Abstract
While biomedical ontologies have traditionally been used to guide the identification of concepts or relations in biomedical data, recent advances in deep learning are able to capture high-quality knowledge from textual data and represent it in graphical structures. As opposed to the top-down methodology used in the generation of ontologies, which starts with the principled design of the upper ontology, the bottom-up methodology enabled by deep learning encodes the likelihood that concepts share certain relations, as evidenced by data. In this paper, we present a knowledge representation produced by deep learning methods, called Medical Knowledge Embeddings (MKE), that encode medical concepts related to the study of epilepsy and the relations between them. Many of the epilepsy-relevant medical concepts from MKE are not yet available in existing biomedical ontologies, but are mentioned in vast collections of epilepsy-related medical records which also imply their relationships. The evaluation of the MKE indicates high accuracy of the medical concepts automatically identified from clinical text as well as promising results in terms of correctness and completeness of relations produced by deep learning.
Introduction
Over the past two decades, the biomedical research community has increased its efforts to produce ontologies encoding biomedical knowledge, justified by the steady increase in biological and biomedical research and the growth of data that is being collected in all areas of biology and medicine. Not only is the number of ontologies increasing and their size growing, but their relevance in biomedical research is also rising as they contribute to the interpretation of the biomedical data and enable complex inference from their encoding. The BioPorta* of the National Center for Biomedical Ontology (NCBO) is the most comprehensive repository of biomedical ontologies in the world (as of this writing it includes 541 ontologies, with almost 8 million classes and almost 40 million indexed records). Many of the ontologies available from the BioPortal became widely used resources, e.g. the Gene Ontology1 (GO), one of the most important resources available in genomics research. The survey published in Huang et al. (2009)2 discusses 68 bioinformatics enrichments tools informed by GO, that have played a very important and successful role contributing to the gene functional analysis of large gene lists for various high-throughput biological studies, evidenced by thousands of publications citing these tools. Moreover, Lependu et al. (2011)3 showed that it is possible to create reference annotation sets for enrichment analysis† using other ontologies than the GO, still available from BioPortal, for example, the Human Disease Ontology (DO). As reported in Noy et al. (2009)4, the ontologies in the BioPortal are publicly available in several formats, including OWL, RDF, OBO format or the Protege frame language. As such, they follow the principles of the OBO Foundry5, forming graph-theoretic structures, with concepts connected by edges representing relations such as ‘Is-A’ or ‘Part-Of’ or others from the OBO Relation Ontology (RO), generating well-principled ontologies for many biomedical domains.
Recently, a new ontology was added in the BioPortal, namely the Epilepsy Syndrome and Seizure Ontology‡ (ESSO), encoding 2,705 classes with an upper ontology targeting epilepsy as a disease and designed to be machine readable and to allow for federated queries across distributed databases and patient data capturing systems. The availability and development of the ESSO ontology answers the recommendations of the Institute of Medicine report6 for promoting the understanding of epilepsy by increasing the power of data in comprehensive, timely, and accurate epilepsy surveillance. Because epilepsy affects an estimated 2.2 million people in the United States, it is one of the most common neurological disorders. However ontological resources for this disorder are only now starting be become available to biomedical researchers. Nevertheless, large clinical datasets relevant to epilepsy are also becoming available. For example, the Temple University Hospital (TUH) EEG Corpus7 assembles over 25,000 sessions of electroencephalography (EEG) of 15,000 patients collected over 12 years. Clinical electroencephalography is an electrophysiological monitoring method used to record electrical activity of the brain, representing the most important investigation in the diagnosis and management of epilepsies. As expected, EEG reports contain a wealth of epilepsy-related knowledge, derived from clinical practice. This knowledge is expressed by clinical language used in the reports and it explicitly mentions many of the concepts that can be linked to the ESSO ontology. Moreover, many implicit relations between these concepts can be inferred from the EEG reports. While biomedical ontologies have traditionally been used to guide the identification of concepts or relations in biomedical data, recent advances in deep learning were able to capture knowledge from textual data enabling an alternative knowledge representation.
This alternate knowledge representation, known as knowledge embeddings (KE), incorporates deep learning to model the interactions between concepts and relations and generate graphical knowledge structures. Knowledge embeddings are defined as multidimensional continuous vector representations of concepts and their relations. The KE methods were inspired, as reported in Weston et al. (2013)8, by the work of Craven et al. (1999)9, which matched the Yeast Protein Database with PubMed abstracts.
In this paper, we aim to investigate the medical knowledge embeddings (MKE) automatically learned from the TUH EEG corpus, encoding multiple EEG findings (e.g. EEG events and activities), associated medical problems, and treatments. Unlike the top-down methodology used in the generation of ontologies, which starts with the principled design of the upper ontology, the bottom-up methodology enabled by deep learning observes the likelihood that concepts share certain relations, as evidenced by data. Specifically, whereas the edges in an ontology graph represent hand-coded “hard” relations between entities, the edges in the knowledge graph are “softer”, i.e. probabilistic in nature. Unlike concepts and relations encoded in the BioPortal ontologies, the MKE associate relations between medical concepts with a probability or likelihood, enabling a probabilistic representation of biomedical knowledge. Thus, the MKE are able to account for the variability and inconsistencies in the way this knowledge is expressed in natural language by assigning more plausible relations a higher likelihood. While previous KE were generated from human curated knowledge bases, our work is unique in that we automatically extract entities and relations from free text in a data-driven approach. To the best of our knowledge, this is the first report of an the development of an embedded medical knowledge graph using free text clinical records.
We learned MKE representing 1,195,927 instances of binary relations between epilepsy-related concepts. These relations involved 2,442 instances of medical concepts. We evaluated the MKE by (1) the quality of the medical concepts identified in EEG reports; (2) assessing the plausibility of the potential relations discovered in EEG reports as well as (3) measuring the knowledge completeness as a form of link prediction10. We believe that the MKE encode medical knowledge that is complimentary to the knowledge available in traditional ontologies and can be used (1) to provide data-driven knowledge that can be linked to ontologies from BioPortal, and (2) as a potential mechanism for enriching existing ontologies using the learned concepts, relations, and probabilities.
Background
Recently, qualified medical knowledge graphs (QMKGs) automatically discerned from medical records have been used successfully in a system designed for patient cohort identification11,12. As reported in Goodwin & Harabagiu (2013)13 the QMKG was generated using big-data techniques applied to a large set of clinical records available to the participants in the TREC Medical Records track (TRECMed), a task developed in 2011 and 2012 as an Information Retrieval challenge pertinent to real-world clinical medicine and evaluated in the annual TExt Retrieval Conference (TREC) hosted by the National Institute for Standards and Technology (NIST). In another TREC special track on Clinical Decision Support (TREC-CDS), the system reported in Goodwin & Harabagiu (2016)14 used a knowledge representation as a Clinical Picture and Therapy Graph (CPTG) which was automatically acquired from the MIMIC-III15 clinical database. The TREC-CDS has addressed the challenge of retrieving bio-medical articles relevant to a medical case when answering one of three generic medical questions: (a) “what is the diagnosis?”; (b) “what test(s) should be ordered?”; and (c) “which treatment(s) should be administered?”. The system described in Goodwin & Harabagiu (2016)14 answered these types of questions by relying on a medical knowledge representation as a factorized Markov network16, suited ideally for answer inference.
Medical knowledge embeddings (MKE) enable a new probabilistic knowledge representation which is differs from the QMKG and the CPTG because (1) the relationships are not informed only by cohesive properties of texts, but by patterns of interactions between medical concepts, as captured by deep learning methods; and (2) similar medical concepts and relations share the same neighborhoods in the multi-dimensional space enabled by the knowledge embeddings. The latter property resolves semantic heterogeneity which arises when disparate terminology is used to refer to the same concepts or relations while identical terms may refer to distinct concepts. As noted in Sahoo et al. (2014)17 a seizure with alteration of consciousness may be referred to as complex partial seizure, dialeptic seizure or focal dyscognitive seizure by different epilepsy experts. An MKE representation should place all these expressions in a similar location of the multi-dimensional space, as it learns that they are involved in the similar relations with other epilepsy-relevant concepts. Thus, unlike the Epilepsy and Seizure Ontology17 (EpSO), the MKE representation does not require reconciliation of semantic heterogeneity, while being used for retrieving patient cohorts from medical records18.
Data
In this work, we used the EEG reports publicly available from the Temple University Hospital (TUH), comprising over 25,000 EEG reports from over 15,000 patients collected over 12 years. Following the American Clinical Neurophysiology Society Guidelines for writing EEG reports, the reports from the TUH EEG Corpus start with a clinical history of the patient, including information about the patient’s age, gender, and conditions prevalent at the time of the recording followed by a list of the medications the patient is currently taking that might modify the EEG (e.g. “Keppra”, “Lamictal”).Both initial sections depict the clinical picture of the patient, containing a wealth of medical concepts, including the medical problems (e.g. “seizures”), signs, and symptoms(e.g. “loss of consciousness”) as well as significant medical events which may be temporally grounded (e.g. “2 years ago”). The following sections of the EEG report target mostly information related to the EEG techniques, findings and interpretation. The introduction section describes the techniques used for the EEG (e.g. “digital video routine EEG”, “standard 10-20 electrode placement system with additional anterior temporal and single lead EKG”), as well as the patients conditions prevalent at the time of the recording (e.g., fasting, sleep deprivation), level of consciousness (e.g. “during wakefulness”), and possible activating procedures that were performed (e.g. “hyperventilation”). The description section is the mandatory part of the EEG report, and it provides a complete and objective description of the EEG signal, noting all observed activity (e.g. “frontocentral beta activity”), patterns (e.g. “K-complexes”) and events (e.g. “eye opening”). The impression section states whether the EEG test is normal or abnormal (i.e. indicating some form of cerebral dysfunction). If the impression is abnormal, then the abnormalities are listed in order of importance. The clinical correlation section explains what the EEG findings mean in terms of clinical interpretation (e.g. “findings are consistent with idiopathic generalized epilepsy”).
Methods
Bottom-up knowledge acquisition methods rely on the automatic identification of concepts and relations from data to enable (i) the population of the knowledge representation and (ii) linking the acquired knowledge to existing ontologies. In learning medical knowledge embeddings (MKE) from EEG reports we do not only perform bottom-up acquisition of medical knowledge from EEG reports, but we also represent the knowledge probabilistically in a multi-dimensional space and perform inference on it. To do so, we followed a methodology which involves the following four steps:
STEP 1: Decide which medical concepts and which relations between them are expressed in the EEG reports;
STEP 2: Automatically generate the Knowledge Graph by extracting medical concepts and relations from the EEG reports; STEP 3: Learn Medical Knowledge Embeddings (MKE) from the associated Knowledge Graph;
STEP 4: Perform inference with MKE.
It is to be noted that the the MKE represent only knowledge available from the EEG reports, which do not discuss the taxonomic organization of medical concepts or their partonymy relations. These forms of relations are encoded in medical ontologies, thus the MKE provide complementary knowledge to medical ontologies. However, many of the concepts represented in the MKE are also encoded in existing medical ontologies, providing a simple mechanism of linking the MKE to various ontologies available in BioPortal. For example, the clinical history and the medication list of EEG reports mention multiple medical concepts already encoded in the Unified Medical Language System (UMLS)19 ontology:
Example 1: CLINICAL HISTORY: This is a 20-year-old female with history of seizures described as generalized tonicclonic with loss of consciousness for a few minutes. Last seizures occurred 2 years ago.
MEDICATIONS: Keppra and Lamictal.
Medical problems such as seizures, and treatments such as “Keppra”, “Lamictal” are encoded in UMLS while concepts such as idiopathic generalized epilepsy will be linked both to UMLS and the ESSO ontology. However, these ontologies do not capture relations between such concepts that are implied in the EEG reports, e.g. which brain activities evidence some epilepsyspecific medical problems. Our four-step methodology aims to capture and represent such relationships, while also providing their probabilistic likelihood, learned automatically from the medical practice evidenced in the large corpus of EEG reports.
STEP 1: Decide which medical concepts and relations between them are expressed in EEG reports
In addition to medical problems and treatments that describe the clinical picture and therapy of a patient, EEG reports mention EEG events, which represent stimuli that activates the EEG (e.g. hyperventilation) and EEG activities, representing brain waves or sequences of waves20. The section of the EEG reports describing the EEG record mention a multitude of EEG activities and events recognized by the neurologist from the analysis of the EEG signal. The following example illustrates mentions of EEG events such as photic stimulation and eye opening, while mentions of EEG activities are beta activity and polyspike discharges:
Example 2: DESCRIPTION OF THE RECORD: …the alpha rhythm was 9-10 Hz infrequency seen in the occipital region, which attenuates with eye opening. … Photic stimulation was performed at multiple flash frequencies and results in a symmetric driving response without any photoparoxysmal response.
EEG activities are also mentioned in the impression section and in the clinical correlation section. Thus we decided to encode in the MKE four types of medical concepts: (1) EEG events; (2) EEG activities; (3) medical problems and (4) treatments. Whenever these concepts are also encoded in other ontologies, we linked to them. For example, medical problems such as idiopathic generalized epilepsy, when identified in an EEG report, with methods developed in the STEP 2 of our methodology, shall be linked to UMLS through its concept unique identifier (CUI). In addition to these four types of concepts, we decided to discern four types of binary relations that are implicit in the EEG reports. Each of these relations operates between a source argument and a destination argument. The relations along with examples of the four types of medical concepts are illustrated in Figure 1. The four binary relation types that we considered were motivated by discussions with several practicing neurologists and surgeons, corresponding to the implicit knowledge they discern from EEG reports. As shown in Figure 1, the EVIDENCES binary relation always has a medical problem as its destination concept, which is always mentioned in the clinical correlation section of the EEG report. The following example shows how the medical problem idiopathic generalized epilepsy, is evidenced by findings such as polyspike discharges, which is a mention of an EEG activity, in the impression section:
Example 3: IMPRESSION: This is an abnormal EEG recording capturing wakefulness through stage II sleep due to generalized spike and wave and polyspike discharges seen during wakefulness.
CLINICAL CORRELATION: The above findings are consistent with idiopathic generalized epilepsy.
Figure 1:
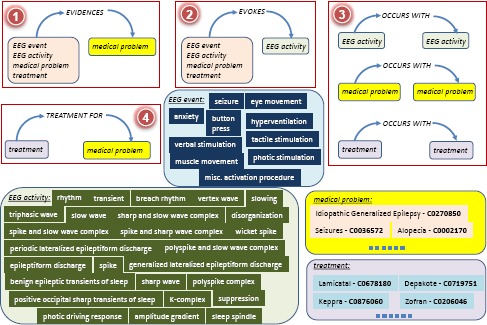
Medical concepts and relations considered forMedical Knowledge Embeddings (MKE)
As shown in Figure 1, the EVIDENCES relation considers EEG events, EEG activities, treatments, and medical problems as providing evidence for the medical problem from the clinical correlation section of the EEG report. The EVOKES binary relation always has an EEG activity as a destination concept, as it attempts to capture the medical concepts that evoke the respective EEG activity. Those medical concepts can be either EEG events, or other EEG activities, medical problems or treatments followed by the patient. The third relation, namely OCCURS-WITH constraints both its arguments to be of the same type, e.g. either EEG activities, medical problems or treatments. The TREATMENT-FOR relation captures the treatments prescribed for certain medical problems. Table 1 illustrates examples of each of the four relations we considered, involving medical concepts illustrated in Figure 1, which lists all the EEG events and EEG activities that we decided to encode in the MKE, while providing several examples of medical problems and treatments, along with their UMLS CUIs. We used the vocabularies of EEG Activities and EEG Events from Maldonado et al. (2017)21 based on the International Federation of Clinical Neurophysiology’s glossary of terms20.
Table 1:
Examples of the Relations and Concepts expressed in EEG reports.
| Evidences | Evokes |
|---|---|
| 〈seizures, EVIDENCES, idiopathic generalized epilepsy〉 | 〈photic stimulation, EVOKES, photic driving response〉 |
| 〈polyspike discharges, EVIDENCES, idiopathic generalized epilepsy〉 | 〈hyperventilation, EVOKES, slowing〉 |
| 〈facial grimacing, EVIDENCES, psychogenic seizure〉 | 〈seizures, EVOKES, periodic lateralized epileptiform discharge〉 |
| 〈toxoplasmosis, EVIDENCES, degenerative brain disorder〉 | 〈shaking, EVOKES, rhythm〉 |
| Treatment For | Occurs With |
| 〈lamictal, TREATMENT-FOR, idiopathic generalized epilepsy〉 | 〈keppra, OCCURS-WITH, lamictal〉 |
| 〈depakote, TREATMENT-FOR, generalized anxiety disorder〉 | 〈encephalopathies, OCCURS-WITH, occipital lobe epilepsy〉 |
| 〈dilantin, TREATMENT-FOR, hematoma, subdural, chronic〉 | 〈cerebral dysgenesis, OCCURS-WITH, recurrent convulsions〉 |
| 〈ampicillin, TREATMENT-FOR, infection of foot〉 | 〈spike and slow wave complex, OCCURS-WITH, polyspike complex〉 |
STEP 2: Automatically generate the Knowledge Graph by extracting medical concepts and relations from the EEG reports
The extraction of medical knowledge from EEG reports consists of (1) automatic identification of medical concepts and (2) binary relation detection. Medical concept identification aims to recognize all the four types of concepts mentioned in EEG reports, along with their inferred polarity and modality. For identifying polarity of medical concepts in EEG reports, we considered that each concept can have either a negative or a positive polarity, depending on whether the medical concept was negated or not in the text. The recognition of the modality, as in Maldonado et al. (2017)21 uses the modality values of factual, possible, and proposed to indicate that medical concepts mentioned in the EEG reports are actual findings, possible findings and findings that may be true at some point in the future, respectively. Through the identification of modality and polarity of the clinical concepts, we aimed to capture the neurologists beliefs about the clinical concepts mentioned in the EEG report. Thus our medical concept identification method needed also to qualify the concepts by their polarity and modality.
Medical Concept Identification was performed by taking advantage of our existing active deep learning methodology21, which is illustrated in Figure 2. This methodology first uses two stacked Long Short-Term Memory (LSTM) networks for detecting the boundaries of medical concepts in the text of the EEG report. The task of identifying spans of text that correspond to mentions of medical concepts is called boundary detection. We relied on one stacked LSTM to identify the boundaries of EEG activities and another stacked LSTM to identify the boundaries of EEG events, medical problems and treatments. The motivation for using two separate stacked LSTM networks is determined by the fact that different features are used to identify the boundaries of EEG activities than boundaries of the other three types of medical concepts, as detailed in Maldonado et al. (2017)21. Once the boundaries of each medical concepts are known, two Deep Rectified Linear Networks (DRLNs) are used to (a) identify the medical concepts and (b) discern their polarity and modality. EEG activities are identified only with their polarity, as their mentions are always factual, as illustrated in Figure 2. Moreover, medical problems and treatments are both normalized into UMLS concepts using MetaMap Lite22.
Figure 2:
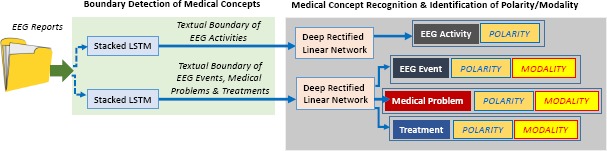
Deep Learning Architectures used for Recognizing Qualified Medical Concepts from EEG Reports.
Detecting Relations between Medical Concepts was possible when pairs of medical concepts identified in the same EEG report were considered. Specifically, we established the four types of relations illustrated in Figure 1 by considering: (1) a potential EVIDENCES relation between any medical concepts from an EEG report and a medical problem identified in its clinical correlation section; (2) a potential EVOKES relation between any medical concept and an EEG activity, provided that the treatments were not identified in the clinical correlation section, as they may indicate possible or recommended treatments; (3) a potential OCCURS-WITH relation between pairs of EEG activities, medical problems and treatments that are identified in the same section of the EEG report; and (4) a potential TREATMENT-FOR relation between any treatment and a medical problem identified in the history section of the EEG report. We discard potential relations involving medical concepts with “negative” polarities and “possible” modalities since these medical concepts, while mentioned, were not actually observed. All these potential relations are indicative of implied relations, that are not always directly stated in the text of the EEG report.
Taken together, the set of medical concepts extracted from the entire TUH EEG corpus along with the collection potential relations between them constitute a Knowledge Graph, G = {V,E} where V is the set of graph vertices and E is the set of graph edges. In our knowledge graph, V is the set of medical concepts and E is the set of relations between them. The medical knowledge embeddings learned in the following step will provide the likelihood of any example of one of these relations.
STEP 3: Learning medical knowledge embeddings (MKE) from the associated knowledge graph
Learning MKE is made possible by relying on the TransE10 method, widely used23-25 for representing multi-relational data corresponding to concepts and relations by modeling concepts as points in a continuous vector space, ℝN, called the embedding space, where N is a parameter indicating the dimensionality of the embedding space. In our use of the TransE framework, relations between medical concepts are represented as translation vectors, also in ℝN, that connect the two points representing the two medical concepts in the embedding space. TransE learns an embedding, ci, for each concept c® and an embedding, r, for each relation type r such that the relation embedding is a translation vector between the two concept embeddings representing its arguments. This means that for any medical concept c®, the concept most likely to be related to c® by the relation r should be the medical concept whose embedding is closest to in the embedding space. By modeling the medical concepts as points in the embedded space and the relations between them as translation vectors, we can measure the plausibility of any potential relation between any pair of concepts using the geometric structure of the embedding space. The plausibility of a relation between a source medical concept and a destination medical concept, represented as a triple, 〈cs, r,cd〉, is inversely proportional to the distance in the embedding space between the point predicted by our model and the point in the embedding space representing the destination argument of the relation, i.e. . In this work, we use Manhattan Distance as our distance function:
| (1) |
where ||·||L1 is the L1 norm. Using this distance function, plausible triples have low value of f (since for plausible triples) and implausible triples have a high value of f.
Neural Network Architecture for learning MKE. To learn the optimal points and translation vectors, we use a neural network that will in fact produce the MKE. Formally, let C be the set of medical concepts found in the EEG reports and L be the set of relation types. Let be the set of m relation triples extracted from the corpus of EEG reports at Step 2; where each is a medical concept and each ri ∂ L is a relation type. The embedding, , for a concept is calculated by first generating a one-hot vector representation of given by v() which is a |C|-dimensional vector of zeros with a one in the dimension corresponding to the index of the concept in the set of concepts C. The embedding is derived by multiplying the one-hot vector v() with the embedding matrix . Each row of E corresponds to a medical concept embedding and the operation v()E corresponds to selecting the v()th row of E. Likewise, the embedding for a relation type ri is given by where w(ri) maps ri to a one-hot vector of size |L| and R is the relation embedding matrix. Consequently, Equation 1 can be computed using:
| 2 |
To learn useful embeddings we must also define a training objective that encodes useful relationships. Inspired by the work of Bordes et al. (2011)26, we use the following training objective: if either the source argument or destination argument from a training triple is removed, the model should be able to correctly predict the correct medical concept. For example, the model should ensure that value of f (keppra, TREATMENT-FOR, idiopathic generalized epilepsy) is less than the value of f (morphine, TREATMENT-FOR, idiopathic generalized epilepsy) since keppra is a treatment for idiopathic generalized epilepsy, but morphine is not. Formally, we wish to learn the values of E and R such that for any training triple , the following two constraints are met:
| (3) |
| (4) |
To learn the optimal embedding matrices E and R, we optimize the objective defined by the constraints outlined in Equations 3-4 by iterating the following process:
-
1.
Randomly select a training triple from X.
-
2.
Create a corrupted version of the triple by selecting a medical concept cneg at random from the set of medical concepts C and randomly replacing either or in xi. such that
-
3.
Update E and R by backpropagating the ranking margin loss23, max(0,γ+ f(xi)-, where γ is the margin parameter that determines how much of a margin should exist between triples in the training set and triples not in the training set.
4. Normalize each row e of E (i.e. )
This process is repeated for each triple in X a fixed number of iterations (200,000 in this work). Our collection of 1,195,927 relation triples extracted from the TUH EEG corpus consisted of |X| = 138,369 unique relation triples. It is important to note that, as reported in Bordes et al. (2013) 10, the normalization in the fourth step prevents the model from trivially minimizing the loss by artificially increasing entity embedding norms.
STEP 4: Performing Inference with the MKE graph
Inference from a knowledge base can be viewed as answering questions using its encoded knowledge. Answering questions like (Q1) “what is the most likely treatment for idiopathic generalized epilepsy?”, (Q2) “what EEG activity is most likely to occur with polyspike discharges?”, and (Q3) “what is the likelihood that a patient with background slowing is diagnosed with cerebral dysfunction?” requires the ability to perform probabilistic inference. The MKE can be used to perform probabilistic inference by (1) representing the question as a relation triple q and (2) measuring the plausibility of q using equation 1 with the embeddings matrices E and R automatically learned from the TUH EEG corpus. We estimated the probability of in terms of the geometric structure of the embedding space. Formally:
| (5) |
For example, answering (Q1) is the result of (Cs TREATMENT-FOR, idiopathic generalized epilepsy); answering (Q2) is the result of (polyspike discharges, OCCURS-WITH, cd); and answering (Q3) is the result of P(background slowing, EVOKES, cerebral dysfunction).
Experiments
To evaluate the MKE, we measure (a) the quality of the medical concepts that were extracted from the EEG reports as well as (b) the quality of the relations learned between them. When evaluating the medical concepts, we relied on the latest performance of our active deep learning annotation methodology21 and found that the quality of boundary detection of EEG activities had an F1-score of 0.9154 while the F1-score for detecting the boundaries of the other three forms of medical concepts was 0.9421. The identification of the medical concept type was performed with an F1-score of. 9532 and the polarity was detected with an accuracy of 0.978 while the modality was recognized with an accuracy of 0.973.
The relations represented in the MKE were evaluated in terms of (a) their plausibility; and (b) their completeness. The plausibility of relations encoded in MKE was assessed in three ways, measuring how well MKE rank triples from a test set T, of 1,000 relation triples held out from the data used to train the MKE. For each triple t in the test set, we randomly remove either the source or destination argument and produce a set of candidate triples by replacing the removed argument with every medical concept c ∈ C. We rank the candidate triples in ascending order according to the distance function f. This allows us to calculate the following metrics using the rankings produced from every triple in the test set:
Mean Reciprocal Rank (MRR) is a standard ranking evaluation that measures how high the first correct triple is ranked according to the model. refers to the rank of the first correct triple in the ranking, where a correct triple is defined as any triple from any of the training, validation, or tests sets.
Precision at 10 (P@ 10) is another standard ranking evaluation that measures the percentage of the top K ranked triples are correct. As with MRR, correct triples are defined as any triple from any of the training, validation, or tests sets. The Precision at 10 evaluation shows how well the MKE ranks the triples about which the model is most confident.
Hits at K (H@10, H@ 100) is a standard evaluation used for knowledge graph embeddings10,26 for evaluating link prediction. Hits at K measures how often the specific test triple t occurs in the K highest ranked triples, as opposed to precision which measures how often any correct triple occurs in the k highest ranked triples. We report both Hits at 10 and Hits at 100 to illustrate how often t is ranked among the most plausible triples, and how often t is ranked in the top 5% of triples.
The evaluation of completeness of the relations from the MKE also used the test set, T. We evaluated how well the MKE can infer new knowledge in the form of new relations from the test set. To measure how well the MKE can model relations of the held out triples from the test set, we consider each test triple, t ∂ T, and a corrupted version of the test triple, z, created by randomly replacing either the source argument or destination argument with a random medical concept and compute the Pairwise Plausibility Accuracy (PPA). The PPA measures the percentage of test triples for which the plausibility, of the test triple t is higher than plausibility, , of the corrupted triple. PPA demonstrates how well the MKE can differentiate between a correct, t, and an incorrect triple, z, even if the model had never encountered t. For these evaluations, the MKE were learned from 137,369 training triples automatically extracted from the TUH EEG corpus as described in the Methods section. We selected the dimension of the embedding space N=50 from [25,50,100,200] and the margin parameter γ =1.0 from [0.1,1.0,5.0,10.0] using grid search on a validation set of 500 relation triples.
Table 2 presents these results. The results for the Pairwise Plausibility Accuracy show that the MKE can correctly distinguish between relations that occur in the data (but that the model has not seen during training) and corrupted relations 88.95% of the time. The micro-averaged Mean Reciprocal Rank of 83.33% indicates that for the majority of triples in the test set, the top ranked candidate triple is correct. While the MRR of the OCCURS-WITH relation is the lowest (62.3%), it should be noted that, on average, there is at least one correct candidate triple ranked in the top two. The Precision at 10 metrics show that 66.73% of the top 10 ranked triples were correct, in general. It is interesting to note that the results for the Hits at 10 metric have the most variability between relation types. For the OCCURS-WITH relation, test triple, t, only occurs within the top 10 ranked triples 27.77% of the time. In contrast, for the EVOKES relation, t occurs within the top 10 ranked triples 84.91% of the time. In general, the Hits at 100 results show that the MKE correctly ranks t in the top 5% of candidate triples 81.3% of the time.
Table 2:
Quality of relations encoded in theMKE, measured using Pairwise Plausibility Accuracy (PPA), Mean Reciprocal Rank (MRR), Precision at 10 (P@10), Hits at 10 (H@10) and Hits at 100 (H@100).
| Relation Type | PPA | MRR | P@10 | H@10 | H@100 |
|---|---|---|---|---|---|
| EVIDENCES | 86:04% | 96:44% | 77:22% | 63:37% | 84:43% |
| EVOKES | 94:22% | 96:10% | 84:62% | 84:91% | 97:17% |
| OCCURS-WITH | 90:00% | 62:30% | 45:58% | 27:77% | 68:36% |
| TREATMENT-FOR | 82:89% | 83:78% | 72:28% | 45:18% | 80:70% |
| >MICRO-AVERAGED | 88:95% | 83:33% | 66:73% | 47:35% | 81:30% |
Discussion
To analyze the correctness of medical knowledge distilled from EEG reports in the MKE, we manually inspected the 30 most plausible triples for each relation type. Specifically, for each triple, we determined whether that triple is consistent with established medical knowledge. Many of the triples in the MKE encode general knowledge which is difficult to judge. For example, consider the triple (Dilantin, TREATMENT-FOR, disease). Determining whether or not dilatinin is a treatment for disease, necessitates considering additional context specifying the disease. In general, we found the EVOKES relation type to have the highest percentage of correct triples, highlighting the ability of the MKE to capture neurological experience from EEG reports. By contrast, the MKE successfully identified a number of unexpected Occurs-With relations, including (hypothyroidism, OCCURS-WITH, turner syndrome), and (infantile spasms, OCCURS-WITH, MELAS Syndrome). Whereas the coincidence of hypothyroidism and Turner Syndrome is fairly well known, the relationship between infantile spasms and MELAS syndrome is relatively obscure. Infantile Spasms, also known as West syndrome, is an exceedingly rare condition, with an estimated incidence in the United States of about 0.25-0.4 per 1000 live births27. The MELAS syndrome is an even rarer inherited disorder of mitochondrial function which may be responsible for 8% of cases of infantile spasms28. That the MKE recognized the connection between these two very rare conditions is quite interesting, and suggests that knowledge graph embedding holds promise for the elucidation of unusual concepts and relations from EEG reports in particular, and perhaps in medical reports more generally.
Owing to the data-driven nature of our technique, we generated some incorrect triples, as might be expected when using noisy free text data. For example, we observed two common types of errors when evaluating the evidences relation: (1) relation inversion, inverting the source and destination arguments of the relation; and (2) relation confusion, confusing one relation type with another. Consider the following example of a triple exhibiting relation inversion: (E1) (liver cirrhosis, EVIDENCES, encephalopathies). As defined in Figure 1, the source argument of the EVIDENCES relation is a medical concept suggesting or supporting the diagnosis listed in the destination argument. By contrast, it could be argued that, for triple (E1), the destination argument encephalopathies more commonly evidences the source argument liver cirrhosis. We believe these types of error could be addressed by incorporating semantic attributes (e.g. temporal information) to contextualize or constrain the arguments allowed for each relation type. Relation confusion is exemplified by the triple (E2) (rifaximin, EVIDENCES, brain diseases, metabolic). The source argument rifaximin is an antibiotic used in the management of the encephalopathy (i.e. the destination argument brain diseases, metabolic) related to severe liver failure. Thus, whereas there is a biologically plausible explanation for (E2), the EVIDENCES relation clearly does not accurately describe the relation; instead, the relation OCCURS-WITH may be preferred. This type of error could be mitigated in future work by introducing constraints into the knowledge embedding framework, as reported in Guo et al. (2015)23. Finally, there were rare cases in which the MKE assigned a high plausibility to triples in which the source argument contradicts the destination argument, i.e. (insulin, TREATMENT-FOR, Diabetes Mellitus, Non-Insulin-Dependent). We believe that these types of error may be resolved by incorporating knowledge from existing ontologies to enforce consistency.
Conclusion
In this paper, we presented the medical knowledge embeddings (MKE) automatically learned from clinical text in EEG reports. Unlike traditional ontologies which encode curated knowledge, the MKE infers probabilistic knowledge by extracting a large number of potential relation triples. Experimental results demonstrate the promise of this approach and highlight the potential of the MKE for bridging the knowledge gaps of existing neurological ontologies. The MKE presented in this paper showcase the way in which deep learning techniques applied to large collections of medical records can supply medical knowledge derived from clinical practice to complement the knowledge already encoded in existing biomedical ontologies. By encoding the plausibility of medical knowledge, the MKE also enable probabilistic reasoning on its knowledge. Future work will consider techniques for learning plausibility thresholds that will allow MKE to be considered for curation and acceptance in existing, expert and community-validated biomedical ontologies.
Acknowledgements
Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under award number 1U01HG008468. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Gene set enrichment (also functional enrichment analysis) is a method to identify classes of genes or proteins that are over-represented in a large set of genes or proteins, and may have an association with disease phenotypes.
References
- 1.Ashbumer M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nature genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LePendu P, Musen MA, Shah NH. Enabling enrichment analysis with the human disease ontology. Journal of biomedical informatics. 2011;44:S31–S38. doi: 10.1016/j.jbi.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic acids research. 2009;37(suppl 2):W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.England MJ, Liverman CT, Schultz AM, Strawbridge LM. Epilepsy across the spectrumml: Promoting health and understanding. A summary of the Institute of Medicine report. Epilepsy & Behavior. 2012;25(2):266–276. doi: 10.1016/j.yebeh.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harati A, Choi S, Tabrizi M, Obeid I, Picone J, Jacobson M. In: Global Conference on Signal and Information Processing (GlobalSIP), 2013 IEEE. IEEE; 2013. The Temple University Hospital EEG Corpus; pp. 29–32. [Google Scholar]
- 8.Weston J, Bordes A, Yakhnenko O, Usunier N. Connecting language and knowledge bases with embedding models for relation extraction. arXiv preprint arXiv:13077973. 2013 [Google Scholar]
- 9.Craven M, Kumlien J, et al. Constructing biological knowledge bases by extracting information from text sources. In: ISMB. 1999;1999:77–86. [PubMed] [Google Scholar]
- 10.Bordes A, Usunier N, Garcia-Duran A, Weston J, Yakhnenko O. Translating embeddings for modeling multi-relational data. In: Advances in neural information processing systems; 2013:2787–2795. [Google Scholar]
- 11.Goodwin T, Harabagiu SM. In: Semantic Computing (ICSC), 2013 IEEE Seventh International Conference on. IEEE; 2013. Automatic generation of a qualified medical knowledge graph and its usage for retrieving patient cohorts from electronic medical records; pp. 363–370. [Google Scholar]
- 12.Goodwin T, Harabagiu SM. In: International Conference of the Cross-Language Evaluation Forum for European Languages. Springer; 2013. The impact of belief values on the identification of patient cohorts; pp. 155–166. [Google Scholar]
- 13.Goodwin T, Harabagiu SM. Graphical induction of qualified medical knowledge. International Journal of Semantic Computing. 2013;7(04):377–405. [Google Scholar]
- 14.Goodwin TR, Harabagiu SM. In: Proceedings of the 25th ACM International on Conference on Information and Knowledge Management. ACM; 2016. Medical Question Answering for Clinical Decision Support; pp. 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Scott DJ, Villarroel M, Clifford GD, Saeed M, Mark RG. In: Engineering in Medicine and Biology Society, EMBC, 2011 Annual International Conference of the IEEE. IEEE; 2011. Open-access MIMIC-II database for intensive care research; pp. 8315–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koller D, Friedman N. MIT press; 2009. Probabilistic graphical models: principles and techniques. [Google Scholar]
- 17.Sahoo SS, Lhatoo SD, Gupta DK, Cui L, Zhao M, Jayapandian C, et al. Epilepsy and seizure ontology: towards an epilepsy informatics infrastructure for clinical research and patient care. Journal of the American Medical Informatics Association. 2014;21(1):82–89. doi: 10.1136/amiajnl-2013-001696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodwin TR, Harabagiu SM. In: AMIA Annual Symposium Proceedings. Vol. 2016. American Medical Informatics Association;; 2016. Multi-modal Patient Cohort Identification from EEG Report and Signal Data; p. 1794. [PMC free article] [PubMed] [Google Scholar]
- 19.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noachtar S, Binnie C, Ebersole J, Mauguiere F, Sakamoto A, Westmoreland B. A glossary of terms most commonly used by clinical electroencephalographers and proposal for the report form for the EEG findings. The International Federation of Clinical Neurophysiology. Electroencephalography and clinical neurophysiology Supplement. 1999;52:21. [PubMed] [Google Scholar]
- 21.Maldonado R, Goodwin TR, Harabagiu SM. American Medical Informatics Association. Active Deep Learning-Based Annotation of Electroencephalography Reports for Cohort Identification. 2017;2017 [PMC free article] [PubMed] [Google Scholar]
- 22.Demner-Fushman D, Rogers W, Aronson A. MetaMap Lite: an evaluation of a new Java implementation of MetaMap. Journal of the American Medical Informatics Association: JAMIA. 2017 doi: 10.1093/jamia/ocw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo S, Wang Q, Wang B, Wang L, Guo L. Semantically Smooth Knowledge Graph Embedding. In: ACL (1) 2015:84–94. [Google Scholar]
- 24.Lin Y, Liu Z, Sun M, Liu Y, Zhu X. Learning Entity and Relation Embeddings for Knowledge Graph Completion. In: AAAI. 2015:2181–2187. [Google Scholar]
- 25.Wang Z, Zhang J, Feng J, Chen Z. Knowledge Graph Embedding by Translating on Hyperplanes. In: AAAI. Citeseer. 2014:1112–1119. [Google Scholar]
- 26.Bordes A, Weston J, Collobert R, Bengio Y. Learning structured embeddings of knowledge bases. In: Conference on artificial intelligence; 2011; EPFL-CONF-192344. [Google Scholar]
- 27.Paciorkowski AR, Thio LL, Dobyns WB. Genetic and biologic classification of infantile spasms. Pediatric neurology. 2011;45(6):355–367. doi: 10.1016/j.pediatrneurol.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadleir L, Connolly M, Applegarth D, Hendson G, Clarke L, Rakshi C, et al. Spasms in children with definite and probable mitochondrial disease. European journal of neurology. 2004;11(2):103–110. doi: 10.1046/j.1351-5101.2003.00724.x. [DOI] [PubMed] [Google Scholar]


