Abstract
Psoriasis is a chronic, debilitating skin condition that affects approximately 125 million individuals worldwide. The cause of psoriasis appears multifactorial, and no unified mitigating signal or single antigenic target has been identified to date. Metabolomic studies hold great potential for explaining disease mechanism, facilitating early diagnosis, and identifying potential therapeutic areas. Here, we present an integrated disease metabolomic biomarker discovery strategy that combines mechanism-based biomarker discovery with clinical sample-based metabolomic profiling. We applied this strategy in identifying and understanding metabolite biomarkers for psoriasis. The key innovation of our strategy is a novel mechanism-based metabolite prediction system, mmPredict, which assimilates vast amounts of existing knowledge of diseases and metabolites. mmPredict first constructed a psoriasis-specific mouse mutational phenotype profile. It then constructed phenotype profiles for a total of 259,170 chemicals/metabolites using known chemical genetics and human metabolomic data. Metabolites were then prioritized based on the phenotypic similarities between disease- and metabolites. We evaluated mmPredict using 150 metabolites identified using our in-house metabolome profiling study of psoriasis patient samples. mmPredict found 96 of the 150 metabolites and ranked them highly (recall: 0.64, mean ranking: 8.73%, median ranking: 2.33%, p-value: 4.75E-44). These results show that mmPredict is consistent with, as well as a complement to, traditional human metabolomic profiling studies. We then developed a strategy to combine outputs from both systems and found that the oxidative product of linoleic acid, 13(S)-hydroxy-9Z,11E-octadecadienoic acid (13- HODE), ranked highly by both mmPredict and our in-house experiments. Our integrated analysis indicates that 13- HODE may be a mechanistic link between psoriasis and cardiovascular comorbidities associated with psoriasis. In summary, we developed an integrated metabolomic prediction system that combines both human metabolomic studies and mechanism-based prediction and demonstrated its application in the skin disease psoriasis. Our system is highly general and can be applied to other diseases when patient-based metabolomic profiling data becomes more increasingly available.
Data is publicly available at: http://nlp.case.edu/public/data/mmPredict_PSO
1. Introduction
The prevalence of skin disease exceeds those of obesity, hypertension or cancer. One in three Americans suffers from a skin disease, and ~2-3% of these individuals have psoriasis. Costs of treating psoriasis patients are believed to exceed $1B, including over $350M in prescriptions alone [1–4]. The pathophysiology of psoriasis is complex, and no unified mitigating signal or single antigenic target has been identified.
Human metabolomics is the comprehensive characterization of all metabolites found in the human body. Human metabolomics has great potential for explaining disease mechanism, facilitating early diagnosis, and identifying potential therapeutic areas. Common technologies for metabolomics include mass spectrometry (MS) and nuclear magnetic resonance spectroscopy (NMR) [5–6]. Human metabolites are highly heterogeneous. The Human Metabolome Database (HMDB), a comprehensive database of small molecule metabolites found in the human body, contains 42,032 metabolites of lipids, small peptides, amino acids, organic acids, vitamins, carbohydrates, nucleic acids, as well as metabolites derived from drugs, environmental contaminants, food additives, toxins, cosmetics, and other xenobiotics such as microbial or fungal symbionts [7]. Profiling the human metabolome is difficult owing to the challenges of reproducibility and knowledge generalization, as the human metabolome is affected by not only intrinsic but also many extrinsic factors such as sample collection, storage, processing and data analysis [8]. The NIH Common Fund Metabolomics Program was launched in 2012 with a primary goal to increase the national capacity to conduct metabolomics research [9]. Currently, the NIH Metabolomics Workbench Metabolite Database (MWMD) contains metabolomics studies for a very limited number of diseases [10]. While current metabolomics profiling strategies can identify clinically significant metabolite biomarkers, it is limited in understanding the underlying mechanistic links between identified metabolites and diseases.
Here, we present a novel mechanism-based metabolomic biomarker discovery system, mmPredict, to complement current patient-based metabolomic profiling studies. We developed an approach to combine outputs from these two complementary strategies and demonstrate its utility in the clinical skin disease psoriasis. The output of this integrated system is a ranked list of metabolomic biomarkers that incorporate both clinical significance and interpretable molecular mechanisms. mmPredict integrates large amounts of data from human disease genetics, chemical genetics, mouse mutational phenomics, human metabolomics, and genetic pathways in order to predict disease metabolomic biomarkers. The underlying rationale for mmPredict is that, if the changes in metabolite- associated genes cause many phenotypes (reflected in mouse models) that are also involved in any given disease (psoriasis in this study), then the metabolite is likely to be involved mechanistically in the disease. In order to test the validity of mmPredict to traditional metabolomic profiling, we compared the output of mmPredict to our inhouse metabolomic profile. Our in-house psoriasis metabolome was identified using a traditional metabolomics comparison study of skin samples obtained from the involved plaques of psoriasis patients (n=12) compared to skin samples obtained from age-matched healthy control subjects (n=9) and identified 150 significantly altered metabolites. mmPredict highly ranked psoriasis metabolites identified from our in-house metabolomic profiling study, indicating that these two strategies are consistent with, as well as complementary to, each other. We then developed a strategy to combine outputs from these two complementary strategies and identified psoriasis- associated metabolite biomarkers likely to have both mechanistic and clinical significance.
mmPredict performs both genome- and phenome-wide matching between metabolites and diseases. We compared mmPredict to a genome-wide- strategy that we recently developed to identify human gut microbial metabolite biomarkers for colorectal cancer (CRC) [11] as well as Alzheimer’s disease [12]. We demonstrated in this study that mmPredict is more effective in identifying clinically relevant metabolite biomarker for psoriasis than the genome-wide approach.
2. Data and methods
2.0. The overview of mmPredict
mmPredict matches psoriasis metabolites based on both genetic and phenotypic relevance. mmPredict consists of the following components: (1) mmPredict constructs mouse mutational phenotype profiles for a given disease (psoriasis in this study) using publicly available disease genetics and genomics databases; (2) it constructs mouse mutational phenotype profiles for a total of 259,170 chemicals/metabolites; (3) mmPredict prioritizes metabolites for a given disease based on the phenotype profile similarities between the disease and metabolites (Fig. 1).
Fig 1:
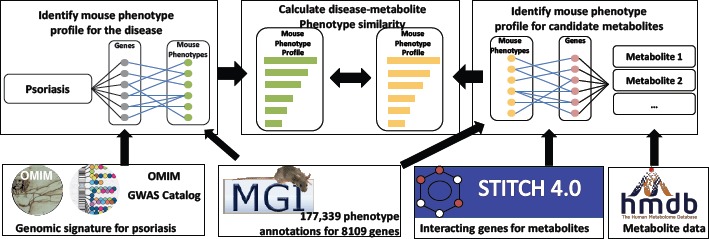
mmPredict: a combined genome- and phenome-wide metabolite prediction system, including three components and five data resources.
We evaluated mmPredict using 150 metabolites identified by comparing involved tissue from psoriasis patients to healthy control skin in our in-house metabolomic study. We developed an approach to combine predictions from mmPredict and from our in-house metabolomic study to identify metabolites likely to have both clinical and mechanistic significance.
For comparison, we implemented a genome-wide metabolite prediction system mmPredict_Gen. The underlying assumption is that if a metabolite is associated with many psoriasis-associated genes, then the metabolite may be associated with the mechanism(s) causing psoriasis. We have previously applied this strategy to identify human gut microbial metabolites associated with colorectal cancer [11] as well as Alzheimer’s disease [12]. The difference between mmPredict and mmPredict_Gen is that mmPredict utilizes both genome- and phenome-wide information, while mmPredict_Gen prioritizes metabolites for diseases based on genetic profile similarities (Fig 2). We demonstrated in our study that mmPredict performed consistently better than mmPredict_Gen.
Fig 2:
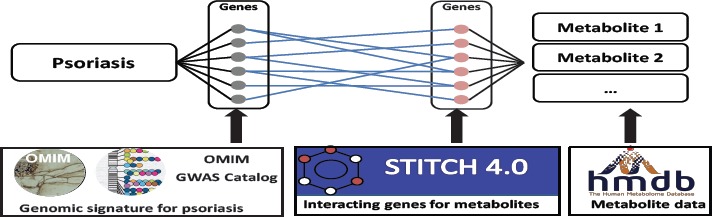
mmPredict_Gen: a genome-wide metabolite prediction system (five data resources are shown).
2.1. Data
We used the disease phenotype knowledge bases that we recently constructed, as well as publicly available data from the human metabolome, disease genetics, chemical genetics, functional protein interactions and signaling pathway databases to identify metabolites associated with psoriasis and its’ comorbidities.
2.1.1. Disease genetics and genomics data
We used two complementary databases to obtain genes associated with psoriasis. We identified 49 genes associated with common complex forms of psoriasis from the Catalog of Published Genome-Wide Association Studies (GWAS catalog), an exhaustive source containing descriptions of disease-/trait-associated single nucleotide polymorphisms (SNPs) from published GWAS studies [13]. We identified 35 genes associated with rare Mendelian forms of psoriasis from the Online Mendelian Inheritance in Man (OMIM) database, the most comprehensive source of disease genetics for Mendelian disorders [14]. We used these two complementary resources of disease genetics to demonstrate the robustness of our algorithms and findings.
2.1.1. The Human Metabolome Database (HMDB)
HMDB is intended for applications in metabolomics, biomarker discovery and other applications and contains detailed information regarding small molecule metabolites found in the human body [7]. Currently, HMDB is the most comprehensive collection of metabolite data, containing 42,032 metabolites. We used HMDB to obtain a list of metabolites found in the human body.
2.1.2. Chemical genetics data
We used the STITCH (Search Tool for Interactions of Chemicals) database [15] to obtain chemical/metabolite-gene associations. We used chemical-gene associations found in the human body (1,466,636 chemical-gene pairs for 259,171 chemicals and 15,620 genes) and metabolites from HMDB to link human metabolites to human genes.
2.1.4. Genome-wide mutational phenotypes in experimental mouse models
We used gene-phenotype associations from the Mouse Genome Database (MGD) [16] to assess the phenotypic effects of metabolites. The Phenotypes/Alleles project in the MGD provides access to spontaneous, induced, and genetically engineered mutations and their specific phenotypic outcomes. Currently, MGD contains 278,553 gene- phenotype associations, representing 41,905 mutant alleles and 10,744 phenotypes. For example, the mutation of 13- HODE-associated gene myeloperoxidase (MPO) in mouse models is associated with 12 phenotypes, including ‘decreased inflammatory response,’ ‘atherosclerotic lesions’ and ‘increased monocyte cell number.’ The mutation of psoriasis-associated gene TRAF3 interacting protein 2 (TRAF3IP2) is associated with 80 phenotypes including ‘decreased inflammatory response,’ ‘increased pruritus,’ and ‘increased monocyte cell number'. We have recently shown that systematic approaches to interrogate human genes to their mouse mutational phenotypes in MGD havegreat potential in understanding both disease mechanisms and drug effects [17–19]. In this study, we leveraged the large number of gene-phenotype associations from MGD and performed genome- and phenome-wide analysis to infer phenotypic effects of metabolites on disease.
2.2. Methods
2.2.1. Construct mouse mutation phenotype profiles for input disease
We identified 35 psoriasis-associated genes from the OMIM database and 49 psoriasis-associated genes from the GWAS Catalog. We mapped these genes to their corresponding mouse gene homologs (e.g., TRAF3IP2 =< Traf3ip2) using human-mouse homolog mapping data from the Mouse Genome Database (MGD) [16]. The mapped mouse genes were then linked to their corresponding mutational phenotypes (e.g., knockout) in mouse models (e.g., Traf3ip2 => ‘increasedpruritus ‘) using gene-phenotype association data from MGD. For each mapped phenotype, we assessed its probability of being associated with the given set of genes (e.g., the phenotype “increased pruritus” is associated with 10 of the 49 psoriasis genes) as compared to its probability associated with the same number of randomly selected genes (e.g., the phenotype “increasedpruritus” is on average associated with 0.5 of 49 randomly selected genes). The random process is repeated 1000 times and a t-test was used to assess the statistical significance.
2.2.2. Construct mutational phenotype profiles for chemicals
We built phenotype profiles for 259,170 chemicals/metabolites from the STITCH database in the same manner as described above (e.g., 13-HODE =<MPO =>Mpo =< ‘increased monocyte cell number’).
2.2.3. Prioritize metabolites for psoriasis and test the performance using in-house metabolomics study
Metabolite prioritization
We calculated the phenotypic similarity of psoriasis and metabolites. We compared four different approaches in calculating the similarities, including cosine similarity, overlap, Jaccard similarity [20], and ontology-based semantic similarity [17-19, 21]. We showed that ranking based on Jaccard similarity performed consistently better than the other three similarity measures used in this study. Therefore, throughout this study, the metabolite ranking values are based on the Jaccard similarity.
Evaluation of recall and ranking
We evaluated mmPredict in identifying and prioritizing metabolite biomarkers for psoriasis using our in-house metabolome study. We identified 150 significantly altered metabolites by comparing global metabolic profiles in skin samples from normal control subjects (n=9) and age matched psoriasis patient involved psoriasis plaques (n=12). We evaluated the algorithm for both identifying and ranking known metabolites using recall, mean ranking, and median rankings. Random expectation that the metabolites would have an average ranking of 50% was used to evaluate the significance of these rankings.
Evaluation of prioritization (precision-recall curve)
We evaluate mmPredict using an 11-point interpolated average precision measure, which is commonly used to evaluate retrieved ranked lists for search engines [22]. For each ranked list, the interpolated precision was measured at the 11 recall levels of 0.0, 0.1, 0.2, …, 1.0. At each recall level, we calculated the arithmetic mean of the interpolated precision. A composite precision-recall curve showing 11 points was then graphed.
2.2.4. Develop a strategy to combine outputs from mmPredict and patient-based metabolomics profiling
We developed an approach to combine rankings from mmPredict and fold changes from the clinical metabolomic study. The output of mmPredict is a list of metabolites ranked by their percent ranking (“ranking_m”). For example, 13-HODE ranked at the top 0.23% among 259,170 chemicals. We then ranked the 150 in-house metabolites by their fold changes and calculated their percent ranking (“ranking_c ”) among all metabolites. For example, 13-HODE was ranked at the top 0.67% (top one) among the 150 in-house generated metabolites. The combined ranking is a balance measure of rankings from both studies and is defined as: ranking_combined = 2*(ranking_m*ranking_c)/(ranking_m + ranking_c). We studied the top one ranked metabolite by performing both genetic pathway enrichment and mutational phenotype enrichment analyses.
3. Results
3.1. Understanding psoriasis genetics, genomics, and phenomics
We used mmPredict to perform both genome- and phenome-wide analyses to prioritize metabolites for psoriasis. To understand both genetic and phenotypic implications of psoriasis-associations genes, we began our study using Ingenuity Pathway Analysis (IPA) [23] to examine the top canonical pathways generated by searching for either rare Mendelian psoriasis genes or common complex psoriasis genes. As shown in Table 1, the top diseases and biological functions identified dermatological diseases and conditions, with immunological and inflammatory response also being identified. Common cellular response pathways and cells (e.g., dendritic cells and janus activated kinase (JAK) signaling) as well as other skin and immunologically prominent diseases (e.g., graft versus host disease (GVHD), Rheumatoid Arthritis (RA) and Diabetes Mellitus (DM)) were also identified.
Table 1.
Top 5 genetic pathways, diseases, and biological functions enriched for psoriasis-associated genes. Skinspecific diseases and pathways are shown in pink and Immune-related diseases and pathways are shown in green.
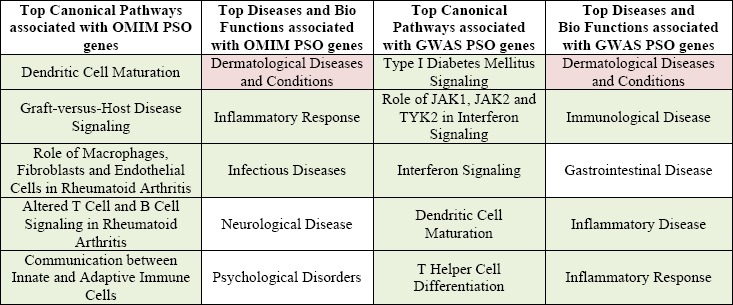 |
Table 2 outlines the top 10 phenotypes associated with psoriasis genes identified through either the OMIM or GWAS databases. As shown in the table, the top phenotypes are enriched for immune-response and skin-related phenotypes, confirming the observational and experimental data supporting psoriasis as a predominantly immune- mediated autoimmune disorder. Both genetic pathway enrichment and mutational phenotype enrichment analyses provided the rationale underlying mmPredict’s combined genome- and phenome-wide analysis. As shown in the following sections, genome-wide prioritization is not as effective as the combined approach.
Table 2.
Top 10 phenotypes enriched for psoriasis genes from the OMIM and the GWAS catalog. Skin-specific diseases and pathways are shown in pink and Immune-related diseases and pathways are shown in green.
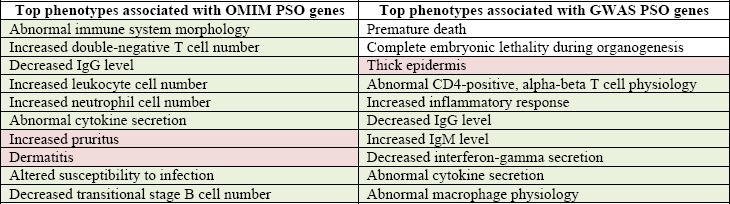 |
3.2. mmPredict ranked clinically derived psoriasis-associated metabolites highly
The output of mmPredict is a ranked list of 259,170 chemicals. We also filtered the list of chemicals from HMDB by their origins. In HMDB, metabolites are classified based on their origins, such as “endogenous”, “food”, “microbial”, “drug”, “plant”, and “toxin/pollutant”. Only 4,057 of the 259,170 chemicals appeared in HMDB, including 2,067 endogenous metabolites. We evaluated the performance of mmPredict using the 150 clinically derived metabolites. As shown in Table 3, mmPredict found 96 of these 150 metabolites (recall: 0.64) and consistently ranked them highly for both the Mendelian form of psoriasis (mean ranking based on Jaccard similarity: 8.73%, median ranking based on Jaccard similarity: 2.33%, p-value: 4.75E-44) and common complex form of psoriasis (mean ranking based on Jaccard similarity: 9.08%, median ranking based on Jaccard similarity: 2.39%, p-value: 1.35E-42). After filtering the chemicals by metabolites from HMDB, the rankings are still significantly higher than random expectation. However, the recall are lower, indicating that the list of chemicals from the STITCH database represents a more complete list of metabolites found in the human body than the list of metabolites from HMDB.
Table 3.
Evaluation of mmPredict with 150 clinically derived psoriasis metabolites. The ranking was based on Jaccard similarity.
| Metabolite source | Psoriasis genetics | Recall | Mean Ranking | Median Ranking | P-value |
|---|---|---|---|---|---|
| STITCH (259,170 chemicals) | OMIM | 0.64 | 8.73% | 2.33% | 4.75E-44 |
| GWAS | 0.64 | 9.08% | 2.39% | 1.35E-42 | |
| HMDB (4057 metabolites) | OMIM | 0.41 | 22.36% | 14.03% | 8.54E-13 |
| GWAS | 0.41 | 23.25% | 14.56% | 8.54E-13 | |
| HMDB (2067 endogeneous metabolites) | OMIM | 0.34 | 19.27% | 11.67% | 2.64E-14 |
| GWAS | 0.34 | 20.42% | 12.63% | 2.16E-13 |
For comparison, we show in Table 4 that genome-wide prioritization alone is less effective than the combined genome- and phenome-wide implementation of mmPredict. mmPredict has consistently better mean and median rankings than mmPredict_Gene across three metabolites data resources.
Table 4.
Evaluation of mmPredict_Gen with 150 psoriasis metabolites. The ranking was based on Jaccard similarity.
| Metabolite source | Psoriasis genetics | Recall | Mean Ranking | Median Ranking | P-value |
|---|---|---|---|---|---|
| STITCH (259,170 chemicals) | OMIM | 0.64 | 46.55% | 53.55% | 0.309 |
| GWAS | 0.64 | 42.50% | 37.92% | 0.039 | |
| HMDB (4057 metabolites) | OMIM | 0.40 | 44.24% | 45.42% | 0.184 |
| GWAS | 0.40 | 39.21% | 40.46% | 0.008 | |
| HMDB (2067 endogeneous metabolites) | OMIM | 0.34 | 39.84% | 36.64% | 0.024 |
| GWAS | 0.34 | 35.09% | 30.93% | 0.001 |
We further show in the plotted 11-point interpolated precision-recall curve that mmPredict is effective in enriching clinically derived psoriatic metabolites at 11 recall cutoffs (Fig. 3). At a recall level of 0.1, the mean precision of the metabolites is 0.156, which represents a 524% enrichment as compared to the precision of 0.025 for all metabolites (recall = 1.0). These results indicate that mmPredict enriched clinically identified metabolites among the top and its predictions are consistent with patient-based metabolomic profiling. The precision was calculated using the clinically derived metabolites and does not represent the true precision measure of mmPredict. The low values of the precision (e.g., 0.156 at recall of 0.1) indicate that many more metabolites are to be discovered and that mmPredict can complement patient-based profiling in identifying metabolites not captured in our in-house metabolomic study.
Fig 3:
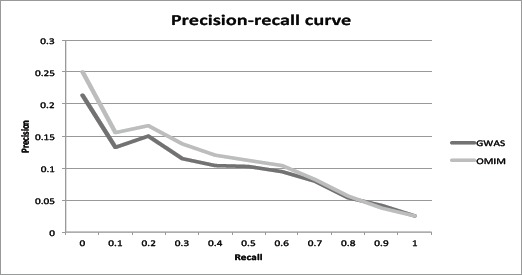
Precision-recall curve using the 150 metabolites altered in psoriatic tissue.
3.2. 13-HODE represents a metabolite biomarker mechanistically linking psoriasis to its cardiovascular comorbidities
mmPredict represents a mechanism-based approach to identify metabolites for psoriasis. Our in-house metabolomics study represents a standard approach to identify altered metabolites in psoriasis patients. Our combined scores from these two complementary studies identified 13-HODE as the highest ranked metabolite, indicating that this metabolite may be involved in psoriasis at both mechanistic and clinical levels. To understand how 13-HODE is mechanistically linked to psoriasis, we identified a total of 78 13-HODE-associated genes from the STITCH database. We performed pathway enrichment analysis using Ingenuity Pathway Analysis (IPA) to examine the top canonical pathways associated with 13-HODE. The top five canonical pathways identified by IPA are Atherosclerosis Signaling, Eicosanoid Signaling, Glutathione Redox Reactions I, Bupropion Degradation, and Acetone degradation I. The top five diseases and biofunctions are Organismal Injury and Abnormalities, Cardiovascular Disease, Gastrointestinal Disease, Inflammatory Disease, and Inflammatory Response. The pathway enrichment analysis shows that 13-HODE is involved in inflammatory response and cardiovascular diseases, common linkages associated with human psoriasis.
We then investigated the phenotypic effects of 13-HODE-associated genes by interrogating the associations of mouse phenotypes with mutations of HODE-associated genes. We identified a total of 1,214 phenotypes associated with mutations of the 78 13-HODE-associated genes. As shown in Table 5, the top ranked phenotypes associated with mutations of 13-HODE associated genes are clearly related to cardiovascular disease, immune response and abnormal skin morphology and function. These results indicate that 13-HODE may be mechanistically involved in both psoriasis and its’ cardiovascular comorbidities.
Table 5.
Top-ranked phenotypes related to CVD, immune system dysfunction, and abnormal skin morphology and function, and their rankings among all 1,214 HODE mutation-associated phenotypes.
| Phenotype Category | Phenotype | Rank (top%) |
|---|---|---|
| CVD phenotypes | Congestive heart failure | 0.41% |
| Cardiac fibrosis | 0.82% | |
| Cardiac hypertrophy | 1.57% | |
| Atherosclerosis | 2.22% | |
| Immune phenotypes | Abnormal neutrophil physiology | 1.15% |
| Abnormal macrophage physiology | 1.81% | |
| Abnormal immune system physiology | 1.98% | |
| Increased monocyte cell number | 3.13% | |
| Skin phenotypes | Abnormal cutaneous collagen fibril morphology | 3.79% |
| Abnormal epidermis stratum corneum morphology | 5.60% | |
| Impaired skin barrier function | 6.43% | |
| Decreased skin tensile strength | 7.50% |
We also categorized the 13-HODE-associated phenotypes (1,214 phenotypes were classified into 54 classes) and analyzed which phenotype categories are highly associated with HODE (the top 11 HODE-associated phenotype classes are shown in Fig. 4). Classes we found relevant to psoriasis and comorbidities include: 1) abnormal hematopoietic system (9.2% of all phenotypes), 2) abnormal skin morphology (5.0% of all phenotypes), 3) abnormal immune system (5.0% of all phenotypes), 4) abnormal cardiovascular system morphology (5.0%), and 5.) abnormal cardiovascular system physiology (4.1%). Another skin-related class, abnormal skin adnexa morphology also ranked highly (3.3%). These results suggest that HODE may be a biomarker linking remote inflammation observed in the skin with cardiovascular diseases, immune system activation, and psoriasis.
Fig 4:
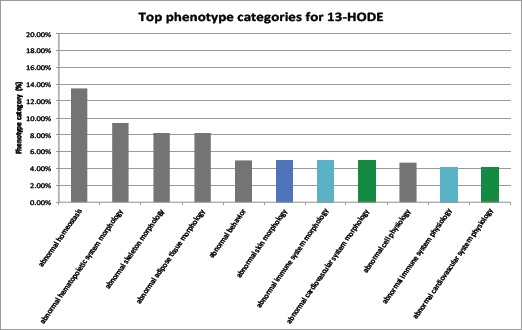
Top-ranked phenotype classes related to CVD (green), immune system dysfunction (blue), and abnormal skin morphology and function (azure).
Discussion and conclusions
We present a novel mechanism-based metabolomic biomarker discovery system, mmPredict, to complement current patient-based metabolomics profiling studies. We developed an approach to combine outputs from these two complementary strategies and demonstrated its utility in the human clinical disease psoriasis. The identification of metabolomic biomarkers and the understanding of their roles in psoriasis and psoriasis-related co-morbidities may provide insight into the basic mechanisms of psoriasis pathogenesis and enable new possibilities for psoriasis diagnosis, prevention, and treatment.
Both mmPredict and our strategy of combining these two complementary strategies for use in studying human metabolomics are general. We demonstrated its utility in psoriasis since we have performed in-house human metabolomic profiling on psoriasis patient and healthy control skin samples. We expect that mmPredict would be equally effective in identifying metabolite biomarkers for other diseases, albeit alternative sources of protein metabolites, such as serum or plasma may be necessary. However, to fully evaluate mmPredict and our integrated strategy in other diseases will require the gold standard of known disease-associated metabolite biomarkers, which are currently lacking due to the paucity of published disease specific metabolomic data. With the vast amounts of knowledge built into mmPredict, the only input needed for the system is a list of genes, therefore we expect that mmPredict can be applied to identify metabolite signatures unique for disease subtypes, disease progression, as well as treatment response given that the involved genes are available.
Computational algorithm design critically depends on the data incorporated into the system. In this study, we integrated disease genetics data with human metabolomics, chemical genetics, genetic pathways, and mouse mutational phenotypes. Our study shows that combined genome-phenome approach performed significantly better than the genome approach alone, indicating interrogating human genes to their functional effects on disease-specific phenotypes has major contribution in the improved performance. Currently, we are further improving mmPredict by incorporating other types of data, including higher-level disease and drug phenotypic data observed in humans.
Author’s contributions
QW and RX have jointly conceived, designed and implemented the algorithms, performed data analysis and algorithm evaluation, and prepared the manuscript. The metabolomics study was performed by TSM, KDC and NW. All authors read, edited and approved the final manuscript.
Acknowledgements
RX is funded by Case Western Reserve University/Cleveland Clinic CTSA Grant (UL1 RR024989), the NIH Director’s New Innovator Award under the Eunice Kennedy Shriver National Institute Of Child Health \& Human Development of the National Institutes of Health (DP2HD084068, Xu), American Cancer Society Research Scholar Grant (RSG-16-049-01 - MPC, Xu), the Landon Foundation-AACR INNOVATOR Award for Cancer Prevention Research (15-20-27-XU), Mary Kay Foundation Grant (057-15, Xu), and Pfizer 2015 ASPIRE Rheumatology and Dermatology Research Award (WI206753, Xu). QW’s effort on this project is partially supported by DP2HD084068. TSM was supported by NIH P30 AR039750-24, R01DE018276-05, R01 AR063437-03, R01 AR062546-02; KDC was supported by P30, AR039750; NW was supported by NIH R21 AR063852, R01 AR062546, R01 AR063437, and P30 AR39750; RC was supported by T32 AR007569-20.
References
- 1.Ackermann C., Kavanaugh A. Economic burden of psoriatic arthritis. Pharmacoeconomics. 2008;26(2):121–129. doi: 10.2165/00019053-200826020-00003. [DOI] [PubMed] [Google Scholar]
- 2.Feldman S.R., Evans C., Russell M.W. Systemic treatment for moderate to severe psoriasis: Estimates of failure rates and direct medical costs in a north-eastern US managed care plan. Journal of dermatological treatment. 2005;16(1):37–42. doi: 10.1080/09546630510025941. [DOI] [PubMed] [Google Scholar]
- 3.Fowler J.F., Duh M.S., Rovba L., Buteau S., Pinheiro L., Lobo F., Kosicki G. The impact of psoriasis on health care costs and patient work loss. Journal of the American Academy of Dermatology. 2008;59(5):772–780. doi: 10.1016/j.jaad.2008.06.043. [DOI] [PubMed] [Google Scholar]
- 4.Hazard E., Cherry S.B., Lalla D., Woolley J.M., Wilfehrt H., Chiou C.F. Clinical and economic burden of psoriasis. Managed care interface. 2006;19(4):20–26. [PubMed] [Google Scholar]
- 5.Nicholson J.K., Wilson I.D. Understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nature Reviews Drug Discovery. 2003;2(8):668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson J.K., Lindon J.C. Systems biology: metabonomics. Nature. 2008;455(7216):1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 7.Wishart D.S., Jewison T., Guo A.C., Wilson M., Knox C., Liu Y., Bouatra S. HMDB 3.0— the human metabolome database in 2013. Nucleic acids research. 2012:gks1065. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guma M., Tiziani S., Firestein G.S. Metabolomics in rheumatic diseases: desperately seeking biomarkers. Nature Reviews Rheumatology. 2016 doi: 10.1038/nrrheum.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sud M., Fahy E., Cotter D., Azam K., Vadivelu I., Burant C., Sumner S. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training and analysis tools. Nucleic acids research. 2015:gkv1042. doi: 10.1093/nar/gkv1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. http://www.metabolomicsworkbench.org/data/index.php.
- 11.Xu R, Wang Q, Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics. 2005 Jun 11;16(Suppl 7):S4. doi: 10.1186/1471-2164-16-S7-S4. Epub 2015 Jun 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu R., Wang Q. Towards understanding brain-gut-microbiome connections in Alzheimer’s disease. BMC Systems Biology. 2016;10(3):63. doi: 10.1186/s12918-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42(D1):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM. org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic acids research. 2015;43(D1):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn M., Szklarczyk D., Pletscher-Frankild S., Blicher T.H., von Mering C., Jensen L.J., Bork P. STITCH 4: integration of protein-chemical interactions with user data. Nucleic acids research. 2013:gkt1207. doi: 10.1093/nar/gkt1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eppig J.T., Blake J.A., Bult C.J., Kadin J.A., Richardson J.E. (Mouse Genome Database Group). The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic acids research. 2015;43(D1):D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Cai XS, Xu R. Annual American Medical Informatics Association Symposium. San Francisco: CA; 2015. Nov, Combing Human Disease Genetics and Mouse Model Phenotypes Towards Drug Repositioning for Parkinson’s Disease. pp. 13–18. [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Xu R. Phenome-based Gene Discovery Provides Information about Parkinson Disease Drug Targets. BMC Genomics. 2016;17:493. doi: 10.1186/s12864-016-2820-1. [9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Cai XS, Gao Z, Wang BC, Xu R. Towards precision medicine-based therapies for glioblastoma: interrogating human disease genomics and mouse phenotypes. BMC Genomics. 2016;17(7):516. doi: 10.1186/s12864-016-2908-7. [10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han J., Kamber M., Pei J. Data mining: concepts and techniques. Elsevier; 2011. [Google Scholar]
- 21.Resnik P. Using information content to evaluate semantic similarity in a taxonomy. arXiv preprint cmp-lg/9511007. 1995 [Google Scholar]
- 22.Manning C.D., Raghavan P., Schütze H. Introduction to information retrieval. Vol. 1. Cambridge: Cambridge university press; 2008. p. 496. [Google Scholar]
- 23. http://www.ingenuity.com/products/ipa.


