Abstract
Models have been developed to predict stroke outcomes (e.g., mortality) in attempt to provide better guidance for stroke treatment. However, there is little work in developing classification models for the problem of unknown time-since-stroke (TSS), which determines a patient’s treatment eligibility based on a clinical defined cutoff time point (i.e., <4.5hrs). In this paper, we construct and compare machine learning methods to classify TSS<4.5hrs using magnetic resonance (MR) imaging features. We also propose a deep learning model to extract hidden representations from the MR perfusion-weighted images and demonstrate classification improvement by incorporating these additional imaging features. Finally, we discuss a strategy to visualize the learned features from the proposed deep learning model. The cross-validation results show that our best classifier achieved an area under the curve of 0.68, which improves significantly over current clinical methods (0.58), demonstrating the potential benefit of using advanced machine learning methods in TSS classification.
1. Introduction
Stroke is the primary cause of long-term disability and the fifth leading cause of death in the United States, with approximately 795,000 Americans experiencing a new or recurrent stroke each year [1]. Several treatments exist for stroke, including intravenous and intra-arterial tissue plasminogen activator (IV/IA tPA), and mechanical throm-bectomy (clot retrieval). Guidelines support selecting tPA treatment administration only within a maximum of 4.5 hours from stroke symptom onset due to the increased risk of hemorrhage for longer times since stroke (TSS). However, about 30% of the population have unknown time since stroke (TSS), making these patients ineligible for treatment with tPA despite the fact that their strokes may have actually occurred within the treatment window [2].
Predictive models have been made in attempt to predict stroke patient outcomes (e.g., mortality) using clinical variables (e.g., age) and imaging features (e.g., lesion volume) [3–5]. Additional algorithms are under development that attempt to predict patient response to a specific treatment [6]. While much work has been done in predicting stroke patient outcome and treatment response, there is limited work in determining TSS. Studies are underway to investigate the use of a simple imaging feature, a mismatch pattern between magnetic resonance (MR) diffusion weighted imaging (DWI), on which stroke pathophysiology is immediately visible, and fluid attenuated inversion recovery (FLAIR) imaging, on which strokes are not visible for 3-4 hours [7–9], to estimate TSS. The mismatch pattern is known as “DWI-FLAIR mismatch.” While this method is the current state-of-the-art for determining eligibility for thrombolytic therapy in cases of unknown TSS, computing mismatch is a difficult task that requires extensive training and for which clinician agreement has been found to be only moderate, leading to less accurate performance [10–12]. Separately, limited work has been done in TSS classification using MR perfusion-weighted image (PWIs) for TSS classification, which may contain information that encodes TSS [13].
Machine learning models have been applied widely and can achieve good classification performance for problems in the healthcare domain because of their ability to learn and utilize patterns from data to make prediction [14]. Recent developments in deep learning [15] have drawn significant research interest because of the technique’s ability to automatically learn feature detectors specific to the data for classification and prediction tasks, achieving state-of-the- art performance in challenging problems (e.g., ImageNet [16], video classification [17], etc.). In this work, we hypothesize that machine learning models can be used to better classify TSS by learning latent representative features from MR images. We developed a deep learning algorithm based on an autoencoder architecture [18] to extract imaging features (i.e., deep features) from PWIs and evaluate the effectiveness of four machine learning classifiers with and without the deep features to classify TSS. We performed retrospective testing on images from stroke patients by censoring the known TSS and comparing performance to published results using DWI-FLAIR mismatch.
2. Related Work
“DWI-FLAIR mismatch” is defined as the presence of visible acute ischemic lesion on DWI with no traceable hyperintensity in the corresponding region on FLAIR imaging (Figure 1) [7]. The work of using DWI-FLAIR mismatch was first introduced by Thomalla et al. [10], in which they used the mismatch pattern to identify stroke patients with less than 3-hour stroke onset. The method achieved a high specificity of 0.93 and a high positive predictive value (PPV) of 0.94, with a low sensitivity of 0.48 and a low negative predictive value (NPV) of 0.43. Aoki et al. [19] and Petkova et al. [20] followed the same method and applied on their datasets. Both achieved a high sensitivity (0.83 and 0. 90 respectively) and a high specificity (0.71 and 0.93 respectively), but Aoki et al. reported a moderate PPV of 0.64.
Figure 1.
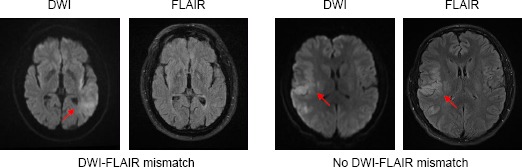
Example of DWI-FLAIR mismatch. LEFT: presence of DWI-FLAIR mismatch (TSS = 1hr); RIGHT: absence of DWI-FLAIR mismatch (TSS = 5.25hrs). Hyperintensities are indicated by the red arrows.
Work has also been done in using DWI-FLAIR mismatch to classify TSS<4.5hrs, which is the current clinical cutoff time for IV tPA treatment. Ebinger et al. [21] developed a mismatch model and it achieved a specificity of 0.79 and a sensitivity of 0.46. Later, a large multicenter study was done by Thomalla et al. [22] to assess the ability of DWI- FLAIR mismatch. The mismatch method achieved a specificity of 0.78 and a PPV of 0.83, with a sensitivity of 0.62 and a NPV of 0.54. The study interobserver agreement of acute ischemic lesion visibility on FLAIR imaging was moderate (kappa = 0.569). Emeriau et al. [23] also investigated the use of mismatch pattern and the model achieved a PPV of 0.88, but a sensitivity of 0.55, a specificity of 0.60, and an NPV of 0.19. The AUC of using mismatch patterns in the identification of TSS was 0.58. There are ongoing large multicenter clinical trials, such as the WAKE-UP trial in the European Union [8] and the MR WITNESS trial in the United States [24], to further investigate the use of DWI- FLAIR mismatch in guiding treatment decisions for patients with unknown TSS.
The above preliminary work using DWI-FLAIR mismatch demonstrates a potential opportunity for using image analysis to classify TSS. However, existing studies all suffer from the use of relatively simplistic features and models [10–12]. Furthermore, it has been proposed that DWI-FLAIR mismatch may be too stringent, and therefore miss individuals who could benefit from thrombolytic therapy [25]. In this work, we develop machine learning models to classify acute ischemic stroke patient TSS using MR imaging features. We proposed a deep learning model, which is based on an autoencoder architecture [18], to extract latent representative imaging features (deep features) from PWIs. We compared the performance of various models (stepwise multilinear regression, support vector machines, random forest, and gradient boosted regression tree) to classify TSS with and without the deep features, and determined the best model for classifying TSS. We also provided a visualization strategy to interpret the deep features, and correlate them to the input images.
3. Methods
3.1. Dataset
In a study approved by the UCLA institutional review board (IRB), clinical stroke data was transferred from our institution’s data repository into a REDCap [26] database. The database holds 1,059 acute stroke patients from 1992 to 2016 who have received at least one or more of the following revascularization treatments: IV tPA, IA tPA, or mechanical thrombectomy. The corresponding patient pre-treatment MR PWIs, apparent diffusion coefficient (ADC) images, DWIs and FLAIR images were obtained from the UCLA Medical Center picture archiving and communication system (PACS).
For this study, we define the following inclusion criteria: 1) patients must experience acute ischemic stroke due to middle cerebral artery (MCA) occlusion; 2) patients must have a recorded time for which the stroke symptoms are first observed; 3) patients must have a recorded time for which the first imaging is obtained before treatment; and 4) patients must have a complete imaging set of PWIs, DWIs, FLAIRs, and ADCs. Patients’ TSS was calculated by subtracting the time at which the stroke symptoms were first observed from the time at which the first imaging was obtained. We followed the existing DWI-FLAIR TSS classification task [23] to binarize the TSS into two classes: positive (1; <4.5hrs) and negative (0; ⩾4.5hrs). After applying the inclusion criteria, 105 patients were obtained (83 positive class; 22 negative class). The patient characteristics are summarized in Table 1. This cohort subset was used to build the models for TSS classification.
Table 1.
Acute Ischemic Stroke patient sub-cohort characteristics.
| Patients (n = 105) | ||
|---|---|---|
| Demographics | Age | 69.6±21.0 |
| Gender | 43 males | |
| Clinical Presentation | Time since stroke (continuous) | 158±108 mins |
| NIHSS† | 12.9±8.08 | |
| Atrial fibrillation | 1 (28); 0 (77) | |
| Hypertension | 1 (62); 0 (43) | |
| Prediction label | Time since stroke (binary) | <4.5hrs (83); ≥4.5hrs (22) |
NIHSS = NIH Stroke Scale International; scale: 0 (no stroke symptoms) - 42 (severe stroke)
3.2. Image Preprocessing
Intra-patient registration of pre-treatment PWI, DWI, ADC and FLAIR images was performed with a six degree of freedom rigid transformation using FMRIB’s Linear Image Registration Tool (FLIRT) [27]. Gaussian filters were applied to remove spatial noise and a multi-atlas skull-stripping algorithm [28] was used to remove skulls. Different tissue type masks (e.g., cerebrospinal fluid (CSF), gray/white matter) were identified using Statistical Parametric Mapping 12 (SPM12) [29] and CSF was excluded from this analysis. The sparse perfusion deconvolution toolbox (SPD) [30] and the ASIST-Japan Perfusion mismatch analyzer (PMA) were used to perform perfusion parameter map generation and arterial input function (AIF) identification (see Section 3.3.2.2).
3.3. Feature Generation
3.3.1. Baseline MR imaging feature
PWIs are spatio-temporal imaging data (4D) that show the flow of a gadolinium-based contrast bolus into and out of the brain over time. They contain concentration time curves (CTCs) for each brain voxel that describe the flow of the contrast (i.e. signal intensity change) over time. Perfusion parameter maps [31] can be derived from PWIs that describe the tissue perfusion characteristics, including cerebral blood volume (CBV), cerebral blood flow (CBF), mean transit time (MTT), time-to-peak (TTP), and time-to-maximum (Tmax). Briefly, CBV describes the total volume of flowing blood in a given volume of a voxel and CBF describes the rate of blood delivery to the brain tissue within a volume of a voxel. By the Central Volume Theorem, CBV and CBF can be used to derive MTT, which represents the average time it takes the contrast to travel through the tissue volume of a voxel. TTP is the time required for the CTC to reach its maximum, which approximates the time needed for the bolus to arrive at the voxel with delay caused by brain vessel structure. Tmax is the time point where the contrast residue function reaches its maximum, which approximates the true time needed for the bolus to arrive at the voxel.
Intensity features (e.g., DWI voxel intensity, CBF voxel value) are often generated for voxel-wise stroke tissue outcome prediction [32]. Yet, generating intensity features based on entire brain MR images may be less descriptive to the stroke pathophysiology and less predictive of TSS because often stroke occurs in only one cerebral hemisphere. Therefore, we generated the imaging features only within regions that have Tmax>6s [33], which capture both the dead tissue core and the salvageable tissue that can possibly be saved by treatments. Feature generation involves two steps: 1) perfusion parameter maps were calculated using the SPD toolbox [30], and the region of interest was defined by Tmax>6s; and 2) the average intensity value was calculated within the region of interest for each image (DWI, ADC, FLAIR, CBF, CBV, TTP, and MTT), resulting in a set of data with seven intensity features. All the features were then standardized independently to zero mean with a standard deviation of 1. These baseline imaging features were used in building the classifiers for TSS classification.
3.3.2. Deep feature Generation
3.3.2.1. Deep Autoencoder (AE)
We hypothesized that a deep learning approach can automatically learn feature detectors to extract latent features from PWIs that can improve TSS classification. We therefore implemented a deep autoencoder (deep AE) that is based on a stacked autoencoder [18] to learn the hidden features from PWIs (Figure 2). Each PWI voxel CTC, with a size of 1 x t (t = time for perfusion imaging), is transformed by the deep AE into k new feature representations that can represent complex voxel perfusion characteristics. The learning of these features is automatic and it is achieved by the hierarchical feature detectors, which are sets of weights that are learned in training via backpropagation. The deep AE consists of an encoder and a decoder. The encoder consists of two components: 1) an input layer; and 2) fully-connected layers, in which input neurons are fully-connected to each previous layer’s output neuron. The encoder is connected to the decoder, which follows reversely the same layer patterns of the encoder. The encoder output (i.e., the middle layer output of the deep AE) is the k new feature representations that can be used for TSS classification (in this work, k = 4).
Figure 2.

Deep AE feature generation. Training patches (with a size of 3 x 3 x 64) were randomly generated from PWIs. Each patch was coupled with its AIF (obtained from PMA toolbox) and the combined matrix was unrolled into a 1D vector that would be fed into the deep network. The proposed deep AE consisted of an encoder and decoder, in which the encoder output would be the new compact representation for the input. The encoder outputs of all PWI voxels were aggregated into the final four AE feature activation maps. A region of interest mask (Tmax>6s) was then applied to the new feature maps to generate the mean intensity values (i.e., deep features).
The proposed network is trained via an unsupervised learning procedure in which the decoder output is the reconstruction of the encoder input. The network is optimized to obtain weights, Θ, that minimize the binary cross entropy loss between the input, I, and the reconstructed output, Î(Θ), across the samples with size n [34]:
| (Eq. 1) |
3.3.2.2. Training data generation
As previous work suggests [35], regional information corresponding to a voxel’s surroundings can improve classification in MR images. Therefore, a small region was included in each training voxel, leading to a size of 3 x 3 x t patch (width x height x time; the z-dimension is omitted; t = 64 in our dataset), where the center of the patch is the voxel of interest for the deep AE feature learning. Each training patch was also coupled with its corresponding arterial input function patch [36], which describes the contrast agent input to the tissue in a single voxel, to improve the learning of hidden features. Each training patch was unrolled into a 1D vector, leading to a size of 1152x1. The 1D data were used to train the deep AE. In total, 105,000 training data were generated by sampling randomly and equally from all the patient PWIs.
3.3.2.3. Deep AE Configuration and Implementation
We observed that standard batch gradient descent did not lead to a good convergence of the deep AE during training. We suspect that this may be due to an inappropriate learning rate (default: 0.01), which typically requires careful tuning. Therefore, we optimized the deep AE using Adam, which computes adaptive learning rates during training and has demonstrated superior performance over other methods [37]. An early-stopping strategy was applied to improve the learning of deep AE weights and prevent overfitting, where the training would be terminated if the performance did not improve over five consecutive epochs (max number of training epochs: 50). The deep AE was implemented in Torch7 [34], and the training was done on two NVIDIA Titan X GPUs and an NVIDIA Tesla K40 GPU. We explored different architectures of the deep AE, including different numbers of encoder hidden layers (from 1-3) and different numbers of hidden units (factor of 2, 4, and 6). Ten-fold patient-based cross-validation was performed to determine the optimal architecture for the deep AE (with an input size of 1152×1 and k = 4 new feature representations). Once the deep AE was trained, we used it to learn four new AE feature maps from each patients’ PWIs by aggregating the deep AE encoder output of all voxels. Then, average intensity values from AE feature maps (denoted as deep features) were generated in the regions of interest following the same procedure as described in Section 3.2.1.
3.4. Machine Learning models for TSS classification
We constructed and compared the performance of four machine learning methods for TSS classification: stepwise multilinear regression (SMR), support vector machine (SVM), random forest (RF), and gradient boosted regression tree (GBRT). Briefly, SMR is a stepwise method for adding and removing features from a multilinear model based on their statistical significance (e.g., F-statistics) to improve model performance [38]. SVM is a supervised learning classification algorithm that constructs a hyperplane (or set of hyperplanes) in a higher dimensional space for classification [39]. RF is an ensemble learning method in which a multitude of decision trees are randomly constructed and the classification is based on the mode of the classes output by individual trees [40]. GBRT is an ensemble learning method similar to RF, in which a multitude of decision trees are randomly generated, yet these trees are added to the model in a stage-wise fashion based on their contribution to the objective function optimization [41].
Different machine learning methods may not perform equally on the same feature set. Also, different model hyperparameter (e.g., a SVM’s hyperparameter, C) contribute differently to the classification. Evaluating model performance without hyperparameter tuning may lead to decreased predictive power due to over-fitting, especially on small and imbalanced datasets. Therefore, we performed leave-one-patient-out validation for evaluation, with a nested crossvalidation for tuning model hyperparameters, following the proposed method [42]. A feature selection method, stability selection [43], was also applied to select the optimal feature subset before cross-validation to determine the best features for modeling (except for SMR because it has built-in feature selection method). This feature selection method produces a fairer feature comparison by aggregating different feature selection results from random subsampling of data and feature subsets. An overview of steps is shown in Figure 3. The SVM and RF were developed using the Python Scikit-learn library [44]. The SMR and GBRT were developed using MATLAB and the XGBoost library [45] respectively.
Figure 3.

An overview of steps to predict TSS (<4.5 hrs). The SPD toolbox was used to generate perfusion parameter maps (e.g., Tmax) from the PWIs. A region of interest mask was defined on Tmax>6s region. Then, the mask was applied to the perfusion maps and MR images (DWI, ADC, and FLAIR) to generate average intensity values. A total of seven baseline average intensity features were used to train the classifiers to predict TSS<4.5hrs. Classifier performances (with and without the addition of deep features) were compared.
4. Results and Discussion
4.1. Deep AE training
The optimal model architecture for the proposed deep AE is 1152-192-4-4-192-1152, with an average mean square error (MSE) of 0.675 ± 0.246 (average deep AEs MSE is 1.29 ± 0.755). The small MSE indicates the reconstruction of input signal is efficient with the encoder-decoder structure, and the encoder output is a compact representation for the input. Figure 4 shows the MSE along epoch of each fold for the optimal deep AE. All the models across folds converged within first ~10 epochs, with minimal weight adjustments in the following epochs as indicated by small changes in the MSE. Most of the models were stopped within 25 epochs (except the model in fold 9).
Figure 4.
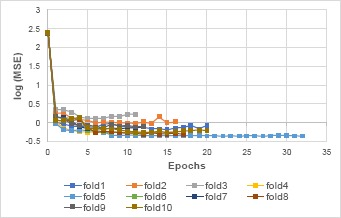
MSE vs. epoch for each fold of the validation for the optimal deep AE (in logarithmic scale for better visualization). All the models across folds converged within ~10 epochs, with minimal adjustments in the following epochs as indicated by small MSE change.
4.2. TSS classification
Leave-one-patient-out validation (see Section 3.3) was performed to evaluate each classifier. The model performance was measured via AUC and model bias via F1-score (Table 2). Youden’s index [46] was used to determine optimal receiver operating characteristic curve (ROC) cutoff points, which were used to calculate the F1-score, sensitivity, specificity, true predictive value (TPV), and negative predictive value (NPV) (Table 3).
Table 2.
The AUC and F1-score of different classifiers on predicting TSS with baseline imaging features and with/without deep features. B (baseline features), B+AE (baseline and deep features).
| AUC | F1-score | |||
|---|---|---|---|---|
| Models | B | B+AE | B | B+AE |
| (1) SMR | 0.570 | 0.683 | 0.608 | 0.765 |
| (2) SVM | 0.470 | 0.640 | 0.632 | 0.859 |
| (3) RF | 0.529 | 0.651 | 0.847 | 0.818 |
| (4) GBRT | 0.526 | 0.623 | 0.862 | 0.681 |
Table 3.
The sensitivity, specificity, TPV, and NPV of different classifiers on predicting TSS with baseline imaging features and with/without deep features. B (baseline features), B+AE (baseline and deep features).
| sensitivity | specificity | TPV | NPV | |||||
|---|---|---|---|---|---|---|---|---|
| Models | B | B+AE | B | B+AE | B | B+AE | B | B+AE |
| (1) SMR | 0.458 | 0.687 | 0.818 | 0.591 | 0.905 | 0.864 | 0.286 | 0.333 |
| (2) SVM | 0.506 | 0.880 | 0.636 | 0.364 | 0.840 | 0.839 | 0.255 | 0.444 |
| (3) RF | 0.857 | 0.750 | 0.333 | 0.667 | 0.837 | 0.900 | 0.368 | 0.400 |
| (4) GBRT | 0.893 | 0.560 | 0.286 | 0.667 | 0.833 | 0.870 | 0.400 | 0.275 |
With the additional deep features, all classifiers (SMR, SVM, RF, and GBRT) showed improvement in AUC. The increase (~10%) of AUC after adding the deep features demonstrated the usefulness of these deep features and their association with the TSS. Figure 5 shows the ROCs of the classifiers trained with the baseline features and the deep features, with a reference ROC based on the DWI-FLAIR mismatch method is shown for comparison [23]. All the classifiers generally performed better with the addition of the deep features. These classifiers performed better than AUC=0.60 (as compared to reference AUC of 0.58), demonstrating the ability of using imaging features with machine learning models to classify TSS. Among all the classifiers, the SMR trained with baseline imaging features and the deep features performed the best, with an AUC of 0.683. Comparing to the reference mismatch method, SMR achieved higher sensitivity (0.69 vs 0.55) and NPV (0.33 vs 0.19) while maintaining similar specificity (0.59 vs 0.60) and TPV (0.86 vs 0.88). Therefore, SMR was determined to be the most suitable classifier for TSS classification.
Figure 5.
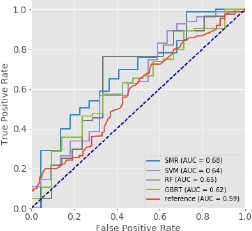
The ROCs of different classifiers trained with both the baseline imaging features and the deep features for TSS prediction. A reference ROC [23] based on the DWI-FLAIR mismatch method is included for comparison. All classifiers demonstrate higher AUC as compared to the reference. The SMR model performed the best with an AUC of 0.68). This result suggests that machine learning models trained with imaging features can be used to better predict TSS.
We observed that the models trained with only the baseline imaging features had low performance. The best model was the SMR with an AUC of 0.570, and other models had lower AUCs (<0.55). This may be due to the insufficient baseline features for classifier construction (i.e., only mean intensity features across MR images and perfusion maps were used). In future work, feature generation techniques, such as descriptive statistics, will be investigated to generate more features for TSS classification. Although these models were less predictive, models trained with additional deep features showed significant improvement in TSS classification. This supports our hypothesis that PWIs contain information encoding TSS, and that the proposed deep AE extracted hidden features in PWIs are predictive of TSS.
4.3. Deep Feature Visualization
To understand what the deep AE learned to extract in the encoder output layer, we applied the visualization technique, top-m selection [47], on the encoder output layer and correlated the results to the four deep AE activation maps (denoted as ae1, ae2, ae3, and ae4). Briefly, every input CTC will cause a hidden neuron unit to output a value (i.e., activation). Some input CTCs will give a higher activation while other inputs will give a lower activation. High activation values indicate the presence of relevant features in the input (e.g., wide and high peak) that can “excite” the hidden units [47]. For each hidden unit of the optimal deep AE encode layer, we performed top-m selection to obtain the top m signals (x*) from the training data that cause the most activations:
| (Eq. 2) |
where Sort(.) is a descending operation, and hi (θ, x) is the activation of the ith hidden unit. Figure 6 shows the four deep AE activation maps and their corresponding average curves of the top-50 (i.e., m=50) input CTCs. Our results show that different hidden units appear to capture different type of input signals. For example, the ae1 map shows higher activations (brighter) in the acute stroke region (left brain; high Tmax), and the corresponding average top-50 CTC (blue curve) has delayed and low concentration, which matches the visualization (that is, the hidden unit detects the stroke-affected CTCs). In contrast, the ae3 map shows low activations (darker) in the acute stroke region and the corresponding average top-50 CTC (red curve) has early sharp and high peak. We also calculated the Pearson correlation coefficient between the deep features generated from these activation maps and TSS. The ae3 deep feature showed statistically significant correlation with TSS (p-value<0.05). These visualization and correlation results demonstrated that the learned features from the optimal deep AE contained information that was predictive of TSS.
Figure 6.
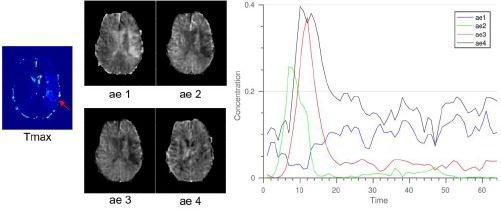
Visualization of the four deep AE activation feature maps from the encoder output layer and the corresponding average curve of the top-50 input CTCs. Based on visual interpretation, different hidden units captured different type of input CTCs. For example, hidden unit 1 (for ae1) captured CTCs with delayed and low concentration (red curves) as indicated by higher activations (brighter) in the acute stroke region (high Tmax; red arrow).
Understanding deep learning representations is challenging because it requires making sense of non-linear computations performed over many network weights [48]. Our visualization result is the first step to attempt to understand what the deep AE is learning. This is important for using deep learning model in medical image analysis because deep learning is often a “black-box” approach that yields superior, but hard-to-interpret results. However, the visualization result in this work is not conclusive. Further research is required to understand what the deep network is learning. One next step we plan to pursue is to apply different visualization techniques (e.g., deconvolution [49]) on the learned networks and perform statistical tests to draw correlations between them to an observation (e.g., small TSS).
5. Conclusion and Future work
In this paper, we showed that SMR, SVM, RF, and GBRT models were able to classify TSS, with SMR achieving the highest AUC. We proposed a deep AE architecture to extract representative features from PWIs and showed that adding deep features boosted the classifiers’ performance, showing the potential application of deep learning feature extraction techniques in TSS classification. In addition, we utilized a visualization method to interpret the features learned in the deep AE and discussed the possible research opportunity in understanding deep learning models for medical images.
We now discuss a few limitations and areas of future work. First, there are roughly 1,100 patients available in our dataset, but the majority of them were missing one or more MR images and were therefore not included in this analysis. Our next step will be looking into multimodal and denoising deep learning frameworks that are capable of handling missing data. Second, we will explore different feature generation techniques (e.g., descriptive and histogram statistics) to enlarge the feature set for training the classifiers. Third, we plan to explore several visualization techniques, such as deconvolution [49] and gated backpropagation [50] in order to understand the deep AE’s features and to draw both visual and statistical correlations to TSS classification. Finally, TSS<4.5hrs is the current clinical cutoff time for IV tPA treatment, yet this may not be an absolute time point in which a stroke patient can benefit from treatment because of changing brain pathophysiology [51]. We therefore plan to investigate and extend our models to more classes (e.g., a ±0.5hr boundary), rather than just TSS<4.5hrs/TSS≥4.5hrs.
6 Acknowledgements
This research was supported by National Institutes of Health (NIH) Grant R01 NS076534, UCLA Radiology Department Exploratory Research Grant 16-0003, and an NVIDIA Academic Hardware Grant.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, Amett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics-2016 update a report from the American Heart Association. Circulation. 2016;133:38–48. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 2.Moradiya Y, Janjua N. Presentation and outcomes of “wake-up strokes” in a large randomized stroke trial: analysis of data from the International Stroke Trial. J Stroke Cerebrovasc Dis. 2013;22(8):e286–e292. doi: 10.1016/j.jstrokecerebrovasdis.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 3.Counsell C, Dennis M, McDowall M, Warlow C. Predicting outcome after acute and subacute stroke: Development and validation of new prognostic models. Stroke. 2002;33(4):1041–7. doi: 10.1161/hs0402.105909. [DOI] [PubMed] [Google Scholar]
- 4.Ho KC, Speier W, El-Saden S, Liebeskind DS, Saver JL, Bui AAT, et al. Predicting discharge mortality after acute ischemic stroke using balanced data. AMIA Annu Symp Proc. 2014 Jan;2014:1787–96. [PMC free article] [PubMed] [Google Scholar]
- 5.Vogt G, Laage R, Shuaib A, Schneider A. Initial lesion volume is an independent predictor of clinical stroke outcome at day 90: An analysis of the Virtual International Stroke Trials Archive (VISTA) database. Stroke. 2012;43(5):1266–72. doi: 10.1161/STROKEAHA.111.646570. [DOI] [PubMed] [Google Scholar]
- 6.Stribian D, Meretoja A, Ahlhelm F, Pitkaniemi J, Lyrer P, Kaste M, et al. Predicting outcome of IV thrombolysis-treated ischemic stroke patients: the DRAGON score. Stroke. 2012;78(6):427–32. doi: 10.1212/WNL.0b013e318245d2a9. [DOI] [PubMed] [Google Scholar]
- 7.Thomalla G, Cheng B, Ebinger M, Hao Q, Tourdias T, Wu O, et al. DWI-FLAIR mismatch for the identification of patients with acute ischaemic stroke within 4.5 h of symptom onset (PRE-FLAIR): A multicentre observational study. Lancet Neurol. 2011;10(11):978–86. doi: 10.1016/S1474-4422(11)70192-2. [DOI] [PubMed] [Google Scholar]
- 8.Thomalla G, Fiebach JB, Østergaard L, Pedraza S, Thijs V, Nighoghossian N, et al. A multicenter, randomized, double-blind, placebo-controlled trial to test efficacy and safety of magnetic resonance imaging-based thrombolysis in wake-up stroke (WAKE-UP) Int J Stroke. 2014;9(6):829–36. doi: 10.1111/ijs.12011. [DOI] [PubMed] [Google Scholar]
- 9.Buck D, Shaw LC, Price CI, Ford GA. Reperfusion therapies for wake-up stroke: systematic review. Stroke. 2014 Jun;45(6):1869–75. doi: 10.1161/STROKEAHA.114.005126. [DOI] [PubMed] [Google Scholar]
- 10.Thomalla G, Rossbach P, Rosenkranz M, Siemonsen S, Krützelmann A, Fiehler J, et al. Negative fluid-attenuated inversion recovery imaging identifies acute ischemic stroke at 3 hours or less. Ann Neurol. 2009;65(6):724–32. doi: 10.1002/ana.21651. [DOI] [PubMed] [Google Scholar]
- 11.Ziegler A, Ebinger M, Fiebach JB, Audebert HJ, Leistner S. Judgment of FLAIR signal change in DWI-FLAIR mismatch determination is a challenge to clinicians. J Neurol. 2012;259(5):971–3. doi: 10.1007/s00415-011-6284-6. [DOI] [PubMed] [Google Scholar]
- 12.Galinovic I, Puig J, Neeb L, Guibernau J, Kemmling A, Siemonsen S, et al. Visual and region of interest-based inter-rater agreement in the assessment of the diffusion-weighted imaging-fluid-attenuated inversion recovery mismatch. Stroke. 2014;45(4):1170–2. doi: 10.1161/STROKEAHA.113.002661. [DOI] [PubMed] [Google Scholar]
- 13.Thomalla G, Gerloff C. Treatment Concepts for Wake-Up Stroke and Stroke with Unknown Time of Symptom Onset. Stroke. 2015;46(9):2707–13. doi: 10.1161/STROKEAHA.115.009701. [DOI] [PubMed] [Google Scholar]
- 14.Bishop CM. Pattern recognition. Mach Learn. 2006;128:1–58. [Google Scholar]
- 15.LeCun Y, Bengio Y, Hinton G. Deep learning. [cited 2016 Jun 9];Nature [Internet] 2015 521(7553):436–44. doi: 10.1038/nature14539. Available from: http://www.nature.com/nature/journal/v521/n7553/abs/nature14539.html. [DOI] [PubMed] [Google Scholar]
- 16.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2014 Sep;115(3):43. [Google Scholar]
- 17.Karpathy A, Leung T. Large-scale Video Classification with Convolutional Neural Networks. Proc 2014 IEEE Conf Comput Vis Pattern Recognit. 2014:1725–32. [Google Scholar]
- 18.Vincent P, Larochelle H, Lajoie I, Bengio Y, Manzagol P-A. Stacked Denoising Autoencoders: Learning Useful Representations in a Deep Network with a Local Denoising Criterion. J Mach Learn Res. 2010 Mar;11(3):3371–408. [Google Scholar]
- 19.Aoki J, Kimura K, Iguchi Y, Shibazaki K, Sakai K, Iwanaga T. FLAIR can estimate the onset time in acute ischemic stroke patients. J Neurol Sci. 2010;293(1-2):39–44. doi: 10.1016/j.jns.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Petkova M, Rodrigo S, Lamy C, Oppenheim G, Touzé E, Mas J-L, et al. MR Imaging Helps Predict Time from Symptom Onset in Patients with Acute Stroke: Implications for Patients with Unknown Onset Time. Radiology. 2010;257(3):782–92. doi: 10.1148/radiol.10100461. [DOI] [PubMed] [Google Scholar]
- 21.Ebinger M, Galinovic I, Rozanski M, Brunecker P, Endres M, Fiebach JB. Fluid-attenuated inversion recovery evolution within 12 hours from stroke onset: A reliable tissue clock? Stroke. 2010;41(2):250–5. doi: 10.1161/STROKEAHA.109.568410. [DOI] [PubMed] [Google Scholar]
- 22.Thomalla G, Cheng B, Ebinger M, Hao Q, Tourdias T, Wu O, et al. DWI-FLAIR mismatch for the identifi cation of patients with acute ischaemic stroke within 4 · 5 h of symptom onset (PRE-FLAIR): a multicentre observational study. 2011;10(November) doi: 10.1016/S1474-4422(11)70192-2. [DOI] [PubMed] [Google Scholar]
- 23.Emeriau S, Serre I, Toubas O, Pombourcq F, Oppenheim C, Pierot L. Can Diffusion-Weighted Imaging-Fluid-Attenuated Inversion Recovery Mismatch (Positive Diffusion-Weighted Imaging/Negative Fluid-Attenuated Inversion Recovery) at 3 Tesla Identify Patients With Stroke at < 4.5 Hours? Stroke. 2013;44(6):1647–51. doi: 10.1161/STROKEAHA.113.001001. [DOI] [PubMed] [Google Scholar]
- 24.Schwamm L. MR WITNESS: A Study of Intravenous Thrombolysis With Alteplase in MRI-Selected Patients (MR WITNESS) 2011 ClinicalTrials.gov. [Google Scholar]
- 25.Odland A, Særvoll P, Advani R, Kurz MW, Kurz KD. Are the current MRI criteria using the DWI-FLAIR mismatch concept for selection of patients with wake-up stroke to thrombolysis excluding too many patients? Scand J Trauma Resusc Emerg Med. 2015;23:22. doi: 10.1186/s13049-015-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris P a., Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research Electronic Data Capture (REDCap) - A metadata driven methodology and workflow process for providing translational research informatict support. J Biomed Inform. 2009;42(2):377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(SUPPL. 1):208–19. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 28.Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C. Multi-Atlas Skull-Stripping. Acad Radiol. 2013;20(12):1566–76. doi: 10.1016/j.acra.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner J, Barnes G, Chen C, Daunizeau J, Flandin G, Friston K, et al. SPM12 Manual The FIL Methods Group (and honorary members) 2014 [Google Scholar]
- 30.Fang R, Chen T, Sanelli PC. Towards robust deconvolution of low-dose perfusion CT : Sparse perfusion deconvolution using online dictionary learning. Med Image Anal. 2013;17(4):417–28. doi: 10.1016/j.media.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho KC, Scalzo F, Sarma VK, EL-Saden S, Arnold WC. A Temporal Deep Learning Approach for MR Perfusion Parameter Estimation in Stroke. In: International Conference of Pattern Recognition. 2016 [Google Scholar]
- 32.Wu O, Koroshetz WJ, Ostergaard L, Buonanno FS, Copen W a, Gonzalez RG, et al. Predicting tissue outcome in acute human cerebral ischemia using combined diffusion- and perfusion-weighted MR imaging. Stroke. 2001;32:933–42. doi: 10.1161/01.str.32.4.933. [DOI] [PubMed] [Google Scholar]
- 33.Olivot JM, Mlynash M, Thijs VN, Kemp S, Lansberg MG, Wechsler L, et al. Optimal tmax threshold for predicting penumbral tissue in acute stroke. Stroke. 2009;40(2):469–75. doi: 10.1161/STROKEAHA.108.526954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collobert R, Kavukcuoglu K, Farabet C. Torch7: A Matlab-like Environment for Machine Learning. BigLearn, NIPS Work. 2011:1–6. [Google Scholar]
- 35.Scalzo F, Hao Q, Alger JR, Hu X, Liebeskind DS. Regional prediction of tissue fate in acute ischemic stroke. Ann Biomed Eng. 2012;40(10):2177–87. doi: 10.1007/s10439-012-0591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calamante F. Arterial input function in perfusion MRI: A comprehensive review. Prog Nucl Magn Reson Spectrosc. 2013;74:1–32. doi: 10.1016/j.pnmrs.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Kingma DP, Ba JL. Adam: a Method for Stochastic Optimization. Int Conf Learn Represent 2015. 2015:1–15. [Google Scholar]
- 38.Draper NR, Smith H. John Wiley & Sons; 2014. Applied regression analysis. [Google Scholar]
- 39.Cortes C, Vapnik V. Support-vector networks. [cited 2016 Jun 8];Mach Learn [Internet] 1995 Sep;20(3):273–97. Available from: http://link.springer.com/10.1007/BF00994018. [Google Scholar]
- 40.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 41.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001:1189–232. [Google Scholar]
- 42.Krstajic D, Buturovic LJ, Leahy DE, Thomas S. Cross-validation pitfalls when selecting and assessing regression and classification models. 2014 doi: 10.1186/1758-2946-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meinshausen N, Bühlmann P. Stability selection. J R Stat Soc Ser B (Statistical Methodol. 2010;72(4):417–73. [Google Scholar]
- 44.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12(Oct):2825–30. [Google Scholar]
- 45.Chen T, Guestrin C. XGBoost: Reliable Large-scale Tree Boosting System. arXiv. 2016:1–6. [Google Scholar]
- 46.Fluss R, Faraggi D, Reiser B. Estimation of the Youden Index and its associated cutoff point. Biometrical J. 2005;47(4):458–72. doi: 10.1002/bimj.200410135. [DOI] [PubMed] [Google Scholar]
- 47.Erhan D, Bengio Y, Courville A, Vincent P. Visualizing higher-layer features of a deep network. Univ Montr. 2009;1341:3. [Google Scholar]
- 48.Li Y, Yosinski J, Clune J, Lipson H, Hopcroft J. Convergent Learning: Do different neural networks learn the same representations? Iclr. 2016;(2014):1–21. [Google Scholar]
- 49.Zeiler MD, Fergus R. In: Computer vision--ECCV 2014. Springer; 2014. Visualizing and understanding convolutional networks; pp. 818–33. [Google Scholar]
- 50.Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M. Striving for Simplicity: The All Convolutional Net. Iclr. 2015:1–14. [Google Scholar]
- 51.Furlan AJ. Endovascular Therapy for Stroke — It’s about Time. N Engl J Med. 2015:1–3. doi: 10.1056/NEJMe1503217. [DOI] [PubMed] [Google Scholar]


