Abstract
A major challenge in using electronic health record repositories for research is the difficulty matching subject eligibility criteria to query capabilities of the repositories. We propose categories for study criteria corresponding to the effort needed for querying those criteria: “easy” (supporting automated queries), mixed (initial automated querying with manual review), “hard” (fully manual record review), and “impossible” or “point of enrollment” (not typically in health repositories). We obtained a sample of 292 criteria from 20 studies from ClinicalTrials.gov. Six independent reviewers, three each from two academic research institutions, rated criteria according to our four types. We observed high interrater reliability both within and between institutions. The analysis demonstrated typical features of criteria that map with varying levels of difficulty to repositories. We propose using these features to improve enrollment workflow through more standardized study criteria, self-service repository queries, and analyst-mediated retrievals.
Introduction
There is a long history of using paper-based health records to identify potential research subjects. The advent of electronic health records (EHRs) has greatly facilitated researchers’ access to relevant patient data, especially when data are transferred to specialized data repositories or warehouses.1 Researchers may use such information for estimating the availability of eligible research subjects in some larger target population (cohort estimation), identifying potential patients for enrollment in a research study (cohort identification), or finding patients whose existing data (whether in summary or detailed form) can be used to explore research questions (data reuse).2 What most such endeavors have in common is a defined, study-specific set of patient characteristics, known as eligibility criteria. These criteria include those that must be present for a patient to be included as a study subject (inclusion criteria) or render the patient unsuitable for inclusion in the study (exclusion criteria).3 The use of EHR data for subject identification (for any purpose) will typically begin with querying a repository to find patients who meet inclusion criteria and then remove those patients with exclusion criteria.
Ideally, a researcher can access a repository in “self-service” mode to complete the search and retrieval processes independently. However, the required queries are often too complex for this approach and may require the assistance of a data analyst who is familiar with the data and tools and can mediate between the researcher and the repository.3,4 Even then, the process may be challenging. Eligibility criteria may be expressed in complex arrangements that are not amenable to automated searching, or they may require data that are either not accessible through query tools or are not recorded in the health record.5 Studies of matching enrollment criteria using EHR repositories have demonstrated success rates ranging from 23%6 to 44%5 due to requirements for temporal restrictions, calculated conditions, or inaccurate medical diagnostic coding.
For example, a study might seek patients with diabetes mellitus on escalating doses of insulin who arrive in the emergency room with elevated blood glucose levels but will be ineligible if they receive treatment prior to enrollment in the study. In this hypothetical case, the researcher or analyst can automatically identify patients with diabetes mellitus who have received insulin previously but can only determine whether they have received escalating doses through complex queries or manual analysis of records of patients who appear to be at least minimally eligible. Finding patients through the repository breaks down completely when a study requires patients who present to the emergency room with new and acute problems, since repository data are only retrospective.
Subject recruitment is further complicated because not only must potential subjects be identified immediately, but they must be enrolled before receiving treatment. It is unreasonable and unethical to expect clinicians to interrupt patient care and delay potentially lifesaving treatment in acute settings. Researchers with such complex, time-sensitive eligibility criteria must often resort to educating front-line health professionals (or posting paper reminders), hoping that clinicians will remember in time to enroll their patients as study subjects. Automated alerts tied to EHRs can be used for simple eligibility criteria7 but will be difficult to implement in more complex situations that are challenging, even for experienced data analysts.
One solution mimics the practice of using an initial screening protocol in which a large set of potentially eligible patients is identified, and each individual record is examined subsequently against more specific criteria. For the EHR repository version of this method, the researcher or analyst performs an initial query to identify patient records that meet some criteria and then reviews full records manually for the remaining criteria, perhaps flagging patients who might be eligible if they appear in care settings (such as the hypothetical patients with diabetes above) for further evaluation. This multi-step process can be facilitated by characterizing eligibility criteria in advance as either being amenable to retrieval with repository tools (“easy”), requiring some more elaborate mechanism (“hard”), or a combination of the two (“mixed”). If a researcher specifies criteria in this manner in a research protocol, repository users can then perform an initial retrieval step (especially when the user is an analyst rather than the researcher) to identify an initial set of patients. This cohort then passes on to the next stage in the process for application of additional study criteria through other means.
The categorization of study criteria by level of difficulty has been previously demonstrated. Three reviewers from a single research institution with combined expertise in clinical medicine, research, EHRs, and querying databases for study cohorts demonstrated high agreement when assigning levels of difficulty to study criteria.8 We have now expanded the original study to include a similar set of expert reviewers from a second research institution. The purpose of this follow-on study is to further characterize the use of these categories and determine if the prior findings are generalizable. Demonstrating inter-institutional reliability and examining differences in categorization between institutions are important steps in determining the overall feasibility of our proposed phased approach for identifying study cohorts.
Methods
We obtained the ClinicalTrials.gov (NCT) identifiers for a convenience sample of ten studies conducted at the University of Alabama at Birmingham (UAB) that were chosen for a separate study of the use of EHR data for cohort prediction (unpublished data). We then incremented each NCT ID successively until the next ID in sequence matched another study in ClinicalTrials.gov. For example, the NCT ID NCT01098981 would be incremented to NCT01098994 to find the next additional study. These twenty studies provided the eligibility criteria that served as the data set for our study (Table 1). The NCT record for each study was examined to identify its eligibility criteria (Figure 1) which were manually copied from a Web browser into a spreadsheet. Unnecessary words were removed from each criterion to make it easier to identify those that were essentially redundant. Duplicate criteria were then removed, and the remaining criteria served as the set to be reviewed for the study.
Table 1.
Research descriptions from ClinicalTrials.gov that were selected for this study. Studies were paired to include ten UAB studies and ten studies that followed them sequentially in the ClinicalTrials.gov database.
| NCT ID | Study Title | URL |
|---|---|---|
| 01098981 | Phase 3, Randomized, Placebo-Controlled, Double-Blinded Trial of the Combined Lysis of Thrombus With Ultrasound and Systemic Tissue Plasminogen Activator (tPA) for Emergent Revascularization in Acute Ischemic Stroke (CLOTBUST-ER) | https://clinicaltrials.gov/ct2/show/NCT01098981 |
| 01098994 | Haptoglobin Phenotype, Vitamin E and High-density Lipoprotein (HDL) Function in Type 1 Diabetes (HAP-E) | https://clinicaltrials.gov/ct2/show/NCT01098994 |
| 01382212 | A Study to Evaluate the Safety of Paricalcitol Capsules in Pediatric Subjects Ages 10 to 16 With Stage 5 Chronic Kidney Disease Receiving Peritoneal Dialysis | |
| 01382225 | Sodium Hyaluronate Ophthalmic Solution, 0.18% for Treatment of Dry Eye Syndrome | https://clinicaltrials.gov/ct2/show/NCT01382225 |
| 01797445 | Study to Evaluate the Safety and Efficacy of E/C/F/TAF (GenvoyaÂ) Versus E/C/F/TDF (StribildÂ) in HIV-1Positive, Antiretroviral Treatment-Naive Adults https://clinicaltrials.gov/ct2/show/NCT01797445 | https://clinicaltrials.gov/ct2/show/NCT01797445 |
| 01797458 | European Study on Three Different Approaches to Managing Class 2 Cavities in Primary Teeth | https://clinicaltrials.gov/ct2/show/NCT0179745 |
| 02121795 | Switch Study to Evaluate F/TAF in HIV-1 Positive Participants Who Are Virologically Suppressed on Regimens Containing FTC/TDF | https://clinicaltrials.gov/ct2/show/NCT01098981 |
| 02121808 | EPO2-PV: Evaluation of Pre-Oxygenation Conditions in Morbidly Obese Volunteer: Effect of Position and Ventilation Mode (EPO2-PV) | https://clinicaltrials.gov/ct2/show/NCT02121808 |
| 01720446 | Trial to Evaluate Cardiovascular and Other Long-term Outcomes With Semaglutide in Subjects With Type 2 Diabetes (SUSTAINâ 6) | https://clinicaltrials.gov/ct2/show/NCT01720446 |
| 01720459 | Effects of Micronized Trans-resveratrol Treatment on Polycystic Ovary Syndrome (PCOS) Patients | https://clinicaltrials.gov/ct2/show/NCT01720459 |
| 01897233 | Study of Lumacaftor in Combination With Ivacaftor in Subjects 6 Through 11 Years of Age With Cystic Fibrosis, Homozygous for the F508del-CFTR Mutation | https://clinicaltrials.gov/ct2/show/NCT01897233 |
| 01897246 | Computer Assisted Planning of Corrective Osteotomy for Distal Radius Malunion | https://clinicaltrials.gov/ct2/show/NCT01098981 |
| 01713946 | A Placebo-controlled Study of Efficacy & Safety of 2 Trough-ranges of Everolimus as Adjunctive Therapy in Patients With Tuberous Sclerosis Complex (TSC) & Refractory Partial-onset Seizures (EXIST-3) | https://clinicaltrials.gov/ct2/show/NCT0171394 |
| 01713953 | Feasibility Study: Ulthera System for the Treatment of Axillary Hyperhidrosis | https://clinicaltrials.gov/ct2/show/NCT01713959 |
| 01567527 | Efficacy, Safety, and Tolerability of an Intramuscular Formulation of Aripiprazole (OPC-14597) as Maintenance Treatment in Bipolar I Patients | https://clinicaltrials.gov/ct2/show/NCT01567527 |
| 01567533 | Pilot Study Evaluating Safety of Sitagliptin Combined With Peg-IFN Alfa-2a + Ribavirin in Chronic Hepatitis C Patients | https://clinicaltrials.gov/ct2/show/NCT01567540 |
| 01936688 | A Study to Evaluate the Efficacy and Safety/Tolerability of Subcutaneous MK-3222 in Participants With Moderate-to-Severe Chronic Plaque Psoriasis (MK-3222-012) | https://clinicaltrials.gov/ct2/show/NCT01936688 |
| 01936701 | Comparison of the Effect of Hydrophobic Acrylic and Silicone 3-piece IOLs on Posterior Capsule Opacification | https://clinicaltrials.gov/ct2/show/NCT01936701 |
| 01833533 | A Study to Evaluate Chronic Hepatitis C Infection in Adults With Genotype 1a Infection (PEARL-IV) | https://clinicaltrials.gov/ct2/show/NCT01833533 |
| 01833546 | A Japanese Phase 1 Trial of TH-302 in Subjects With Solid Tumors and Pancreatic Cancer | https://clinicaltrials.gov/ct2/show/NCT01833546 |
Figure 1.
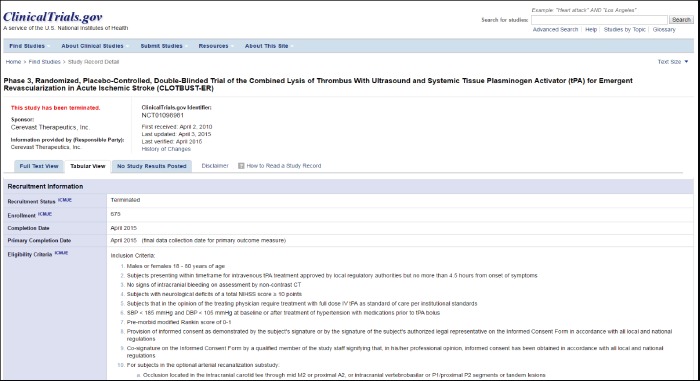
Sample ClinicalTrials.gov record, showing Eligibility Criteria in Tabular View. (Screen has been edited somewhat to allow inclusion of header and tabular information in the same view.)
The authors of this study served as experts for rating the eligibility criteria. We included three experts each from medical schools within two academic research institutions, University of Alabama School of Medicine (UASOM) and Northwestern University Feinberg School of Medicine (NU), to improve the generalizability of our findings and explore factors that might be institution- or repository-specific.
JJC is a physician who was the principle architect of the National Institutes of Health’s Biomedical Translational Research Information System (BTRIS)9 and has extensive experience using it to match patients to research criteria.10-13
WJL is a physician (formerly at UASOM) who is a research fellow with extensive experience with electronic health records and moderate experience with UASOM’s i2b2 (Informatics for Integrating Biology to the Bedside)14 repository.
MCW is a systems analyst architect with responsibility for and experience using both i2b2 and Powerlnsight (Cerner Corporation, Kansas City, MO), the clinical data warehouse attached to the EHR used by UASOM.
LVR is a clinical research associate at NU who leads the implementation of the i2b2 instance within the Northwestern Medical Enterprise Data Warehouse (NMEDW) and trains faculty and staff on its proper use.
AYW is a physician with extensive experience developing and implementing health information standards and moderate experience using EHRs and using the i2b2 repository within NMEDW.
DGF is a research fellow at NU with considerable experience researching and using cohort selection queries for clinical trial recruitment.
Each author reviewed the resulting criteria and rated them with an ordinal scale from one to four (Table 2) that was assessed whether a typical clinical data repository, such as i2b2, BTRIS or Powerlnsight, could identify study subjects matching each criterion based on information available in a typical EHR system. Given the complexity of some criteria, in which multiple clinical concepts or properties are represented together (e.g., classes of diagnoses, severity of disease in addition to presence of disease), ratings were assigned based on the highest level of difficulty. For example, if a criterion named a diagnosis and then specified its severity against a separate evaluation scale and a rater assigned 1 and 3 respectively for each sub-criterion, the overall rating for this item would be 3. Reviewers were instructed to leave the rating blank if there was not a clear answer. The ratings were compiled and summarized to determine the characteristics of criteria that could be readily used with EHR repositories versus those that could not.
Table 2.
Rating Scale for Eligibility Criteria. Each criterion was assessed by each rater for its suitability for retrieval from an electronic health record data repository.
| 1 – “Easy” – The criterion could be used easily to identify subjects in an EHR data repository, assuming the data were present, using the repository’s user interface |
| 2 – “Mixed” – The criterion could be partly retrieved using the repository’s user interface but then would require further manual review |
| 3 – “Hard” – Matching the criterion would require manual review of the record |
| 4 – “Impossible” “Time of enrollment” – The repository would not be likely to include sufficient information to match patients to the criterion, either because the source EHR would not be likely to contain the information or the repository would not include the information in a timely manner |
Since this study involved a fully crossed design, in which each criterion was rated by multiple coders using an ordinal coding system, we calculated the intra-class correlation score (ICC).15 This analysis was performed using R v3.3.1 and the irr package v0.84,16 with results integrated using StatTag v3.0.17 Additionally, we analyzed the number of “near agreement” results across institutions. We defined “near agreement” within an institution as a criterion in which two of the three ratings matched exactly, and the discrepant score was only offset by one category. Likewise, “near agreement” across all raters was classified as at least four of the six raters having matching scores, and the discrepant scores were only offset by one category each.
Results
Criteria Data Set
The 20 clinical trials used in this study are listed in Table 1. The ClinicalTrials.gov records for these twenty studies were cleaned to remove blank spaces, formatting, bullets, numbering, and other redundant text which did not affect meaning. Leading clauses with age and gender criteria (e.g., “women” in “women who are pregnant”) were removed, as these criteria are indicated separately in ClinicalTrials.gov and are already straightforward to retrieve. Other text, such as “with” in “with diabetes mellitus” and “taking” in “taking aspirin”, was removed to yield distinct concepts (e.g., diseases or medications). This process yielded a set of 301 criteria, of which 292 were found to be conceptually unique and were reviewed by each rater (1,752 ratings total). Example ratings are shown in Table 3.
Table 3.
Examples of criteria ratings. A, B and C are co-authors at UAB and NU; 1-4 correspond to categories from Table 2. * indicates criteria for which there was complete or near agreement, included in further analysis. Raters used "x" or "?" to indicate that the meaning of the criterion was unclear or that the categorization was unknown.
| UAB Raters | NU Raters | Criteria | ||||
|---|---|---|---|---|---|---|
| A | B | C | A | B | C | |
| 1 | 1 | 1 | 4 | 4 | 4 | Abnormal liver function as defined in the protocol at Screening * |
| 1 | 1 | 1 | 1 | 1 | 1 | absolute neutrophil count <1,000/mm3 * |
| 1 | 1 | 1 | 3 | 3 | 1 | allergy to starch powder or iodine. * |
| 1 | 1 | 1 | 1 | 1 | 1 | Anti-cancer treatment prior to trial entry * |
| 1 | 1 | 1 | 1 | 1 | 1 | BMI 40 - 80 kg / m2 * |
| 1 | 1 | 2 | 1 | 1 | 1 | digoxin within 6 months of starting treatment. * |
| 1 | 1 | 3 | 1 | 1 | 1 | immunosuppressants within 6 months of starting treatment |
| 1 | 1 | 3 | 1 | 1 | 1 | medication that is either moderate or strong inhibitor or inducer of cytochrome P450 (CYP)3A4 or is a sensitive substrate of other cytochrome P450 |
| 1 | 1 | 2 | 1 | 1 | 2 | <Anti-diabetic drug naïve, or treated with one or two oral antidiabetic drug (OADs), or treated with human Neutral Protamin Hagedorn (NPH) insulin or long-acting insulin analogue or pre-mixed insulin, both types of insulin either alone or in combination with one or two OADs |
| 1 | 2 | 2 | 2 | 2 | 2 | currently being treated for secondary hyperparathyroidism * |
| 2 | 3 | 1 | 2 | 2 | 4 | Chronic heart failure New York Heart Association (NYHA) class IV |
| 1 | 4 | x | 4 | 4 | 4 | For women, effective contraception during the trial and a negative pregnancy test (urine) before enrollment |
| 2 | 2 | 2 | 2 | 2 | 2 | Active or untreated latent tuberculosis (TB) * |
| 2 | 2 | 2 | 2 | 2 | 4 | Pregnancy or lactation period * |
| 2 | 2 | 4 | 4 | 4 | 4 | Women of childbearing potential that are pregnant, intend to become pregnant, or are lactating |
| 2 | 3 | 4 | 2 | 2 | 1 | Dermal disorder including infection at anticipated treatment sites in either axilla. |
| 2 | 3 | 4 | 4 | 4 | 4 | Evidence of lens opacity or cataract at the Screening |
| 3 | 1 | 4 | 4 | 4 | 1 | Presence of corneal and conjunctival staining. |
| 3 | 3 | 3 | 1 | 1 | 1 | HDSS score of 3 or 4. * |
| 3 | 3 | 3 | 4 | 4 | 3 | Prior treatment with any investigational drug within the preceding 4 weeks prior to study entry.* |
| 3 | 3 | 3 | 4 | 4 | 4 | Screening genotype report must show sensitivity to elvitegravir, emtricitabine, tenofovir DF * |
| 3 | 3 | 4 | 4 | 4 | 4 | person deprived of liberty by judicial or administrative decision * |
| 3 | 4 | 4 | 3 | 3 | 4 | Able to swallow tablets * |
| 3 | 4 | 4 | 3 | 3 | 3 | good health * |
| 3 | 4 | 4 | 3 | 3 | 3 | good overall physical constitution * |
| 3 | 4 | 4 | 3 | 3 | 3 | Legal incapacity or limited legal capacity * |
| 3 | 4 | 4 | 4 | 4 | 4 | Living conditions-suggesting an inability to track all scheduled visits by the protocol * |
| 3 | 4 | 4 | 4 | 4 | 4 | Maintenance of a diet consisting of 40 g of carbohydrate per day within 3 months of screening* |
| 3 | ? | 3 | 4 | 4 | 4 | must have completed Part 2 of the base study |
| 4 | 4 | 4 | 4 | 4 | 4 | Ability to understand and sign a written informed consent form, which must be obtained prior to initiation of study procedures * |
| 4 | 4 | 4 | 4 | 4 | 4 | Facial hair * |
| 4 | 4 | 4 | 4 | 4 | 4 | Parents/children who refuse to participate in the study * |
| 4 | 4 | 4 | 4 | 4 | 4 | Unwillingness/inability to limit antioxidant supplement use to study-provided supplements * |
| 4 | 4 | 4 | 4 | 4 | 4 | Willing to be examined * |
| 4 | 4 | 4 | 4 | 4 | 4 | Current alcohol or substance use judged by the investigator to potentially interfere with study compliance |
Aggregate Criteria Ratings
Aggregate ratings are summarized in Table 4. For UAB raters, the ICC was 0.91 (95% CI: 0.88, 0.93) for 286 criteria (excluding 6 criteria with blank responses). Of 286 criteria, 232 (81.12%) demonstrated complete or near-complete agreement (2 reviewers at a site agreed completely while the third differed by only one category). For NU raters, the ICC for agreement was 0.92 (95% CI: 0.90, 0.93) for 292 criteria. Of 292 criteria, 262 (89.73%) demonstrated complete or near-complete agreement.
Table 4.
Individual and combined aggregate ratings for University of Alabama at Birmingham (UAB) and Northwestern University (NU).
| UAB | NU | UAB and NU | |
|---|---|---|---|
| Raters | 3 | 3 | 6 |
| Initial Criteria Reviewed | 292 | 292 | 292 |
| Criteria with Null Ratings | 6 | 0 | 6 |
| Criteria analyzed | 286 | 292 | 286 |
| Complete or Near Agreement | 232 (81.12%) | 262 (89.73%) | 181 (63.3%) |
| Complete Agreement | 142 (49.65%) | 159 (54.45%) | 88 (30.77%) |
| Intra-class Correlation (ICC) | 0.91 | 0.92 | 0.92 |
| 95% Confidence Interval | 0.88, 0.93 | 0.90, 0.93 | 0.90, 0.93 |
| Statistical Significance | p<0.00001 | p<0.00001 | p<0.00001 |
For all raters from UAB and NU collectively, (including 286 criteria), the ICC for consistency was 0.92 (95% CI: 0.90, 0.93) and the ICC for agreement was 0.92 (95% CI: 0.90, 0.93). Of the 286 criteria where all raters provided a response, 181 (63.3%) indicated complete or near-complete agreement (at least 4 reviewers agreed completely while the others differed by only one category). All ICC results were statistically significant (p < 0.00001).
Characteristics of Easy Criteria
These criteria were defined as those that can be used easily to identify subjects in an EHR data repository, assuming the data were present, using the repository’s user interface. In general, these were easily related to the categories of data that are provided by repositories like i2b2 and BTRIS, such as laboratory results, vital signs, diagnoses, procedures and allergies.
The raters generally rated as “easy” criteria that contained defined, single concepts, such as diseases like “glaucoma” or individual laboratory tests combined with discrete values, such as “hemoglobin ≥ 9 g/dL.” They also selected “easy” for general descriptions that could be addressed through class-based queries, such as “Abnormal laboratory tests”, “Anti-cancer treatment prior to trial entry”, and “History of solid organ or hematological transplantation.” There was general agreement that criteria involving complex phenotypes were considered “easy” if they could be addressed through an assemblage of individual easy criteria, such as “infection with human immunodeficiency virus (HIV), Hepatitis B, or Hepatitis C” or “Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) ≤ 5 × the upper limit of the normal range (ULN).”
Characteristics of Mixed Criteria
Mixed criteria were defined as having elements of easy and hard criteria, and these criteria can be partly retrieved using the repository’s user interface but then would require further manual review. These criteria generally included some mention of an “easy” criterion, coupled with some restriction or co-occurring state or procedure that either was not itself an easy criterion, or the relationship between the two components can only be carried out by a manual review.
Most mixed criteria involved the presence of some medical condition with restrictions on the state, such as severity (e.g., “Severe cardiovascular disease (defined by NYHA ≥3)”), chronicity (e.g., “current manic episode with a duration of > 2 years”), or failure to respond to treatment (e.g., “Active or untreated latent tuberculosis (TB).” Some criteria consisted of an “easy” condition paired with a “hard” criterion, such as “pregnant or breast feeding” and “Seizure disorders requiring anticonvulsant therapy”. There were also cases of findings that would be expected to appear only as narrative text in a diagnostic procedure report. In these text reports, the procedure could be readily identified, but detecting the findings or test results would require manual review, such as in “At least one primary molar tooth with caries into dentine involving two dental surfaces (diagnosed according to International Caries Detection and Assessment System - ICDAS, codes 3 to 5)”, “homozygous for the F508del-CFTR mutation”, and “Normal electrocardiogram (ECG).” Finally, there were some criteria that combined “easy” laboratory findings with “hard” ones, such as “Estimated glomerular filtration rate (eGFR) ≥ 50 mL/min according to the Cockcroft-Gault formula for creatinine clearance.”
Characteristics of Hard Criteria
Hard criteria were defined as those which would require manual review of the patient record. These criteria span the breadth of EHR findings that typically appear in clinical notes and procedure reports. Some hard criteria were encountered as parts of “mixed” criteria, such as “females who are breastfeeding”. Others were statements of patient condition that would not be reflected in a typical problem list, such as “prisoner”. Scale-based assessments, such as “Pre-morbid modified Rankin score of 0-1” and “Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1” were common. Finally, some criteria might be found in the Assessment portion of clinical notes or require the researcher to use the clinical record to make the assessment, such as “Candidate for phototherapy or systemic therapy” and “Any other clinical condition or prior therapy that, in the opinion of the Investigator, would make the individual unsuitable for the study or unable to comply with dosing requirements.”
Characteristics of Impossible (Point of Enrollment) Criteria
Criteria were “impossible” or “point of enrollment” if the repository would not be likely to include sufficient information to match patients to the criterion. While some of the “impossible” criteria related to the time factor of the data (such as “Parents/children who refuse to participate in the study” and “presenting within timeframe for intravenous tPA treatment approved by local regulatory authorities but no more than 4.5 hours from onset of symptoms”), other cases depended on the raters’ opinions about whether relevant data are reliably found in EHRs.
Researchers may be surprised that physical findings they would consider basic were considered by the raters to be generally absent, such as “Facial Hair”, and “good health.” Similarly, the raters agreed that information a researcher would typically obtain from a prospective subject or caregiver, such as “patients with impaired decision making ability”, and “Able to swallow tablets”, would likely not be documented in routine clinical care.
Differences in Ratings between Institutions
There were systemic differences in ratings between UAB and NU (Table 3). For example, for criteria specifying that a disorder, medication, or laboratory result must be present at screening, UAB reviewers unanimously rated these criteria as “easy” while NU reviewers rated them as “impossible.” There were additional criteria which the NU reviewers consistently deemed more challenging than the UAB reviewers. There were other criteria that UAB reviewers found more difficult than NU reviewers.
General Recommendations
The data presented above can be summarized by separating out “easy”, “mixed”, “hard”, and “impossible” criteria into explicit lists (see Table 5). This list, in turn, can be used by those developing eligibility criteria such that they can anticipate the workflow needed to identify potential research subjects using repository query tools, manual record review, and real-time interventions at recruitment sites. Potential uses for this list are explored further in the Discussion section, below.
Table 5.
Summary of features of criteria suggesting difficulty retrieving from typical EHR repository.
| Criterion is likely to be easy to retrieve from an EHR repository if: |
|
| Criterion is likely to be hard to retrieve from an EHR repository if: |
|
| Criterion is likely to be impossible to retrieve from an EHR repository if: |
|
Discussion
The use of EHR data for clinical research is a rapidly evolving phenomenon.* The development of methods for “large pragmatic trials” is especially dependent on EHR data.18 However, the methods for defining eligibility criteria remain largely unchanged in practice, with little apparent attention paid to the fact that criteria elicited from patients completing questionnaires might differ fundamentally from those elicited from EHR.
Much informatics research has examined the semantics of clinical research eligibility criteria. Projects such as the Ontology of Clinical Research (OCRe)19 and the Agreement on Standardized Protocol Inclusion Requirements for Eligibility (ASPIRE)20 have defined broad classes of criteria (such as demographics, disease-specific features, functional status, etc.) with the intent of making them computable for comparison across multiple studies. The semantic complexity of these classes of criteria has been studied in depth by Ross and colleagues.21 Weng and colleagues provided a comprehensive review of this work22 and extended it with classification of criteria through use of the National Library of Medicine’s Unified Medical Language System (UMLS) semantic types,23 UMLS semantic network,24 and National Institute of Health’s (NIH) Common Data Elements.25 To our knowledge, however, there have been no systematic studies of how such criteria translate operationally into the potential capabilities of EHR repository query tools.2-4
We did not set out to create a reusable criteria rating scale, but rather sought to find a specific set of criteria that could be used for further analysis. Nevertheless, we experienced high interrater correlation in the use of our scale within and between our two institutions. The sets of researchers have diverse backgrounds (medicine, informatics, and information systems) and draw on experience with three very different repositories (PowerInsight, i2b2 and BTRIS). This result suggests the raters were tracking a real and meaningful concept and that a standardized and generalizable scoring metric could be generated and applied at other institutions. Examples of criteria on which we agreed and disagreed are included in Table 3 for the reader to judge whether our rating method appears generalizable.
We identified characteristics that affect the difficulty of applying different criteria. Based on these findings, we can develop strategies to convert mixed, difficult, or impossible criteria into easier categories. For example, many criteria consist of combinations of simpler criteria or are bound by restrictions such as severity or time constraints. Separating combined criteria into individual components would likely make them easier to apply. Also, specifying clinical trial criteria using controlled terminologies (and increasing the use of structured data entry in EHRs) can promote improved automated patient matching. A shared library of shared queries for criteria can improve the querying process.
There were interesting systemic differences in ratings between the two institutions. Some criteria consistently or even unanimously received completely opposite ratings at the two institutions. In particular, criteria specifying a condition that must be present at the time of screening were unanimously rated as “easy” at UAB but “impossible” at NU. This difference illustrates that the wording of inclusion and exclusion criteria may be ambiguous, subject to individual interpretation, and a source of systemic inconsistency. UAB reviewers may have interpreted “screening” to indicate the time a researcher performs an initial query on the EHR repository, and the correct information would be searchable in the EHR. NU reviewers may have interpreted “screening” to mean the time that a patient presents for an initial screening encounter with a study coordinator. Because the relevant EHR data are retrospective, they may not be current at the time of a screening visit. This discrepancy does not invalidate the use of our categories for criteria. Instead, it highlights that criteria must be written so that their meanings are expressed clearly and unambiguously. It would be revealing to examine other systemic differences in ratings and differences in repository management and query processes at the two institutions to uncover other possible reasons for these differences.
Our examination of a randomly collected set of enrollment criteria suggests that a majority can be readily classified as having some components retrievable from EHR repositories (i.e. rated “easy” or “mixed”). While not necessarily representative, it does imply a large degree of inclusion and exclusion criteria are accessible via structured data. Our preliminary work in the adjacent field of cohort selection queries leads us to suggest that a significant amount of potential participant identification could be accomplished with intelligent use of these structured data.
The most interesting category may be those criteria widely rated as “mixed”, in which a “first pass” or approximate identification may be possible using structured data. This rating therefore represents an obvious break point in complexity. We suggest future research focus on such criteria, ideally looking to separate or better explicate what can be solved with structured data and EHR retrieval and what might must be retrieved with either full chart review or patient screening.
We do not make any claims, given our sample size and sampling method, about the actual ratios between easy, hard and impossible (time of enrollment) criteria across the spectrum of clinical trials. But, it seems logical to take advantage of the knowledge that such distinctions exist for developing the methods to be used for matching patients to criteria when EHR data repositories are being used for at least part of the process. A potential next step is to develop an interactive “wizard” with a graphical user interface that prompts researchers to enter criteria in structured ways that progress in order of increasing complexity. For example:
The wizard would ask first for easy elements that can be addressed readily with a repository search tool:
What laboratory test result would you like, and with what range of values?
What class of medications should the patient be taking (or not taking) and for what duration?
What procedure should have already been or not been performed?
Are diagnoses from billing data sufficient or are higher-quality sources (such as problem lists) required?
The wizard would then address more complex elements needing manual review of records found with initial criteria:
What calculation would you like performed on the previously specified laboratory test result?
What should the indication for the previously specified medication be?
What findings should be present in, or absent from, the text report for the previously requested procedure?
What complex phenotypic pattern should the patient have?
The application would then proceed to elements that can only be ascertained at the time of enrollment:
What acute event will the otherwise eligible patient have (or not have) at the time of presentation?
What will the patient have to agree to do (or not do) in order to be eligible for enrollment?
This wizard would then produce a coherent set of eligibility criteria that could specify a multi-staged enrollment process that starts with a set of data queries, proceeds to manual review of full records, and then specifies what additional information will be needed on specific potential subjects through direct contact, perhaps by flagging them in an EHR’s alerting system to notify the researcher when they appear in a patient care setting.7
The findings of our study are a building block toward the development of an approach to improve the specification of eligibility criteria. An understanding of the pragmatics of interfacing with actual clinical data in repositories can inform previous work on the semantics and interoperability of criteria. A set of questions, such as those listed above, guided by knowledge of criteria semantics, will help to establish a dialogue between the investigator and the data analyst (or provide an opportunity for introspection if the investigator is accessing a repository in self-service mode). Further study can lead to refinements in the categories and further strategies and tools for prioritizing criteria. Improving self-service queries will likely require an iterative process of extensions, corrections and refinements.
Conclusion
The findings of this study support the hypothesis that clinical research eligibility criteria fall into stereotypical categories with respect to ease of querying data available in EHR repositories. Describing these categories explicitly supports the potential development of a structured process using a data entry form or interactive “wizard.” Such an application may encourage researchers to express their criteria in ways that promote realistic expectations for the recruitment process and improve data retrieval by analysts and researchers.
Acknowledgements
This research was supported in part by the University of Alabama School of Medicine Informatics Institute. The authors also thank the University of Alabama at Birmingham Center for Clinical and Translational Science, funded by the National Institutes of Health, National Center for the Advancement of Translational Science (NCATS) (1TL1TR001418-01); the Northwestern University Clinical and Translational Sciences (NUCATS) Institute, funded by NCATS (TL1TR001423); and the National Institute of General Medical Sciences (R01GM105688).
Footnotes
A search of PubMed on March 10, 2016 for articles with the phrase “electronic health record data for research” yields 722 citations from 1996 to 2005 and 10,303 citations from 2006 to the present.
References
- 1.Cao H, Markatou M, Melton GB, Chiang MF, Hripcsak G. Mining a clinical data warehouse to discover disease-finding associations using co-occurrence statistics. AMIA Annu Symp Proc. 2005:106–110. [PMC free article] [PubMed] [Google Scholar]
- 2.Danciu I, Cowan JD, Basford M, Wang X, Saip A, Osgood S, et al. Secondary use of clinical data: the Vanderbilt approach. J Biomed Informatics. 2014;52:28–35. doi: 10.1016/j.jbi.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE—An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc. 2009 Nov 14;2009:391–5. [PMC free article] [PubMed] [Google Scholar]
- 4.Cimino JJ, Ayres EJ, Beri A, Freedman R, Oberholtzer E, Rath S. Developing a self-service query interface for re-using de-identified electronic health record data. Stud Health Technol Inform. 2013;192:632–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Deshmukh VG, Meystre SM, Mitchell JA. Evaluating the informatics for integrating biology and the bedside system for clinical research. BMC Med Res Methodol. 2009;9(1) doi: 10.1186/1471-2288-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson EK, Broder-Fingert S, Tanpowpong P, Bickel J, Lightdale JR, Nelson CP. Use of the i2b2 research query tool to conduct a matched case-control clinical research study: advantages, disadvantages and methodological considerations. BMC Med Res Methodol. 2014;14(1) doi: 10.1186/1471-2288-14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenders RA, Hripcsak G, Sideli RV, DuMouchel W, Zhang H, Cimino JJ, et al. Medical decision support: experience with implementing the Arden Syntax at the Columbia-Presbyterian Medical Center. Proc Annu Symp Comput Appl Med Care. 1995:169–173. [PMC free article] [PubMed] [Google Scholar]
- 8.Cimino JJ, Lancaster WJ, Wyatt MC. Classification of clinical research study eligibility criteria to support multi-stage cohort identification using clinical data repositories. Stud Health Technol Inform (accepted for publication) [PubMed] [Google Scholar]
- 9.Cimino JJ, Ayres EJ, Remennik L, Rath S, Freedman R, Beri A, et al. The National Institutes of Health’s Biomedical Translational Research Information System (BTRIS): design, contents, functionality and experience to date. J Biomed Informatics. 2014;52:11–27. doi: 10.1016/j.jbi.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimino JJ. Normalization of Phenotypic Data from a clinical data warehouse: case study of heterogeneous blood type data with surprising results. Stud Health Technol Inform. 2015;216:559–563. [PMC free article] [PubMed] [Google Scholar]
- 11.Manning JD, Marciano BE, Cimino JJ. Visualizing the data - using lifelines2 to gain insights from data drawn from a clinical data repository. AMIA Jt Summits Transl Sci Proc. 2013 Mar 18;2013:168–72. [PMC free article] [PubMed] [Google Scholar]
- 12.Cimino JJ. The false security of blind dates: chrononymization’s lack of impact on data privacy of laboratory data. Appl Clin Inform. 2012 Oct 24;3(4):392–403. doi: 10.4338/ACI-2012-07-RA-0028.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimino JJ, Farnum L, DiPatrizio GE, Goldspiel BR. Improving adherence to research protocol drug exclusions using a clinical alerting system. AMIA Annu Symp Proc. 2011;2011:257–266. [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, et al. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc. 2007:548–552. [PMC free article] [PubMed] [Google Scholar]
- 15.Hallgren KA. Computing inter-rater reliability for observational data: an overview and tutorial. Tutor Quant Methods Psychol. 2012;8(1):23–34. doi: 10.20982/tqmp.08.1.p023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamer M, Lemon J, Fellows I, Singh P. Package irr: various coefficients of interrater reliability and agreement. [cited 2017 Mar 8];2015 Available from https://cran.r-proiect.org/web/packages/irr/irr.pdf. [Google Scholar]
- 17.Welty LJ, Rasmussen LV, Baldridge AS. StatTag. Chicago: Galter Health Sciences Library. 2016 doi: 10.18131/G3K76. [DOI] [Google Scholar]
- 18.Lurie JD, Morgan TS. Pros and cons of pragmatic clinical trials. J Comp Eff Res. 2013;2(1):53–58. doi: 10.2217/cer.12.74. [DOI] [PubMed] [Google Scholar]
- 19.Sim I, Tu SW, Carini S, Lehmann HP, Pollock BH, Peleg M, et al. The Ontology of Clinical Research (OCRe): An informatics foundation for the science of clinical research. J Biomed Informatics. 2014;52:78–91. doi: 10.1016/j.jbi.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sim I, Niland JC. Chapter 9: Study Protocol Representation. In: Richesson RL, Andrews JE, editors. Clincial Research Informatics. London: Spring-Verlag; 2012. pp. 155–174. [Google Scholar]
- 21.Ross J, Tu S, Carini S, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits Transl Sci Proc. 2010;2010:46–50. [PMC free article] [PubMed] [Google Scholar]
- 22.Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: A literature review. J Biomed Informatics. 2010;43(3):451–467. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Z, Yetisgen-Yildiz M, Weng C. Dynamic categorization of clinical research eligibility criteria by hierarchical clustering. J Biomed Informatics. 2011;44(6):927–935. doi: 10.1016/j.jbi.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011 Dec;18 Suppl 1:i116–24. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Z, Miotto R, Weng C. A human-computer collaborative approach to identifying common data elements in clinical trial eligibility criteria. J Biomed Informatics. 2013;46(1):33–39. doi: 10.1016/j.jbi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


