech Recognition with Automatic Detection of Sentence Boundaries and Disfluencies. IEEE/ACM Trans Audio Speech Lang Process. 2006:1–15. [Google Scholar]
- 4.Wang W, Tur G, Zheng J, Ayan NF. Automatic Disfluency Removal for Improving Spoken Language Translation. In: Proc IEEE Int Conf Acoust Speech Signal Process; 2010. pp. 5214–7. [Google Scholar]
- 5.Bechet F, Favre B. ASR Error Segment Localization for Spoken Recovery Strategy. In: Proc IEEE Int Conf Acoust Speech Signal Process; 2013. pp. 6837–41. [Google Scholar]
- 6.Ghannay S, Esteve Y, Camelin N. Word Embeddings Combination and Neural Networks for Robustness in ASR Error Detection. In: Proc Eur Signal Process Conf EUSIPCO; IEEE. 2015. pp. 1671–5. [Google Scholar]
- 7.He J, Marin A, Ostendorf M. Effective Data-driven Feature Learning for Detecting Name Errors in Automatic Speech Recognition. In: SLT Workshop Spok Lang Technol; IEEE. 2014. pp. 230–5. [Google Scholar]
- 8.Pellegrini T, Trancoso I. Improving ASR Error Detection with Non-decoder Based Features. In: Interspeech. 2010:1950–3. [Google Scholar]
- 9.Kalgaonkar K, Liu C, Gong Y. Estimating Confidence Scores on ASR Results Using Recurrent Neural Networks. In: Proc IEEE Int Conf Acoust Speech Signal Process; 2015. pp. 4999–5003. [Google Scholar]
- 10.Ogawa A, Hori T. ASR Error Detection and Recognition Rate Estimation Using Deep Bidirectional Recurrent Neural Networks. In: Proc IEEE Int Conf Acoust Speech Signal Process; pp. 4370–4. [Google Scholar]
- 11.Angel Del-Agua M, Piqueras S, Gimenez A, Sanchis A, Civera J, Juan A. ASR Confidence Estimation with Speaker-adapted Recurrent Neural Networks. In: Interspeech. 2016:3464–8. [Google Scholar]
- 12.Lafferty J, Mccallum A. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. In: Proc Int Conf Mach Learn; 2001. pp. 282–9. [Google Scholar]
- 13.Voll K, Atkins S, Forster B. Improving the Utility of Speech Recognition through Error Detection. J Digit Imaging. 2008;21(4):371–7. doi: 10.1007/s10278-007-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voll K. In: Advances in Artificial Intelligence. Springer; 2007. A Hybrid Approach to Improving Automatic Speech Recognition Via NLP; pp. 514–25. [Google Scholar]
- 15.Schreitter S, Klein A, Matiasek J, Trost H. Using Domain Knowledge about Medications to Correct Recognition Errors in Medical Report Creation. In: Proc of the NAACL HLT; Association for Computational Linguistics. 2010. pp. 22–8. [Google Scholar]
- 16.Ratcliff JW, Metzener DE. Pattern Matching: The Gestalt Approach. Dr Dobbs J. 1988;13(7):46. [Google Scholar]
- 17.Transcribed Medical Transcription Sample Reports and Examples - MTSamples [Internet] [cited 2015 Apr 30]. Available from: http://mtsamples.com/
- 18.i2b2: Informatics for Integrating Biology & the Bedside [Internet] [cited 2015 Sep 1]. Available from: https://www.i2b2.org/
- 19.SNOMED CT [Internet] U.S. National Library of Medicine. [cited 2015 May 1]. Available from: https://www.nlm.nih.gov/healthit/snomedct/
- 20.RxNorm [Internet] U.S. National Library of Medicine. [cited 2017 Jan 1]. Available from: https://www.nlm.nih.gov/research/umls/rxnorm/
- 21.The SPECIALIST LEXICON [Internet] [cited 2017 Jan 1]. Available from: https://lsg3.nlm.nih.gov/LexSysGroup/Projects/lexicon/current/web/index.html.
- 22.Brown PF, DeSouza P V, Mercer RL, Pietra VJ Della, Lai JC. Class-based N-gram Models of Natural Language. Assoc Comput Linguist. 1992 Dec;18(4):467–79. [Google Scholar]
- 23.Mikolov T, Corrado G, Chen K, Dean J. Efficient Estimation of Word Representations in Vector Space. arXiv Prepr. 2013:1–12. [Google Scholar]
- 24.Moore RC, Lewis W. Intelligent Selection of Language Model Training Data. In: Proc Conf Assoc Comput Linguist; 2010. pp. 220–4. [Google Scholar]
- 25.Kneser R, Ney H. Improved Backing-off for M-gram Language Modeling. In: Proc IEEE Int Conf Acoust Speech Signal Process; Detroit, Michigan, USA. 1995. pp. 181–4. [Google Scholar]


