Abstract
In biomedicine, high-quality metadata are crucial for finding experimental datasets, for understanding how experiments were performed, and for reproducing those experiments. Despite the recent focus on metadata, the quality of metadata available in public repositories continues to be extremely poor. A key difficulty is that the typical metadata acquisition process is time-consuming and error prone, with weak or nonexistent support for linking metadata to ontologies. There is a pressing need for methods and tools to speed up the metadata acquisition process and to increase the quality of metadata that are entered. In this paper, we describe a methodology and set of associated tools that we developed to address this challenge. A core component of this approach is a value recommendation framework that uses analysis of previously entered metadata and ontology-based metadata specifications to help users rapidly and accurately enter their metadata. We performed an initial evaluation of this approach using metadata from a public metadata repository.
Introduction
Reproducibility of biomedical discoveries has become a major challenge in science. Investigations in a variety of fields have shown alarmingly high levels of failure when attempting to reproduce published studies.1,2 To help address this issue, many funding agencies and journals are now demanding that experimental data be made publicly available—and that those data have associated descriptive metadata.3 In the last few years, the biomedical community has met this challenge by driving the development of metadata standards, which scientists use to inform their annotation of experimental results. For example, the MIAME standard4 describes metadata about microarray experiments. The overarching goal when defining these standards is to provide sufficient metadata about experimental data to allow the described experiment to be reproduced. Community-based groups have defined an array of standards describing metadata for a variety of scientific experiment types. A large number of standards-conforming repositories have been built, greatly enhancing the ability of scientists to discover scientific knowledge.5
Despite the increasing use of these standards, the quality of metadata deposited in public metadata repositories is often very low.6 A central problem is that metadata authoring process itself can be extremely onerous for scientists.7 A typical submission requires spreadsheet-based entry of metadata—with metadata frequently spread over multiple spreadsheets—followed by manual assembly of multiple spreadsheets and raw data files into an overall submission package. Validation is often post-submission and weak. A secondary problem is that metadata standards are typically written at a high level of abstraction. For example, while a standard may require capturing the organism associated to a biological sample, it typically will not specify how the value of the organism must be supplied. Little use is made of the large number of controlled terminologies currently available in biomedicine. Submission repositories reflect this lack of precision and usually have weak or nonexistent mechanisms for linking terms from controlled terminologies to submissions. Faced with this lack of standardization, users often provide ad hoc values or simply omit many values. These difficulties combine to ensure that typical metadata submissions are sparsely populated and poorly described, and thus require significant post-processing to extract semantically useful content.
In this paper, we describe the development of a methodology and associated tools that aim to improve the metadata acquisition process. We outline a recommender framework that provides an intuitive and principled approach to metadata entry. The framework uses analyses of previously entered metadata combined with ontology-based metadata specifications to help guide users to rapidly and accurately enter their metadata. The recommender framework is part of the CEDAR Workbench (cedar.metadatacenter.net), an end-to-end metadata acquisition and management system under development by the Center for Expanded Data Annotation and Retrieval (CEDAR).8 The ultimate goal is to speed up the creation of metadata submitted to public repositories and to increase the quality of that metadata.
Related Work
Browser-based auto-fill and auto-complete functionality has a long history on the Web. Common auto-fill examples include the automatic population of address and payment fields by Web browsers in standard HTML forms. Auto-complete suggestions are commonly made for page URLs, where browsers typically maintain a history of visited pages and suggest likely pages based on a simple frequency analysis of previously visited pages. More advanced auto-complete functionality can be seen in search engines from major Web search vendors, where suggestions are based on analyses of both Web content and searches made by users.
A variety of auto-fill and auto-complete recommendation systems have been developed that perform more substantial analyses of previously entered content. A common approach is to process raw form content to extract high-level semantic concepts that drive the recommendation process. A system called Carbon9 presents auto-complete suggestions based on an analysis of Web forms previously filled in, combined with semantic information from those forms. The system uses this information to help users fill in structurally different forms. A related system called iForm10 was developed to assist form completion by analyzing both previously filled versions of a form and free text to extract likely values for fields. The approach focused on performing a semantic analysis of data-rich input text to automatically select text segments and then associating the text segments with fields in a form.
Several recommender systems that support auto-fill and auto-complete with ontology terms have been described in the literature. RightField,11 which is distributed as an Excel plugin, provides mechanisms for embedding ontology-based value fields in spreadsheets. Users populating the resulting spreadsheets are presented in real time with suggestions restricted to terms from subsets of specified controlled vocabularies. Ontology-based systems that specifically address the metadata acquisition challenge include Annotare12, which is used to submit experimental data to the ArrayExpress metadata repository,13 and ISA-Tools,14 which provides a generic spreadsheet-based tool chain for metadata authoring. Both systems provide strong support for using controlled terms and allow users to link metadata to controlled terminologies. None of them provides value-recommendation functionality, however.
By combining the analysis-driven and ontology-based recommendation strategies used by these systems, we can generate more powerful suggestions than is possible with each approach alone. We believe the combination of the two techniques can provide the speed of analysis-driven recommendations coupled with the added precision of ontology-based suggestions. This paper advances our preliminary work on metadata prediction15,16 by outlining the development of a methodology and associated tools that demonstrate this combination.
Methods
We designed an approach for metadata recommendation that simplifies the metadata authoring process in the CEDAR Workbench. The CEDAR Workbench is a suite of Web-based tools and REST APIs for metadata authoring and management, centered on the use of metadata-acquisition forms called metadata templates (or simply templates). In the CEDAR Workbench, templates are used to formally encode metadata standards and to create highly-interactive interfaces for acquiring metadata conforming to those standards. Templates define the data attributes (called template fields or fields) needed to describe experimental data. For example, an experiment template may have a disease field containing the name of the disease studied by a particular experiment. Our approach simplifies metadata authoring by suggesting the most appropriate values for template fields when acquiring metadata. We outline our approach and then explain how we implemented it in the CEDAR Workbench.
Description of the approach
Let t be a metadata template, which contains a set of template fields f1..fn. Now suppose that a user is filling out the template t with metadata. Our approach generates a ranked list of suggested values for fields f1..fn based on: (1) the template instances previously authored for the template; and (2) the field values already entered by the user for the current template, which we call the recommendation context.
For a template field being filled out, our approach retrieves all values previously entered into that field and calculates a relevancy score in the interval [0,1]. This score represents the likelihood of the value occurring again based on previously created template instances and on the recommendation context. The relevancy score for a field-value pair p in a template instance s, derived from a template t, is calculated as:
where instances(t) are all previously authored instances of the template t, and matchingInstances(A) returns the instances that contain all the field-value pairs in A. The matchingInstances(A) function is defined as:
Here, fv(s) represents all the field-value pairs for an instance s:
where fields(s) are the fields in the template t, from which s is derived, values(s) are all the values in the instance s, and value(f) is the value assigned to a field f in the instance s. While the instance is being created fv(s) represents the recommendation context.
Example: Suppose we have a template t with disease and tissue fields and have four instances of t with values as shown in Table 1.
Table 1.
Field names and values for four sample instances of a template with disease and tissue fields.
| Field | instance 1 | instance 2 | instance 3 | instance 4 |
|---|---|---|---|---|
| Disease | liver cirrhosis | liver cirrhosis | liver cirrhosis | breast cancer |
| Tissue | liver | liver | blood | Breast |
Suppose that the user is populating a new instance of t and has entered liver cirrhosis as a value for the disease field. The relevancy scores for the values liver, blood, and breast of the tissue field can be calculated as follows:
Our method takes advantage of previously populated fields to generate a context-sensitive estimate of the values for an unpopulated field. When there is no context (i.e., when no other fields in a template have been filled out), it simply computes the frequencies of all the values found for the unpopulated field and ranks the values accordingly. The method does not impose any restriction on the order in which the fields must be filled out. The analysis for a field can be performed on all instances in the repository and using all the values previously entered for other fields, independently of the order they were filled out. This ability of the method to consider contextual information enables it to go beyond simple one-cause, one-effect relationships and to consider the combined effects that previously entered values may have on the target field.
The basic approach can be extended to deal with fields whose values have been constrained to particular ontologies, ontology branches, or lists of ontology terms (e.g., a disease field could be constrained to contain diseases from the Disease Ontology17). When dealing with these ontology-based field values, our method calculates the frequencies of the underlying term identifiers independently of the display value used. For example, suppose that the repository contains template instances that refer to hypertension in different ways, such as HTN, increased blood pressure, and high blood pressure, and that those instances have been linked to the identifier of the term hypertension in the Disease Ontology (http://purl.obolibrary.org/obo/DOID_10763). In this case, our analysis approach would use the term identifier to calculate the frequency of the Disease Ontology term, effectively aggregating the frequencies of all synonyms of hypertension.
The generated recommendation scores can be used to produce a ranked list of suggested values for a target template field. Each recommendation consists of the suggested value and a number in the interval [0,1] that represents the frequency of the value in previously populated instances. For plain text metadata, the system suggests textual values. For ontology-based metadata, the system suggests ontology term identifiers. These recommendations can be presented to the user using a user-friendly preferred label for the ontology term defined in its source ontology (e.g., hypertension is the preferred label for http://purl.obolibrary.org/obo/DOID_10763 in the Disease Ontology). The recommendations for a particular field can be calculated in real time as a template is being filled in. The metadata repository’s recommendation index can be updated whenever a template instance is saved, allowing other metadata instance creators to immediately use the updated recommendation values.
Implementation
We implemented our approach for metadata recommendation as a Web service called the Value Recommender, and integrated it into the CEDAR Workbench. The CEDAR Workbench provides two core tools that form a metadata authoring pipeline: the Template Designer and the Metadata Editor. The Template Designer allows users to interactively create metadata templates in much the same way as they would create survey forms. Using live lookup to BioPortal, the Template Designer allows template authors to find terms in ontologies to annotate their templates, and to constrain the values of template fields to specific ontology terms18. The Metadata Editor uses a template specification generated by the Template Designer to automatically generate a forms-based metadata acquisition interface for that template. The generated interfaces allow users to populate metadata templates with metadata.
We modified both tools and several other CEDAR components to work with the Value Recommender service (see Figure 1). We extended the Template Designer to allow users to specify the fields for which value recommendations are enabled. We enhanced CEDAR’s template specification model to store this preference. This preference is used to signal to CEDAR’s metadata indexing engine that field-level metadata in its metadata repository should have additional analysis steps applied to it. Fields marked for value recommendation are indexed by CEDAR’s Elasticsearch-based engine (https://www.elastic.co) such that their values can be compared with other value recommended fields. These statistics are used in real time by the newly developed Value Recommender component. Note that users can also use standard CEDAR functionality to constrain fields to contain values from controlled terminologies held in the BioPortal server. Both constraint types can be specified simultaneously for a field.
Figure 1.
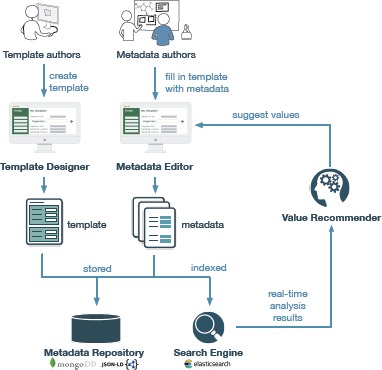
Workflow of the Value Recommender service in the CEDAR Workbench. Template authors use the Template Designer to create templates. Metadata authors fill in templates with metadata. The Value Recommender uses the Search Engine to analyze the metadata stored in the Metadata Repository and to provide metadata authors with suggestions.
We extended the Metadata Editor to use the Value Recommender service to suggest appropriate values for metadata fields during field entry (see Figure 2). Users entering metadata using the Metadata Editor are prompted in real time with drop-down lists, auto-completion suggestions, and verification hints supplied by the Value Recommender service. Recommendations for unfilled fields are updated in real time as users incrementally complete metadata acquisition forms. The editor presents a drop-down list for value-recommended fields containing suggested values ranked in order of likelihood. The editor can also be configured to indicate whether suggested values are ontology terms, in which case it also shows the BioPortal acronym for that ontology.
Figure 2.
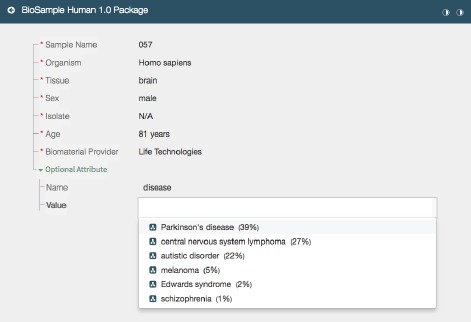
Screen shot of the CEDAR Metadata Editor showing recommended values for a particular field. In this case the editor shows suggestions for disease values. It presents a drop-down list containing suggested values ranked in order of likelihood. Ontology-based terms are indicated with an ontology icon. The relevancy score for each suggested value is presented as a percentage.
Evaluation
We analyzed the performance of our framework when suggesting appropriate metadata values using both plain text metadata and metadata represented using ontology terms. We constructed an evaluation pipeline to drive the analysis workflow (see Figure 3). The main steps of our evaluation workflow are as follows.
Figure 3.
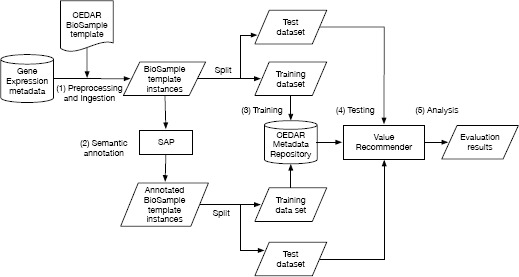
Evaluation workflow. (1) Design a template for the BioSample repository, and populate it with metadata from the Gene Expression Omnibus (GEO); (2) Annotate the template instances obtained with terms from biomedical ontologies; (3) Upload the training set to the CEDAR Workbench; (4) For each of the test instances, generate suggestions for the disease, sex, and tissue fields; (5) Compare the suggestions obtained using the Value Recommender with the suggestions obtained using the baseline method.
1. Preprocessing and ingestion
We used the CEDAR Workbench to design a metadata template targeted to the BioSample metadata repository.19 This repository, which is provided by the National Center for Biotechnology Information (NCBI), captures descriptive information about biological materials used in scientific experiments. BioSample defines several packages that represent specific types of biological samples, and specifies the list of attributes by which each sample should be described. The BioSample Human package,20 for example, is designed to capture metadata from studies involving human subjects, and includes attributes such as tissue, disease, age, and treatment. We used this package specification to develop a BioSample template in CEDAR to describe human samples.
For the purpose of our evaluation, we populated BioSample template instances using metadata from the Gene Expression Omnibus (GEO),21 a database of gene expression data which contains experimental metadata largely authored by original data submitters. The GEO database currently contains over 2 million records, and includes over 80,000 studies, each of which contains metadata for related biological samples.
We downloaded metadata from the GEO repository using GEOmetadb,22 and extracted all corresponding metadata elements for all human samples. We chose the fields title, sample_id, series_id, status, submission_date, last_update, type, sources_name, organism, and characteristics (including disease and tissue). Then, we picked the human samples that contained both disease and tissue metadata (35,157 samples), and transformed them into BioSample template instances conforming to CEDAR’s JSON-based model.23
2. Semantic annotation
We define semantic annotation (or simply annotation) as the process of finding a correspondence or relationship between a term in plain text and an ontology term that specifies the semantics of the term in plain text. We used a component of the CEDAR system, called the Semantic Annotation Pipeline (SAP),24 to automatically annotate all the fields values in 35,157 BioSample instances using biomedical ontologies. Table 2 shows the BioSample fields used in our evaluation. It presents both the number of plain text values and the number of ontology terms resulting from the semantic annotation process. The annotation ratio represents the mean number of plain text values per ontology term. For instance, we observed that the concept female was represented in plain text using values such as female, Female, f, F, and FEMALE.
Table 2.
Comparison between the number of plain text values for the fields disease, sex, and tissue, and the number of ontology terms resulting from applying our Semantic Annotation Pipeline (SAP) to the plain text values.
| Field | Description | Plain text | Ontology terms | Annotation ratio | ||
|---|---|---|---|---|---|---|
| Values | Examples | Values | Examples (preferred labels) | |||
| disease | Disease diagnosed | 1,064 | Lung carcinoma, carcinoma of lung | 261 | lung carcinoma | 4.07 |
| sex | Sex of sampled organism | 16 | female, Female, f, F, FEMALE | 2 | female | 8 |
| tissue | Type of tissue the sample was taken from | 604 | liver, Liver, liver tissue, liver biopsy, Liver biopsy tissue | 171 | liver | 3.53 |
3. Training
We partitioned the sample data—both for plain text values and for values annotated with ontology terms—into two sets. We used 80% (28,126 instances) of the sample data for training and 20% (7,031 instances) of the data for testing. We uploaded the training set to the CEDAR Workbench using the CEDAR API. We then indexed the training set with Elasticsearch.
4. Testing
For each of the test instances, we used the Value Recommender to generate value recommendations for the disease, sex, and tissue fields. The Value Recommender suggested values for each field based on the values of the other fields (e.g., suggestions for tissue were generated using the values for disease and sex). We used the majority vote as our baseline, which means picking the value with more occurrences in the training data. This process differs from the Value Recommender in that it ignores co-occurring values. For each field, we compared the suggestions provided by both the Value Recommender and the baseline with the expected value. We considered that the expected value for a field was the value for the field contained in the instance.
The Value Recommender produces a list of suggested values ranked by relevance. We assessed the performance of our method using the mean reciprocal rank (MRR) statistic. This statistic is commonly used for evaluating processes that produce a list of possible responses to a query, ordered by probability of correctness. We limited the output to the top three recommendations, and calculated the reciprocal rank (RR) as the multiplicative inverse of the rank of the first correct recommendation.
For example, if the correct value were ranked in the 3rd position, the reciprocal rank would be calculated as 1/3. Then, MRR was calculated as the mean of all RRs obtained. Table 3 shows the RR statistic for some example recommendation results. The MRR of the three values suggested in the table can be calculated as the mean of the RR value for all three rows: (1 + 1/2 + 1/3) / 3 = 0.61.
Table 3.
Example of recommended values and reciprocal rank (RR) for the disease field.
| Expected value | Recommended values | RR |
|---|---|---|
| asthma | 1) asthma | 1 |
| 2) lung cancer | ||
| 3) lipid metabolism disorder! | ||
| lymphoma | 1) rheumatoid arthritis | 1/2 |
| 2) lymphoma | ||
| 3) acute myeloid leukemia | ||
| lung cancer | 1) rheumatoid arthritis | 1/3 |
| 2) asthma | ||
| 3) lung cancer |
5. Analysis of results
Figure 4 compares the mean reciprocal ranks obtained using our recommendation framework with the majority value baseline. It shows results both for plain text values (left) and values annotated with ontology terms (right).
Figure 4.
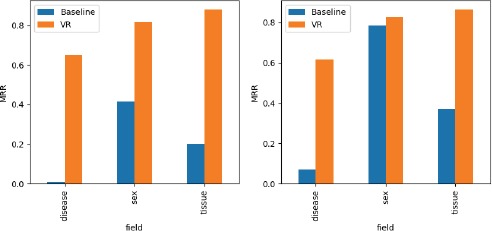
Mean reciprocal rank for BioSample instances with plain text values (left) and with ontology-terms (right), both for the baseline and the Value Recommender (VR).
The results indicate that our framework performs considerably better than the baseline for the three fields. Our context-sensitive recommendation method obtained an average MRR of 0.78 for plain text values, and 0.77 for ontology terms, compared to the baseline method’s average MRR of 0.21 for plain text values and 0.41 for ontology terms. By examining the results we see that, for example, our method correctly suggested asthma as a value for the disease field when the tissue was epithelium of bronchus. However, the baseline method suggested hepatocellular carcinoma, a disease that is not related to that kind of tissue. Our approach performs consistently well both for plain text values (0.78) and for ontology-based metadata (0.77). The importance of contextual information is particularly evident when analyzing the results obtained for plain text values, where there are substantially more values for each field than for ontology terms (for example, as shown in Table 2, there are 1,064 plain values versus 261 ontology terms for the disease field). The average MRRs of the baseline are considerably lower for plain text values (0.21) than for ontology terms (0.41).
Finally, we investigated more closely the effect of the context on the recommendations. The best results were obtained for the tissue field, with MRRs of 0.88 for plain text and 0.86 for ontology terms, illustrating the strong influence of the context on that field. Once disease and sex field values are provided, our approach is able to identify the appropriate value for the tissue field in most cases. Figure 5 shows the top suggestions provided by the Value Recommender for the disease field, with increasing levels of context. The figure shows how lung cancer can be much more clearly suggested as a likely choice when sex and tissue have been specified. This example reflects the increase in discrimination that is possible when more contextual information is available.
Figure 5.
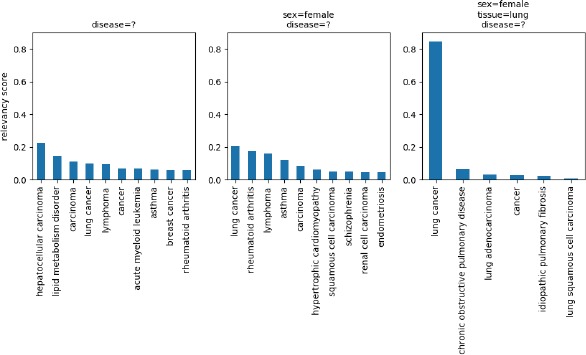
Top 10 suggestions provided by the Value Recommender for the disease field of the BioSample template, with increasing levels of context. The last plot shows 6 different values because only 6 suggestions were returned.
Discussion
We developed and deployed a metadata recommender service as part of an end-to-end metadata management system called the CEDAR Workbench. We found significant improvements could be obtained by considering contextual information when making recommendations. Our evaluation suggests that, by adding our recommendation capabilities on top of already-offered user interface optimizations, the CEDAR Workbench can provide major enhancements in both speed of metadata creation, and accuracy of those metadata.
A limitation of our evaluation is that we did not use all values in GEO, restricting our analysis to human samples only. We plan to carry out further analyses using all samples in GEO to determine how our method generalizes to additional sample types. We also plan to perform similar analyses on data from the BioSample repository, which contains metadata on 2,787,750 public biological samples used in scientific experiments. Additionally, we plan to study how the metadata recommender service performs with templates that contain a greater number, and more diverse fields. While disease, sex, and tissue are relevant metadata for biological samples, they are a small and relatively simple set of fields and generalizability may be a challenge.
The Value Recommender system is the first of a planned set of intelligent authoring components in the CEDAR system. Future efforts will concentrate on deeper analyses of metadata to discover more complex relationships among metadata fields, which will then drive tools to assist users when entering metadata. As a first step, we plan to extend the recommender to derive and use more in-depth knowledge of correlations among values in the dataset. Specifically, we will apply our previous research on association rule mining25 to identify degrees of correlation among metadata items. This approach will strengthen the positive associations that our current recommendation engine provides, and will allow us to point out possible errors by identifying unlikely values. With sufficient levels of accuracy, our system may be used to automatically fill in missing values for a significant number of metadata fields.
Our work also has implications for scientists focused on retrospective augmentation of metadata. For example, the system could be used to interactively help curate previously submitted data by suggesting more specific values for populated fields, in addition to suggesting values for empty fields. It could also assist curators with suggestions for correcting incorrectly entered element values. In particular, the system could be targeted to both retrospective and real-time quality assurance by detecting unlikely field combinations. Strong discrepancies detected between the element value entered by a user and the predicted value could be highlighted to human curators for review. By rapidly providing highly interactive recommendation, the system could also help curators quickly deal with greater volumes of metadata submissions, and help address the problem of curation scalability faced by many repositories.
Conclusion
We have described the development of a recommendation framework that focuses on helping biomedical investigators annotate their experimental data with high quality metadata. The framework takes advantage of associations among the values of multiple fields in existing metadata to recommend context-sensitive metadata values. A key focus is on interoperation with ontologies. Using formal ontology-based specifications and interactive look-up services linked to the BioPortal ontology repository,26 the system tunes its recommendations to target controlled terminologies. We outlined an initial evaluation of the framework using metadata from the GEO repository,21 and provided an implementation of the system in a metadata management system called the CEDAR Workbench.
These tools aim to provide a series of intelligent authoring functions that lower the barrier to the creation and population of metadata templates, and help ensure that the resulting metadata acquired using these templates is of high quality. The ultimate goal is to provide the ability for investigators to easily create metadata that are comprehensive, standardized, and make the corresponding data sets conform to FAIR principles.27
Acknowledgements
CEDAR is supported by the National Institutes of Health through an NIH Big Data to Knowledge program under grant 1U54AI117925. NCBO is supported by the NIH Common Fund under grant U54HG004028. The CEDAR Workbench is available at https://cedar.metadatacenter.net and on GitHub (https://github.com/metadatacenter).
References
- 1.Roche DG, Kruuk LEB, Lanfear R, Binning SA. Public Data Archiving in Ecology and Evolution: How Well Are We Doing? PLoS Biol. 2015;13(11) doi: 10.1371/journal.pbio.1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 3.Borgman CL. The conundrum of sharing research data. J Am Soc Inf Sci Technol. 2012;63(6):1059–1078. [Google Scholar]
- 4.Brazma A. Minimum Information About a Microarray Experiment (MIAME)--successes, failures, challenges. Sci World J. 2009;9:420–423. doi: 10.1100/tsw.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McQuilton P, Gonzalez-Beltran A, Rocca-Serra P, et al. BioSharing: curated and crowd-sourced metadata standards, databases and data policies in the life sciences. Database J Biol databases curation. 2016;2016 doi: 10.1093/database/baw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bui Y, Park J-R. An assessment of metadata quality: A case study of the National Science Digital Library Metadata Repository. Proc CAIS/ACSI 2006. 2006;13 [Google Scholar]
- 7.Tenopir C, Allard S, Douglass K, et al. Data sharing by scientists: Practices and perceptions. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musen MA, Bean CA, Cheung KH, et al. The center for expanded data annotation and retrieval. J Am Med Informatics Assoc. 2015;22(6):1148–1152. doi: 10.1093/jamia/ocv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Araujo S, Gao Q, Leonardi E, Houben GJ. Carbon: Domain-independent automatic web form filling. Lect Notes Comput Sci. 2010;6189 LNCS:292–306. [Google Scholar]
- 10.Toda GA, Cortez E, Silva AS Da, Moura E De, Silva da AS, Moura de E. A Probabilistic Approach for Automatically Filling Form-Based Web Interfaces. Proc VLDB Endow. 2010;4(3):151–160. [Google Scholar]
- 11.Wolstencroft K, Owen S, Horridge M, et al. RightField: Embedding ontology annotation in spreadsheets. Bioinformatics. 2011;27(14):2021–2022. doi: 10.1093/bioinformatics/btr312. [DOI] [PubMed] [Google Scholar]
- 12.Shankar R, Parkinson H, Burdett T, et al. Annotare-a tool for annotating high-throughput biomedical investigations and resulting data. Bioinformatics. 2010;26(19):2470–2471. doi: 10.1093/bioinformatics/btq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson H, Sarkans U, Shojatalab M, et al. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33(Database issue):D553–D555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocca-Serra P, Brandizi M, Maguire E, et al. ISA software suite: Supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26(18):2354. doi: 10.1093/bioinformatics/btq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panahiazar M, Dumontier M, Gevaert O. Context Aware Recommendation Engine for Metadata Submission. Proc 1st Int Work Cap Sci Know (collocated with K-CAP ’15). 2015:3–7. [Google Scholar]
- 16.Posch L, Panahiazar M, Dumontier M, Gevaert O. Predicting structured metadata from unstructured metadata. Database (Oxford). 2016;2016:1–9. doi: 10.1093/database/baw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schriml LM, Arze C, Nadendla S, et al. Disease ontology: A backbone for disease semantic integration. Nucleic Acids Res. 2012;40(D1) doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Romero M, O’Connor MJ, Dorf M, et al. Supporting ontology-based enrichment of biomedical metadata in the CEDAR Workbench. Proc Int Conf Biom Ont. 2017 [Google Scholar]
- 19.Barrett T, Clark K, Gevorgyan R, et al. BioProject and BioSample databases at NCBI: Facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40(D1) doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.BioSample Human Package. The National Center for Biotechnology Information. version 1.0. [Accessed March 8, 2017]. https://www.ncbi.nlm.nih.gov/biosample/docs/packages/Human.E0/
- 21.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Davis S, Stephens R, Meltzer PS, Chen Y. GEOmetadb: Powerful alternative search engine for the Gene Expression Omnibus. Bioinformatics. 2008;24(23):2798–2800. doi: 10.1093/bioinformatics/btn520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connor MJ, Martinez-Romero M, Egyedi AL, Willrett D, Graybeal J, Musen MA. An open repository model for acquiring knowledge about scientific experiments. Proc 20th Int Conf Knowl Eng Knowl Manag (EKAW). 2016;10024:762–777. [Google Scholar]
- 24.Shankar R, Martinez-Romero M, Connor MJO, Graybeal J, Khatri P, Musen MA. SAP - A CEDAR-based pipeline for semantic annotation of biomedical metadata. BD2K All-Hands Meet. 2016 [Google Scholar]
- 25.Panahiazar M, Dumontier M, Gevaert O. Predicting Biomedical Metadata in CEDAR: a Study of Gene Expression Omnibus (GEO) J Biomed Inform. 2017 doi: 10.1016/j.jbi.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whetzel PL, Noy N, Shah N, et al. BioPortal: Ontologies and integrated data resources at the click of a mouse. CEUR Workshop Proc. 2011;833:292–293. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson MD, Dumontier M, Aalbersberg IjJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]


