Abstract
Background
Transporter proteins mediate the translocation of substances across the membranes of living cells. Many transport processes are energetically expensive and the cells use 20 to 60% of their energy to power the transportomes. We hypothesized that there may be an evolutionary selection pressure for lower energy transporters.
Results
We performed a genome-wide analysis of the compositional reshaping of the transportomes across the kingdoms of bacteria, archaea, and eukarya. We found that the share of ABC transporters is much higher in bacteria and archaea (ca. 27% of the transportome) than in primitive eukaryotes (13%), algae and plants (10%) and in fungi and animals (5–6%). This decrease is compensated by an increased occurrence of secondary transporters and ion channels. The share of ion channels is particularly high in animals (ca. 30% of the transportome) and algae and plants with (ca. 13%), when compared to bacteria and archaea with only 6–7%. Therefore, our results show a move to a preference for the low-energy-demanding transporters (ion channels and carriers) over the more energy-costly transporter classes (ATP-dependent families, and ABCs in particular) as part of the transition from prokaryotes to eukaryotes. The transportome analysis also indicated seven bacterial species, including Neorickettsia risticii and Neorickettsia sennetsu, as likely origins of the mitochondrion in eukaryotes, based on the phylogenetically restricted presence therein of clear homologues of modern mitochondrial solute carriers.
Conclusions
The results indicate that the transportomes of eukaryotes evolved strongly towards a higher energetic efficiency, as ATP-dependent transporters diminished and secondary transporters and ion channels proliferated. These changes have likely been important in the development of tissues performing energetically costly cellular functions.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4816-5) contains supplementary material, which is available to authorized users.
Keywords: Energetic efficiency, Evolution, Cellular membrane, Mitochondria, Transporters
Background
The expansion of life on Earth has involved competition and also cooperation among organisms for the utilisation of resources which have been accessible to them [1, 2]. Arguments have been made in favour of growth rate over growth efficiency in organisms competing within a specific niche [3], which implies a natural selection towards an improved ability to capture and utilize the available free energy sources for survival and reproduction [4, 5]. Darwinian evolutionary theory originally covered only phenotypic improvements at the organismal level, but we nowadays also recognize the importance of molecular and cellular events such as the acquisition of mitochondria by eukaryotes. This enabled an increase of eukaryotic cell size and complexity due to a more “efficient” generation of the cellular fuel ATP [6, 7]. Cells need to allocate considerable resources to energize their transportomes. For example, brain neurons account for approximately 20% of the basal metabolic rate in humans, mostly for the movement of ions across neuronal membranes [8]. In general, a metabolic cost of up to 60% of the total ATP requirement in organisms is estimated for the activity of their transportomes [9, 10]. Thus, it would be reasonable to imagine that an improved energetic performance of the transportome has contributed to a higher fitness over the course of evolution.
Despite the importance of cellular transportomes (also reported as the second largest component of the human membrane proteome [11]), transporters are surprisingly understudied [12]. Additionally, the presence of substrate-binding proteins as the partners of a subset of membrane transporters [13] makes the cellular transport machinery more complicated than if we consider only the transporters themselves. The tightening of porous and leaky primordial envelopes such that they did not let in (and could learn to efflux) all kinds of substances [14, 15] has been proposed as a turning point for membrane transporters to co-evolve together with lipid bilayer membranes [16]. Different classes of transporters, each including several transporter superfamilies, share a common ancestral family of peptides which carry 1–4 transmembrane domains and form homo- and hetero-oligomer channels [17–20]. Intragenic duplication and triplication have been the major events promoting the diversification of transporter proteins [18, 20]. Classical evolutionary theory on the basis of natural selection proposed by Charles Darwin [4, 5] explains how the random variability of the genome as the diversification force has given the chance for specialisation, improved performance, and adaptation to the continuously changing biosphere. Here, we annotated the cellular transportomes of bacteria, archaea and eukarya and analysed their composition with a focus on the energetic efficiency. The analyses include all the three classes of transporters, i.e., ion channels, secondary transporters, and ATP-dependent transporters. To translocate substrates, ATP-dependent transporters bind and hydrolyse ATP [21], ion channels form pores for selective diffusion of ions and small molecules, and the secondary transporters shuttle substrate molecules across biological membranes either through energy independent facilitated diffusion or via exploiting membrane electrochemical potential gradients through uniport, symport and antiport [22].
Results
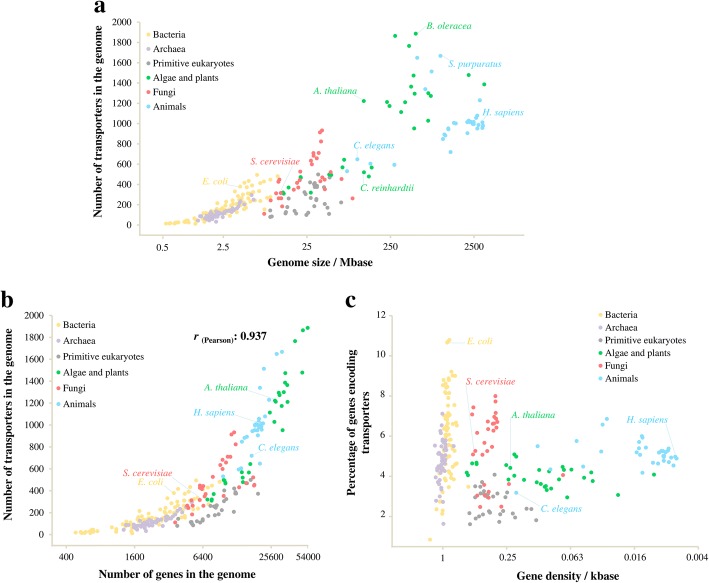
To study the compositional changes of transportomes, we analysed the transportomes of 249 evolutionarily distant species (of which 222 were annotated in this study) from archaea, bacteria and eukarya. These included 126 prokaryotic species (from 16 taxonomic phyla and 60 taxonomic orders), 30 primitive eukaryotes (different species from Alveolata, Kinetoplastida and Amoebozoa), 30 algal and plant species, 30 fungal species, and 33 animal species (See Additional file 2: Data S1). The transportomes were annotated using the Transporter Automatic Annotation Pipeline at TransportDB [23]. Notably, the species had large differences in the size of both their genomes and their transportomes (Fig. 1a, b and Additional file 2: Data S1).
Fig. 1.
Transportomes differ in size among species and evolutionarily distant domains of life. (a) The number of membrane transporters per organism in relation to the genome size. (b) The number of membrane transporters per genome in relation to the number of total genes. (c) Percentage of transporter-coding genes in relation to gene density
We found that the sizes of an organism’s transportome tends to increase with its genome size, though there are considerable intra-kingdom variations (Fig. 1a) (see also [24]). In particular, primitive eukaryotes have small transportomes with 100–500 members, relative to their genome sizes of tens of Mb when compared to the bacteria with modestly sized genomes (less than 10 Mb). Ion channels, secondary transporters, and primary active transporters were found in each of the analysed domains of life (See Additional file 2: Data S1). This indicates a very early appearance for these three classes of transporters, possibly in a common ancestor.
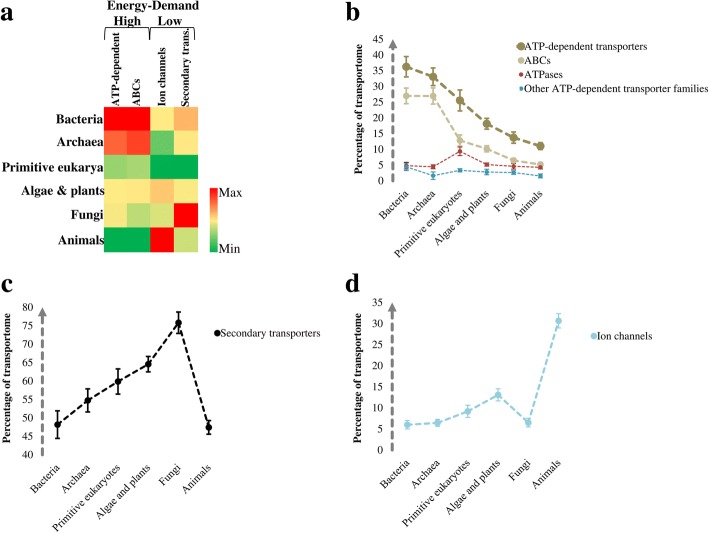
In agreement with previous reports [25, 26], the genome size had a higher rate of enlargement than did the gene number, resulting in a decreased gene density over the course of evolution (Fig. 1c). The transportome enlargement was found to be well correlated with the increase in the gene number (r (Pearson) = 0.937) with the exception of primitive eukarya, where the transportome-encoding proportion of genes was the lowest (Fig. 1b, c). Of most interest, the composition of the transportomes changed from prokaryotes to eukaryotes and also among the eukaryotic kingdoms along with the transportome enlargement (Fig. 2a). Specifically, we found higher intra-genomic frequencies (frequency relative to the total number of genes in the genome) of ATP-dependent transport classes in prokaryotes than eukaryotes. An opposing trend was found for low-energy-demanding transport classes. This indicates different rates of gene proliferation among the evolved transporter classes; low-energy-demanding transporter families have expanded at a higher rate. Notably, the transportomes of primitive eukaryotes also represent a transition state between prokaryotic and higher eukaryotic kingdoms (Fig. 2a).
Fig. 2.
The evolutionary dynamics of transportomes composition. (a) Heat-map representation of the changes in the number of members of the transporter classes. To calculate the intra-genomic frequencies, the numbers of transporter members are normalized to the total number of genes per genome. The heat-map is drawn for each class of ion channels, secondary transporters, and ATP-dependent transporters and therefore colors are not comparable between the classes. (b) The fraction of ATP-dependent transporters in the transportomes. All of the variations of ATP-dependent transporters and ABC superfamily except the difference between bacteria and archaea are significant with a p-value less than 0.001. (c) The fraction of secondary transporters in the transportomes. Only the difference between bacteria and animals is not statistically significant (p = 0.635). (d) The fraction of ion channels in the transportomes. All differences, except among fungi, bacteria and archaea, are significant with a p-value less than 0.001. The values on panels b-d are shown as mean +/− t-test based 99% confidence intervals. The variations were also confirmed on the arc sin √x transformed data (See Additional file 2: Data S1). The family names of the transporters can also be found in the Additional file 2: Data S1
We further compared the prevalence of secondary transporters, ion channels, and ATP-dependent transporters within the transportomes of each species, and averaged these over the larger taxonomic groups (Fig. 2b-d). In general, we observed compositional changes that indicate a positive adaptive selection of the no- to low-cost-flux equilibrative ion channels and carriers, and negative selection of the energetically more expensive ABC transporters. More specifically, we found significant variations in each of the transporter classes (Fig. 2b-d). While 27% of all the bacterial and archaeal transporters are ABC transporters, this fraction decreases to 13% in primitive eukaryotes, 10% in algae and plants and a mere 5–6% in fungi and animals (Fig. 2b). On the other hand, an increased contribution to the cellular transportome was found for secondary transporters in eukaryotes, particularly in fungi (Fig. 2c). Ion channels accounted for only 6–7% of bacterial and archaeal transportomes, but were more abundant in algae and plants (≈ 13%) and particularly in animals (≈ 30%) (Fig. 2d). During evolution, and in parallel with the genome enlargement and gene pool expansion, each of the three classes of transporters had a chance to contribute proportionally to the expansion of transportomes. By contrast, our results show a preference for the low-energy demanding transporters (ion channels and carriers) over the energy-costly transporter classes (ATP-dependent families, and ABCs in particular) in the transition from prokaryotes to eukaryotes.
We defined the energy usage efficiency of a transportome (EUE) as the average required energy per single substrate translocation. We calculated the EUE values for the transportomes studied (more details in the Methods section). The EUE describes the overall energetic performance of transportomes at organismal level and most importantly it does not indicate the total energy consumption by the cellular transportome, because the latter depends also on the flux through individual transporters that is largely unknown. In contrast to the total energy requirements, the EUE is therefore not subjected to spatiotemporal variations. By comparing the average EUEs of the transportomes across the different domains of life, we found that the EUE has improved in eukaryotes by reductions of up to 0.50 ATP in the average ATP consumption per single transport event mediated by transporters (Table 1).
Table 1.
Improvement in the energy-usage efficiency (ΔEUE) calculated as changes in the average ATP-usage per single transport cycle
| Domains of life | Bacteria | Archaea | Primitive eukaryotes | Algae and plants | Fungi | Animals |
|---|---|---|---|---|---|---|
| Bacteria | 0 | |||||
| Archaea | −0.03 | 0 | ||||
| Primitive eukaryotes | −0.16 | − 0.14 | 0 | |||
| Algae and plants | −0.27 | −0.25 | − 0.11 | 0 | ||
| Fungi | −0.30 | −0.27 | − 0.13 | −0.02 | 0 | |
| Animals | −0.49 | −0.46 | − 0.32 | −0.21 | − 0.19 | 0 |
The changes (ΔEUE) are calculated as ATP-usagedomain of life in the matrix-row – ATP-usagedomain of life in the matrix-column (see methods)
Negative changes represent the reduction in ATP-usage and improved EUE of transportomes
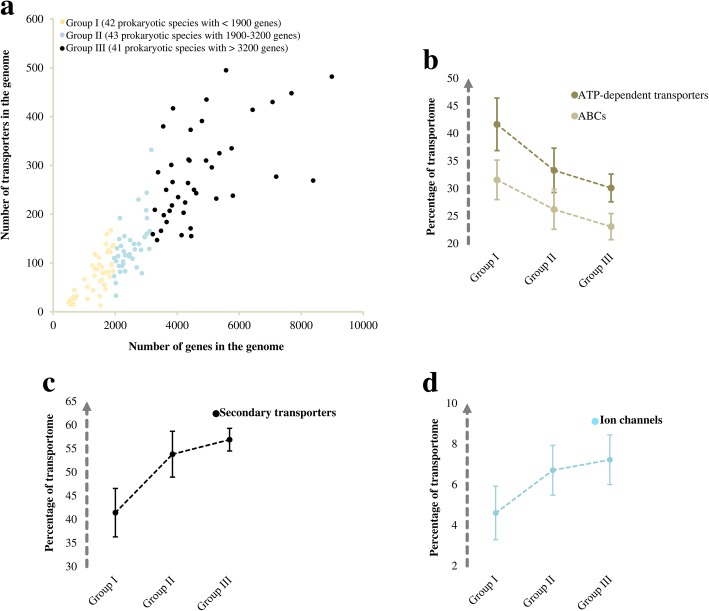
Furthermore, the data suggest that animals have mainly relied on the diversification of ion channels, fungi on secondary transporters, and finally, algae and plants on both transporter classes for the evolution of their transportomes (Fig. 2b-d). For energetically efficient transportomes, organisms therefore adopted different strategies, likely due to their specialisations and different requirements. In fact, this recalls cooperative evolution at tissue and molecular levels (see also [2, 27]). The trend of expansion of ion channels and secondary transporters at the expense of ATP-dependent transporters also holds true for the prokaryotic transportomes (Fig. 3), even though they did not undergo the kind of intensive developmental specialisation as was required in multicellular eukaryotes.
Fig. 3.
The compositional changes in the transportomes of prokaryotes with different genome sizes. (a) Comparison between the transportome size and total gene number among prokaryotes including bacteria and archaea. All of the 126 studied species are clustered into three groups based on the total number of the genes. (b) The fraction of ATP-dependent transporters in the transportomes. (c) The fraction of secondary transporters in the transportomes. (d) The fraction of ion channels in the transportomes. The values on panels b-d are shown as mean +/− t-test based 99% confidence interval. The variations were also confirmed on the arc sin √x transformed data (See Additional file 2: Data S1). Group I and III differ significantly for all of the transporter classes with a p-value of < 0.001
The analyses also indicate family member expansions for secondary transporters (from 60 to 100 in prokaryotes to more than 400 in animals and 600 in plants on average) and ion channels (from 10 members in prokaryotes to more than 300 members in animals). To find prokaryotic- and eukaryotic-specific families, we further enriched our analysis by including the publicly-available data for the secondary transporters and ion channels of prokaryotes found at TransportDB 2.0 (http://www.membranetransport.org/transportDB2/index.html). This extended our prokaryotes to 2637 transportomes (Additional file 1: Table S1). We also included two eukaryotic transportomes from diatoms found at TransportDB 2.0 (Additional file 1: Table S1). When comparing the organisms for the presence of different transporter families, we found that eight secondary transporter families were completely lost in the passage to eukaryotes (Fig. 4). This includes the 2-hydroxycarboxylate transporter family, the p-aminobenzoyl-glutamate transporter family, the short chain fatty acid uptake (AtoE) family, the monovalent cation (K+ or Na+):proton antiporter-3 family, the branched chain amino acid:cation symporter family, the NhaB Na+:H+ antiporter family, the riboflavin transporter family, and the Na+-dependent bicarbonate transporter family (See Additional file 2: Data S1). Additionally, we found that six new families of secondary transporters had emerged in eukaryotes (Fig. 4, See also Additional file 2: Data S1). Specifically, we did not find any prokaryotic hits in GenBank for these eukaryotic families, that must have diverged massively [28] from some ancestral genes. Our results are mostly in agreement with the intra-family speciation of transporters reported by Ren and Paulsen [29]. In contrast, we also found that ion channels had evolved through both intra-family expansions and the substantial appearance of new families. While there were only seven ion channel families specific to the prokaryotes, there are 18 eukarya-specific families (Fig. 4, See also Additional file 2: Data S1). Interestingly, the mitochondria-specific solute carriers, i.e., solute carrier family SLC25 [30, 31], are absent from the transportomes of all 143 archaeal species (See Additional file 1: Table S1 for the list of organisms). Among the bacteria, including 259 alpha-proteobacteria, of which 69 belonged to the order Rickettsiales, we found only seven bacterial genomes that encoded members of the mitochondrial transporter family. These are Neorickettsia risticii, Neorickettsia sennetsu, Legionella pneumophila, Legionella longbeachae, Acidaminococcus intestini, Cardinium endosymbiont and Butyrivibrio proteoclasticus. The presence of the mitochondrial transporter family members indicates that these seven bacterial species are possible origins of the mitochondrion in eukaryotes. The first two species are Gram-negative obligatory intracellular bacteria from the order Rickettsiales, an order that in previous studies was proposed (based on different evidence) as the most likely origin of the mitochondrion in eukaryotes [28, 30–35]. The present findings add significant evidence to this proposal.
Fig. 4.
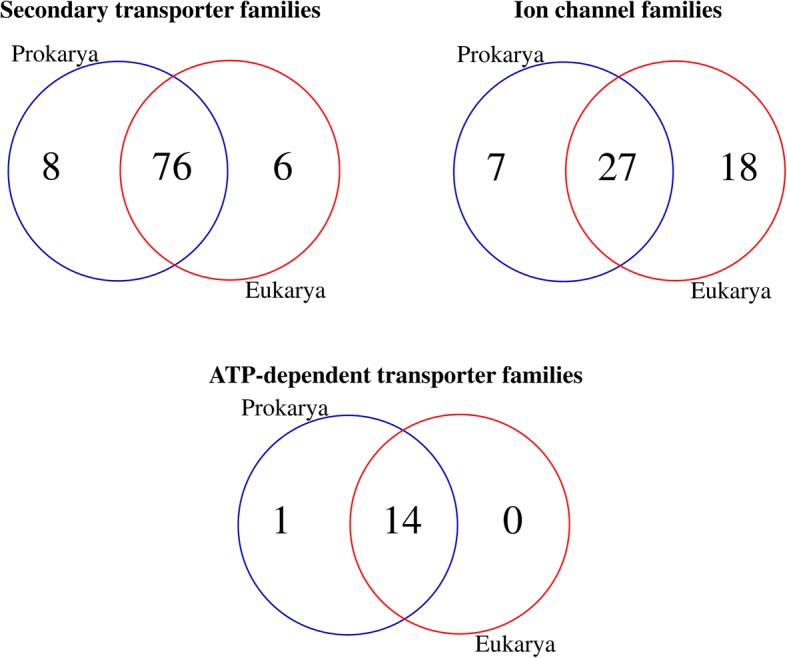
Number of transporter families that are shared between or are specific for prokaryotes and eukaryotes
Discussion
Our genome-wide analysis on the compositional reshaping of the transportomes across the kingdoms of bacteria, archaea, and eukarya suggests an evolutionary preference for energetically efficient transportomes. Our analyses also mapped some ancestral mitochodrial carrier members of SLC25 into the bacterial genomes and excluded the archaeal species. These ancesters include two bacteria from the order Rickettsiales. Rickettsiales are considered to be the mitochodrial origin [32–35]. It seems the eukaryotic members of the SLC25 family, all with 6 transmembrane domains, have diverged massively from their bacterial ancesters with 4–5 transmembrane domains. In agreement with Calvo et al. [36], the absence of SLC25 homologues in the genome of Rickettsia prowazekii, a species from the Rickettsiales, was also confirmed in this study; we did not find any ancestral SLC25 coding genes among the 10 different strains of Rickettsia prowazekii that were included in the analysis.
A higher ratio of secondary transporters to primary ATP-driven transporters was previously reported for yeast when compared to prokaryotes [37]. This is additionally complemented by a relatively large genomic survey on 141 species, of which 9 species were eukaryotic [29]. Our results on the general compositional changes of transportomes are in agreement with the findings of Ren and Poulsen [29]. The publication by Ren and Poulsen, however, did not report the energetics of transportomes or the detailed analysis of transporter classes. Resource depletion and inadequate nutrition affect survival but can also increase energetic efficiency, that is fitness, in the course of evolution [38]. For example, the adaptation to chronic energy stress has been hypothesized to be partly responsible for the general capacity of archaea to out-compete bacteria [39]. In addition, energetic evolution of gene expression has also been discussed by addressing the preference for major codons in highly expressed genes, which alleviates the costly processes of proofreading and removal of dysfunctional proteins [40]. Another environmental stress reported to affect codon usage is the low availability of nitrogen. Marine microorganisms faced with low levels of nitrogen show reduced levels of nitrogen in amino acid sequences, especially in highly expressed proteins, which reduces the total cellular nitrogen budget by up to 10% [41]. There are also observations on the energetic evolution at a phenotypic level. For example, an energetic trade-off between maximum population density and body size has been reported in [42]. Energetic constraints have also been proposed for the number of neurons, which determines the brain size [43]. At a molecular level, although both the solute carrier SLC2 family of facilitated glucose transporters and the SLC5 family of energy-dependent sodium/glucose cotransporters participate in glucose uptake, the human brain has largely been dependent on SLC2 transporters for the energetically free supply of glucose [44]. Positive selection and gene expression adjustments, such that they allocate higher energy fluxes to the brain, have been reported for the SLC2 members in the human branch that has a larger brain compared to the chimpanzees and macaques [45]. This indicates the importance of the energetic evolution of membrane transporters, even at the single family level, and highlights the possibly huge impacts of the energetic evolution of the entire transportomes on organismal adaptation and speciation. On the other hand, the higher dependency of bacterial and archaeal species on the ATP-driven transporters is also interesting. It could be related to the higher affinities of ATP transporters that allowed the cells to capture rare substrates more efficiently. It is also possible that ATP transporters have diversified from the early ATP synthases and this partly explains why they are therefore so abundant in bacteria and archaea.
All of the eight prokarya-specific secondary transporter families (Additional file 2: Data S1) were energy-dependent, i.e., dependent on a proton or sodium gradient, or utilizing a combination of electro- and chemical potential of the membrane [46–51]. A surprising insight here was related to the six evolutionarily younger secondary transporter families found only in eukaryotes (See Additional file 2: Data S1): four of these, about which we have experimental information, were energy-independent and low-energy-demanding transporter families for bile acid, choline, silicate, and vitamin A. The last two belong to the 4 TMS multidrug endosomal transporter and chloroplast maltose exporter families. The choline transporter-like family is involved in choline influx [52]. The ‘birth’ of a cheap and sodium-independent transporter for choline is important because of the broad cellular usage of choline. Choline is an essential precursor for membranes and for the neuromodulator acetylcholine [53, 54]. Another recently evolved transport family was the organic solute transporter family, which is involed in the facilitated diffusion of bile acids from enterocytes into the blood [55]. The silicon transporter family is also energetically cheap and has a silicate:sodium symport stoichiometry of 1:1 [56]. Finally, the animal-specific vitamin A receptor/transporter (STRA6) mediates costless influx and efflux of vitamin A derivatives by a mechanism not seen in any other transporter class [57, 58]. Of particular interest, the animal visual system is dependent on vitamin A and has a substantial energetic cost, e.g., up to 15% of resting metabolism in the Mexican fish Astyanax mexicanus [59, 60]. Such photo-detection involves the single-photon-triggered isomerization of 11-cis-retinal to all-trans-retinal, which must be recycled back through efflux and influx steps of these isomers between the retinal pigmented epithelial and photoreceptor cells [57, 61–63]. Thus, the evolved energy-independent membrane translocation of the vitamin A isomers seems to be an adaptive trait for higher energetic performance. This is in line with the positive selection reported for STRA6 in different mammalian phyla [64]. Additionally, we found a higher representation of ion channels in the animal kingdom when compared to the other domains of life (Fig. 2d). The membrane transport of ions is important for highly energy-demanding sensory tissues [8, 65] and it can therefore be hypothesized that the extensive diversification of ion channels and the costless transport of vitamin A in animals are trade-offs between the benefits of the evolved nervous and visual systems and their high energy requirements. Overall, our findings demonstrate a clear and unequivocal change in the energetic efficiency of transportomes during the course of evolution, a very significant finding.
Conclusions
The present inter-kingdom comparison of transportomes provides genome-scale molecular evidence for their evolution towards an improved energetic efficiency. This has likely been very influential due to the high energy demand of the cellular transport machinery and also for the development of tissues performing energetically costly functions. The present data also strengthen members of the previously reported bacterial order Rickettsiales as the origin of the mitochondrion by recognising Neorickettsia risticii and Neorickettsia sennetsu as the sole species in this order whose genomes harbor putative mitochondria-specific solute carrier (SLC25) coding genes. Since other evidence had also suggested Rickettsiales as candidates for this, our transoportome findings strengthen considerably the case for such a lineage.
Methods
The publicly-available membrane transporter data on ion channels and secondary transporters were extracted from TransportDB (http://www.membranetransport.org/transportDB2/) [23] . The transportomes of 126 prokaryotic species (78 bacteria and 48 archaea) and 96 eukaryotic species (22 primitive eukaryotes, 24 algae and plants, 23 fungi, and 27 animals) (See Additional file 2: Data S1) were annotated using the Transporter Automatic Annotation Pipeline at TransportDB [23]. We also included 27 eukaryotic species including the transportomes of 8 primitive eukaryotes, 6 algae and plants, 7 fungi, and 6 animals publicly available at TransportDB (See Additional file 2: Data S1). To study the compositional changes of transportomes, we did not include any transportomes from prokaryotes publicly available at the TransportDB. This is due to the incomplete information on the ABC transporters. The majority of ABC transporters in prokaryotes are coded by different genes of an operon, where each gene codes for different subunits [66], and these should be excluded from the data and considered as single transporters in our analyses on the transportome composition. However, this information on the ABC coding genes and operons is not provided for the publicly-available transportomes of prokaryotes.
To predict the transportomes, the proteomes of organisms were downloaded from the Genbank and Ensembl databases (http://www.ensembl.org/index.html, http://fungi.ensembl.org/index.html, http://protists.ensembl.org/index.html, http://plants.ensembl.org/index.html) [67]. All proteins with fewer than 100 amino acids were excluded. Taken together, we analysed 78 bacterial, 48 archaeal, 30 primitive eukaryotes, 30 algal and plant, 30 fungal, and 33 animal transportomes, each representing one independent biological replicate (per species). The list of organisms with their genome size and total number of transporters is shown in Additional file 2: Data S1 and Additional file 1: Table S1. The annotated transportomes were manually filtered for multi-prediction hits as well as the alternative isoforms of transport proteins before the analysis. Taken together, our analyses on the transporter families included all of the 15 ATP-dependent transporter families, 51 ion channel families, and 90 secondary transporter families which were present in the studied organisms. We used Student’s t-test to examine the possible differences among the samples, i.e., domains of life. The data were in the format of counts and percentages. Therefore, to exclude a possible divergence from normality, we also performed the analyses on the arc sin √x transformed data. All the analyses can be found in Additional file 2: Data S1.
To make an approximation of the energetic performance of the cellular transportomes, we calculated the inter-transportome changes in energy-usage efficiency (EUE), which we defined as the average free energy demand for a single transport event. The following assumptions were made when calculating EUE values. Equilibrative ion channels require no free energy for the transport action. While some of the secondary transporters also act in an energy-independent manner known as equilibrative transport or facilitated diffusion, others exploit the electrochemical potential established by ATPases across membranes [14, 22, 68–70]. For active transport, secondary transporters are considered to use membrane electrochemical potential gradients, coupled to a varying stoichiometry (0.5 to 3) of ions per turnover [69, 71–92]. Considering the stoichiometry of ≈ 2–3 ions pumped per ATP hydrolysed by ATPases [93–102], even concentrative secondary transporters would not use more than 0.5 ATP equivalent per substrate translocation across a membrane on average. The average of two substrates per ATP tends, therefore, to be conservative for secondary transporters and a higher rate of substrate translocation can also be expected. In contrast, the ATP-dependent members belonging to the ABC superfamily, and also the mitochondrial protein translocase, the type III secretory pathway, the chloroplast envelope protein translocase, and the arsenite-antimonite efflux families, generally show a 1:2 stoichiometry of substrate:ATP hydrolysis [103–109]. To calculate the inter-transportome variations in the energy-usage efficiency (ΔEUE), we therefore applied the equation:
where ADT, ST, and IC are ATP-dependent transporters, secondary transporters, and ion-channels, respectively. The F/V/A-type ATPases and ATP synthases were excluded from the ATP-dependent transporters. This is because they provide the energy as ATP or membrane electrochemical potential for the rest of the transporters. It is worthy of note that the efficiency of energy usage indicates the average energy demand per unit of action. So the expression and activity levels of transporters are not taken into the account when calculating the EUE. A more general example would compare two organisms with exactly the same transportome but with differences in the expression levels of transportome members among them. Here, the EUE of transportome machineries of the two organisms are equal since they use exactly same transportome machinery. This is thus irrespective of the differences in activity levels of transporters, which define the total energy demand for the given transportome.
Additional files
Table S1. The list of organisms with publicly-available data on the ion channels and secondary transporters. (DOCX 168 kb)
Data S1. Genomic and transportome data on the organisms included in the study. (XLSX 255 kb)
Acknowledgements
The authors thank Dr. Gavin H. Thomas from the University of York for discussions and advice. Shahin Noeparvar from the Aarhus University is acknowledged for discussions and participation into the genomic data processing.
Funding
B.D. and I.B. acknowledge the financial support by the Novo Nordisk Foundation (grant number NNF10CC1016517). The project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant Agreement no. 757384). D.B.K. thanks the Biotechnology and Biological Sciences Research Council (grants BB/M006891/1, BB/M017702/1 and BB/P009042/1) for financial support.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files (Additional file 2: Data S1 and Additional file 1: Table S1). The raw genomic data are publicly-available at http://www.ensembl.org/index.html, http://fungi.ensembl.org/index.html, http://protists.ensembl.org/index.html, and http://plants.ensembl.org/index.html.
Abbreviations
- ADT
ATP-dependent transporters
- EUE
energy-usage efficiency
- IC
ion-channels
- ST
secondary transporters
Authors’ contributions
BD, DBK, and IB conceived the study. BD performed the analyses by consulting DBK and IB. BD, DBK, and IB wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4816-5) contains supplementary material, which is available to authorized users.
Contributor Information
Behrooz Darbani, Email: behroozdarbani@gmail.com.
Douglas B. Kell, Phone: +44-161 306 4492, Email: dbk@manchester.ac.uk
Irina Borodina, Phone: +45-2179 8657, Email: irbo@biosustain.dtu.dk.
References
- 1.Demetrius L. Thermodynamics and Evolution. J Theor Biol. 2000;206:1–16. doi: 10.1006/jtbi.2000.2106. [DOI] [PubMed] [Google Scholar]
- 2.Nowak MA. Five rules for the evolution of cooperation. Science. 2006;314:1560–1563. doi: 10.1126/science.1133755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westerhoff HV, Hellingwerf KJ, Van Dam K. Thermodynamic efficiency of microbial growth is low but optimal for maximal growth rate. Proc Natl Acad Sci U S A. 1983;80:305–309. doi: 10.1073/pnas.80.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darwin C. On the origin of species by means of natural selection, or, the preservation of Favoured races in the struggle for life. J. Murray. 1859; [PMC free article] [PubMed]
- 5.Stanley SM. A Theory of evolution above the species level. Proc Natl Acad Sci. 1975;72:646–650. doi: 10.1073/pnas.72.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 7.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 8.Niven JE, Laughlin SB. Energy limitation as a selective pressure on the evolution of sensory systems. J Exp Biol. 2008;211:1792–1804. doi: 10.1242/jeb.017574. [DOI] [PubMed] [Google Scholar]
- 9.Verduyn C, Stouthamer AH, Scheffers WA, van Dijken JP. A theoretical evaluation of growth yields of yeasts. Antonie Van Leeuwenhoek. 1991;59:49–63. doi: 10.1007/BF00582119. [DOI] [PubMed] [Google Scholar]
- 10.Skou JC. The identification of the sodium–potassium pump (Nobel lecture) Angew Chem Int Ed. 1998;37:2320–2328. doi: 10.1002/(SICI)1521-3773(19980918)37:17<2320::AID-ANIE2320>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Almén MS, Nordström KJ, Fredriksson R, Schiöth HB. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.César-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, et al. A call for systematic research on solute carriers. Cell. 2015;162:478–487. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Bosdriesz E, Magnúsdóttir S, Bruggeman FJ, Teusink B, Molenaar D. Binding proteins enhance specific uptake rate by increasing the substrate–transporter encounter rate. FEBS J. 2015;282:2394–2407. doi: 10.1111/febs.13289. [DOI] [PubMed] [Google Scholar]
- 14.Kell DB, Oliver SG. How drugs get into cells: tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front Pharmacol. 2014;5:231. [DOI] [PMC free article] [PubMed]
- 15.Kell DB, Swainston N, Pir P, Oliver SG. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 2015;33:237–246. doi: 10.1016/j.tibtech.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Mulkidjanian AY, Galperin MY, Koonin EV. Co-evolution of primordial membranes and membrane proteins. Trends Biochem Sci. 2009;34:206–215. doi: 10.1016/j.tibs.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perland E, Fredriksson R. Classification Systems of Secondary Active Transporters. Trends Pharmacol Sci. 2017;38:305–315. doi: 10.1016/j.tips.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Saier MH. Tracing pathways of transport protein evolution. Mol Microbiol. 2003;48:1145–1156. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- 19.Saier MH. Transport protein evolution deduced from analysis of sequence, topology and structure. Curr Opin Struct Biol. 2016;38:9–17. doi: 10.1016/j.sbi.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu T, Mitsuke H, Noto K, Arai M. Internal gene duplication in the evolution of prokaryotic transmembrane proteins. J Mol Biol. 2004;339:1–15. doi: 10.1016/j.jmb.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 21.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 22.Forrest LR, Krämer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim Biophys Acta. 2011;1807:167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Elbourne LDH, Tetu SG, Hassan KA, Paulsen IT. TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 2017;45:D320–D324. doi: 10.1093/nar/gkw1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kell DB, Dobson PD, Oliver SG. Pharmaceutical drug transport: the issues and the implications that it is essentially carrier-mediated only. Drug Discov Today. 2011;16:704–714. doi: 10.1016/j.drudis.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 26.Hou Y, Lin S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. PLoS ONE. 2009;4:e6978. [DOI] [PMC free article] [PubMed]
- 27.Frank SA. Perspective: repression of competition and the evolution of cooperation. Evol Int J Org Evol. 2003;57:693–705. doi: 10.1111/j.0014-3820.2003.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 28.Höglund PJ, Nordström KJV, Schiöth HB, Fredriksson R. The solute carrier families have a remarkably long evolutionary history with the majority of the human families present before divergence of Bilaterian species. Mol Biol Evol. 2011;28:1531–1541. doi: 10.1093/molbev/msq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren Q, Paulsen IT. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput Biol. 2005;1:e27. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Asp Med. 2013;34:465–484. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Hediger MA, Clémençon B, Burrier RE, Bruford EA. The ABCs of membrane transporters in health and disease (SLC series): introduction. Mol Asp Med. 2013;34:95–107. doi: 10.1016/j.mam.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, et al. The genome sequence of rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 33.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray MW. Mosaic nature of the mitochondrial proteome: implications for the origin and evolution of mitochondria. Proc Natl Acad Sci U S A. 2015;112:10133–10138. doi: 10.1073/pnas.1421379112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Wu M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep. 2015;5:7949. doi: 10.1038/srep07949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulsen IT, Sliwinski MK, Nelissen B, Goffeau A, Saier MH. Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 1998;430:116–125. doi: 10.1016/S0014-5793(98)00629-2. [DOI] [PubMed] [Google Scholar]
- 38.Parsons PA. Environments and evolution: interactions between stress, resource inadequacy and energetic efficiency. Biol Rev. 2005;80:589–610. doi: 10.1017/S1464793105006822. [DOI] [PubMed] [Google Scholar]
- 39.Valentine DL. Opinion: Adaptations to energy stress dictate the ecology and evolution of the archaea. Nat Rev Microbiol. 2007;5 nrmicro1619 [DOI] [PubMed]
- 40.Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- 41.Grzymski JJ, Dussaq AM. The significance of nitrogen cost minimization in proteomes of marine microorganisms. ISME J. 2011;6 ismej201172 [DOI] [PMC free article] [PubMed]
- 42.Brown JH, Maurer BA. Evolution of species assemblages: effects of energetic constraints and species dynamics on the diversification of the north American avifauna. Am Nat. 1987;130:1–17. doi: 10.1086/284694. [DOI] [Google Scholar]
- 43.Fonseca-Azevedo K, Herculano-Houzel S. Metabolic constraint imposes tradeoff between body size and number of brain neurons in human evolution. Proc Natl Acad Sci. 2012;109:18571–18576. doi: 10.1073/pnas.1206390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szablewski L. Glucose transporters in brain: in health and in Alzheimer’s disease. J Alzheimers Dis. 2017;55:1307–1320. doi: 10.3233/JAD-160841. [DOI] [PubMed] [Google Scholar]
- 45.Fedrigo O, Pfefferle AD, Babbitt CC, Haygood R, Wall CE, Wray GAA. Potential role for glucose transporters in the evolution of human brain size. Brain Behav Evol. 2011;78:315–326. doi: 10.1159/000329852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobczak I, Lolkema JS. Alternating access and a pore-loop structure in the Na+−citrate transporter CitS of Klebsiella pneumoniae. J Biol Chem. 2004;279:31113–31120. doi: 10.1074/jbc.M404283200. [DOI] [PubMed] [Google Scholar]
- 47.Bolla JR, Su C-C, Delmar JA, Radhakrishnan A, Kumar N, Chou T-H, et al. crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology. Nat Commun. 2015;6:6874. doi: 10.1038/ncomms7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swartz TH, Ito M, Ohira T, Natsui S, Hicks DB, Krulwich TA. Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J Bacteriol. 2007;189:3081–3090. doi: 10.1128/JB.00021-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun PR, Al-Younes H, Gussmann J, Klein J, Schneider E, Meyer TF. Competitive inhibition of amino acid uptake suppresses chlamydial growth: involvement of the chlamydial amino acid transporter BrnQ. J Bacteriol. 2008;190:1822–1830. doi: 10.1128/JB.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura T, Fujisaki Y, Enomoto H, Nakayama Y, Takabe T, Yamaguchi N, et al. Residue aspartate-147 from the third transmembrane region of Na(+)/H(+) antiporter NhaB of Vibrio alginolyticus plays a role in its activity. J Bacteriol. 2001;183:5762–5767. doi: 10.1128/JB.183.19.5762-5767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H, et al. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. J Biol Chem. 2002;277:18658–18664. doi: 10.1074/jbc.M112468200. [DOI] [PubMed] [Google Scholar]
- 52.Taguchi C, Inazu M, Saiki I, Yara M, Hara N, Yamanaka T, et al. Functional analysis of [methyl-(3)H]choline uptake in glioblastoma cells: influence of anti-cancer and central nervous system drugs. Biochem Pharmacol. 2014;88:303–312. doi: 10.1016/j.bcp.2014.01.033. [DOI] [PubMed] [Google Scholar]
- 53.Traiffort E, O’Regan S, Ruat M. The choline transporter-like family SLC44: properties and roles in human diseases. Mol Asp Med. 2013;34:646–654. doi: 10.1016/j.mam.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 54.Picciotto MR, Higley MJ, Mineur YS. Acetylcholine as a neuromodulator: cholinergic signaling shapes nervous system function and behavior. Neuron. 2012;76:116–129. doi: 10.1016/j.neuron.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, et al. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem. 2005;280:6960–6968. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knight MJ, Senior L, Nancolas B, Ratcliffe S, Curnow P. Direct evidence of the molecular basis for biological silicon transport. Nat Commun. 2016;7:11926. doi: 10.1038/ncomms11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawaguchi R, Zhong M, Kassai M, Ter-Stepanian M, Sun H. STRA6-catalyzed vitamin a influx, efflux, and exchange. J Membr Biol. 2012;245:731–745. doi: 10.1007/s00232-012-9463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Clarke OB, Kim J, Stowe S, Kim Y-K, Assur Z, et al. Structure of the STRA6 receptor for retinol uptake. Science. 2016;353 [DOI] [PMC free article] [PubMed]
- 59.Zhong M, Kawaguchi R, Kassai M, Sun H. Retina, retinol, retinal and the natural history of vitamin a as a light sensor. Nutrients. 2012;4:2069–2096. doi: 10.3390/nu4122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moran D, Softley R, Warrant EJ. The energetic cost of vision and the evolution of eyeless Mexican cavefish. Sci Adv. 2015;1:e1500363. doi: 10.1126/sciadv.1500363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saari JC. Vitamin a metabolism in rod and cone visual cycles. Annu Rev Nutr. 2012;32:125–145. doi: 10.1146/annurev-nutr-071811-150748. [DOI] [PubMed] [Google Scholar]
- 62.Kiser PD, Golczak M, Maeda A, Palczewski K. Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochim Biophys Acta BBA - Mol Cell Biol Lipids. 2012;1821:137–151. doi: 10.1016/j.bbalip.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sahu B, Maeda A. Retinol dehydrogenases regulate vitamin a metabolism for visual function. Nutrients. 2016;8 [DOI] [PMC free article] [PubMed]
- 64.Wu J, Xiang H, Qi Y, Yang D, Wang X, Sun H, et al. Adaptive evolution of the STRA6 genes in mammalian. PLoS One. 2014;9:e108388. doi: 10.1371/journal.pone.0108388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glynn IM. Annual review prize lecture All hands to the sodium pump. J Physiol. 1993;462:1–30. doi: 10.1113/jphysiol.1993.sp019540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moussatova A, Kandt C, O’Mara ML, Tieleman DP. ATP-binding cassette transporters in Escherichia coli. Biochim Biophys Acta BBA - Biomembr. 2008;1778:1757–1771. doi: 10.1016/j.bbamem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 67.Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, et al. Ensembl genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016;44:D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baldwin SA. Mammalian passive glucose transporters: members of an ubiquitous family of active and passive transport proteins. Biochim Biophys Acta. 1993;1154:17–49. doi: 10.1016/0304-4157(93)90015-G. [DOI] [PubMed] [Google Scholar]
- 69.Monné M, Palmieri F. Antiporters of the mitochondrial carrier family. Curr Top Membr. 2014;73:289–320. doi: 10.1016/B978-0-12-800223-0.00008-6. [DOI] [PubMed] [Google Scholar]
- 70.Mendes P, Oliver SG, Kell DB. Fitting transporter activities to cellular drug concentrations and fluxes: why the bumblebee can fly. Trends Pharmacol Sci. 2015;36:710–723. doi: 10.1016/j.tips.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC cl- channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 72.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 73.Deng D, Yan N. GLUT, SGLT, and SWEET: structural and mechanistic investigations of the glucose transporters. Protein Sci Publ Protein Soc. 2016;25:546–558. doi: 10.1002/pro.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steel A, Nussberger S, Romero MF, Boron WF, Boyd CA, Hediger MA. Stoichiometry and pH dependence of the rabbit proton-dependent oligopeptide transporter PepT1. J Physiol. 1997;498(Pt 3):563–569. doi: 10.1113/jphysiol.1997.sp021883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boron VF, Hediger MA, Boulpaep EL, Romero MF. The renal electrogenic Na+:HCO-3 cotransporter. J Exp Biol. 1997;200:263–268. doi: 10.1242/jeb.200.2.263. [DOI] [PubMed] [Google Scholar]
- 76.Bröer S, Bröer A, Schneider HP, Stegen C, Halestrap AP, Deitmer JW. Characterization of the high-affinity monocarboxylate transporter MCT2 in Xenopus laevis oocytes. Biochem J. 1999;341(Pt 3):529–535. doi: 10.1042/0264-6021:3410529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jennings ML, Adame MF. Direct estimate of 1:1 stoichiometry of K(+)-cl(−) cotransport in rabbit erythrocytes. Am J Physiol Cell Physiol. 2001;281:C825–C832. doi: 10.1152/ajpcell.2001.281.3.C825. [DOI] [PubMed] [Google Scholar]
- 78.Kahle KT, Rinehart J, Lifton RP. Phosphoregulation of the Na-K-2Cl and K-cl cotransporters by the WNK kinases. Biochim Biophys Acta. 2010;1802:1150–1158. doi: 10.1016/j.bbadis.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao C, Noskov SY. The molecular mechanism of ion-dependent gating in secondary transporters. PLoS Comput Biol. 2013;9:e1003296. doi: 10.1371/journal.pcbi.1003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forster IC, Hernando N, Biber J, Murer H. Phosphate transporters of the SLC20 and SLC34 families. Mol Asp Med. 2013;34:386–395. doi: 10.1016/j.mam.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 81.Schweikhard ES, Ziegler CM. Amino acid secondary transporters. toward a common transport mechanism Curr Top Membr. 2012;70:1–28. doi: 10.1016/B978-0-12-394316-3.00001-6. [DOI] [PubMed] [Google Scholar]
- 82.Bürzle M, Hediger MA. Functional and Physiological role of vitamin C transporters. Curr Top Membr. 2012;70:357–375. doi: 10.1016/B978-0-12-394316-3.00011-9. [DOI] [PubMed] [Google Scholar]
- 83.Forster IC, Hernando N, Biber J, Murer H. Phosphate transport kinetics and structure-function relationships of SLC34 and SLC20 proteins. Curr Top Membr. 2012;70:313–356. doi: 10.1016/B978-0-12-394316-3.00010-7. [DOI] [PubMed] [Google Scholar]
- 84.Adijanto J, Philp NJ. The SLC16A family of monocarboxylate transporters (MCTs)--physiology and function in cellular metabolism, pH homeostasis, and fluid transport. Curr Top Membr. 2012;70:275–311. doi: 10.1016/B978-0-12-394316-3.00009-0. [DOI] [PubMed] [Google Scholar]
- 85.Terada T, Inui K. Recent advances in structural biology of peptide transporters. Curr Top Membr. 2012;70:257–274. doi: 10.1016/B978-0-12-394316-3.00008-9. [DOI] [PubMed] [Google Scholar]
- 86.Raja M, Puntheeranurak T, Hinterdorfer P, Kinne R. SLC5 and SLC2 transporters in epithelia-cellular role and molecular mechanisms. Curr Top Membr. 2012;70:29–76. doi: 10.1016/B978-0-12-394316-3.00002-8. [DOI] [PubMed] [Google Scholar]
- 87.Markovich D. Sodium-sulfate/carboxylate cotransporters (SLC13) Curr Top Membr. 2012;70:239–256. doi: 10.1016/B978-0-12-394316-3.00007-7. [DOI] [PubMed] [Google Scholar]
- 88.Cordat E, RAF R. Structure, Function, and trafficking of SLC4 and SLC26 anion transporters. Curr Top Membr. 2014;73:1–67. doi: 10.1016/B978-0-12-800223-0.00001-3. [DOI] [PubMed] [Google Scholar]
- 89.Hendus-Altenburger R, Kragelund BB, Pedersen SF. Structural dynamics and regulation of the mammalian SLC9A family of Na+/H+ exchangers. Curr Top Membr. 2014;73:69–148. doi: 10.1016/B978-0-12-800223-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 90.Schnetkamp PPM, Jalloul AH, Liu G, Szerencsei RT. The SLC24 family of K+ −dependent Na+–Ca2+ exchangers: structure–function relationships. Curr Top Membr. 2014;73:263–87. [DOI] [PubMed]
- 91.Schweigel-Röntgen M, Kolisek M. SLC41 transporters—molecular identification and functional role. Curr Top Membr. 2014;73:383–410. [DOI] [PubMed]
- 92.Döring B, Lütteke T, Geyer J, Petzinger E. The SLC10 carrier family. transport functions and molecular structure Curr Top Membr. 2012;70:105–168. doi: 10.1016/B978-0-12-394316-3.00004-1. [DOI] [PubMed] [Google Scholar]
- 93.Sitsel O, Grønberg C, Autzen HE, Wang K, Meloni G, Nissen P, et al. Structure and function of cu(I)- and Zn(II)-ATPases. Biochemistry (Mosc) 2015;54:5673–5683. doi: 10.1021/acs.biochem.5b00512. [DOI] [PubMed] [Google Scholar]
- 94.Davies JM, Hunt I, Sanders D. Vacuolar H(+)-pumping ATPase variable transport coupling ratio controlled by pH. Proc Natl Acad Sci U S A. 1994;91:8547–8551. doi: 10.1073/pnas.91.18.8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Finbow ME, Harrison MA. The vacuolar H+-ATPase: a universal proton pump of eukaryotes. Biochem J. 1997;324(Pt 3):697–712. doi: 10.1042/bj3240697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hernández A, Cooke DT, Clarkson DT. Effects of abnormal-sterol accumulation on Ustilago maydis plasma membrane H+-ATPase stoichiometry and polypeptide pattern. J Bacteriol. 1998;180:412–415. doi: 10.1128/jb.180.2.412-415.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cross RL, Müller V. The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio. FEBS Lett. 2004;576:1–4. doi: 10.1016/j.febslet.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 98.Tomashek JJ, Brusilow WS. Stoichiometry of energy coupling by proton-translocating ATPases: a history of variability. J Bioenerg Biomembr. 2000;32:493–500. doi: 10.1023/A:1005617024904. [DOI] [PubMed] [Google Scholar]
- 99.Warncke J, Slayman CL. Metabolic modulation of stoichiometry in a proton pump. Biochim Biophys Acta. 1980;591:224–233. doi: 10.1016/0005-2728(80)90154-1. [DOI] [PubMed] [Google Scholar]
- 100.Perlin DS, San Francisco MJ, Slayman CW, Rosen BP. H+/ATP stoichiometry of proton pumps from Neurospora crassa and Escherichia coli. Arch Biochem Biophys. 1986;248:53–61. doi: 10.1016/0003-9861(86)90400-5. [DOI] [PubMed] [Google Scholar]
- 101.Sosa A, Celis H. H+/PPi stoichiometry of membrane-bound pyrophosphatase of Rhodospirillum rubrum. Arch Biochem Biophys. 1995;316:421–427. doi: 10.1006/abbi.1995.1056. [DOI] [PubMed] [Google Scholar]
- 102.Petersen J, Förster K, Turina P, Gräber P. Comparison of the H+/ATP ratios of the H+-ATP synthases from yeast and from chloroplast. Proc Natl Acad Sci U S A. 2012;109:11150–11155. doi: 10.1073/pnas.1202799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Patzlaff JS, van der Heide T, Poolman B. The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J Biol Chem. 2003;278:29546–29551. doi: 10.1074/jbc.M304796200. [DOI] [PubMed] [Google Scholar]
- 104.Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 105.Rensing C, Ghosh M, Rosen BP. Families of soft-metal-ion-transporting ATPases. J Bacteriol. 1999;181:5891–5897. doi: 10.1128/jb.181.19.5891-5897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang H-C, Fu H-L, Lin Y-F, Rosen BP. Pathways of arsenic uptake and efflux. Curr Top Membr. 2012;69:325–358. doi: 10.1016/B978-0-12-394390-3.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Balsera M, Soll J, Buchanan BB. Redox extends its regulatory reach to chloroplast protein import. Trends Plant Sci. 2010;15:515–521. doi: 10.1016/j.tplants.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 108.Minamino T, Shimada M, Okabe M, Saijo-Hamano Y, Imada K, Kihara M, et al. Role of the C-terminal cytoplasmic domain of FlhA in bacterial flagellar type III protein export. J Bacteriol. 2010;192:1929–1936. doi: 10.1128/JB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Herrmann JM. Converting bacteria to organelles: evolution of mitochondrial protein sorting. Trends Microbiol. 2003;11:74–79. doi: 10.1016/S0966-842X(02)00033-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The list of organisms with publicly-available data on the ion channels and secondary transporters. (DOCX 168 kb)
Data S1. Genomic and transportome data on the organisms included in the study. (XLSX 255 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files (Additional file 2: Data S1 and Additional file 1: Table S1). The raw genomic data are publicly-available at http://www.ensembl.org/index.html, http://fungi.ensembl.org/index.html, http://protists.ensembl.org/index.html, and http://plants.ensembl.org/index.html.