Abstract
Human mesenchymal stem cells (hMSCs) are encapsulated in synthetic matrix metalloproteinase (MMP) degradable poly(ethylene glycol)-peptide hydrogels to characterize cell-mediated degradation of the pericellular region using multiple particle tracking microrheology. The hydrogel scaffold is degraded by cell-secreted enzymes and cytoskeletal tension. We determine that cell-secreted enzymatic degradation is the main contributor to changes in the pericellular region, with cytoskeletal tension playing a minimal role. Measured degradation profiles for untreated and myosin II inhibited hMSCs have the highest cross-link density around the cell. We hypothesize that cells are also secreting tissue inhibitor of metalloproteinases (TIMPs) to inhibit MMPs, which allow cell spreading and attachment prior to motility. We develop a Michaelis–Menten competitive enzymatic inhibition model which accurately describes the degradation profile due to MMP–TIMP unbinding.
Keywords: cytoskeletal tension, degradation, matrix metalloproteinase, multiple particle tracking microrheology
Graphical Abstract
Synthetic hydrogel scaffolds can control and manipulate 3D encapsulated cellular processes using environmental cues. This ability to control cellular processes has the potential to enhance their use as wound healing and tissue regeneration platforms.1–5 The design of these materials draws inspiration from cellular and developmental biology and regenerative medicine, slowly increasing the scaffold complexity to recreate in vivo environments in vitro.6,7 These scaffolds recapitulate aspects of the native extracellular matrix (ECM) and present precisely engineered chemical and physical microenvironments to encapsulated cells. In particular, poly(ethylene glycol) (PEG) has been widely used in scaffolds due to its biocompatibility and nonadsorptive protein properties.8 Incorporation of adhesion ligands and enzymatically degradable peptide cross-linkers into the scaffold creates an initially well-defined microenvironment that is re-engineered by encapsulated cells during basic processes.2,6,9 These scaffolds respond temporally and spatially to cellular interactions, continually presenting new cues in the microenvironment to cells throughout this process. To use this complex interplay between cells and their microenvironment to control cellular processes, we must first understand the extent and mechanism of cellular degradation of our synthetic scaffolds. In this work, degradation is defined as cleavage of the peptide cross-linker.
Our work focuses on the characterization of the changing rheological properties in the pericellular region. From this characterization, we determine the main mechanism of degradation by 3D encapsulated human mesenchymal stem cells (hMSCs) prior to motility in PEG-peptide scaffolds. hMSC-mediated degradation occurs through two processes: cell-secreted enzymatic degradation and cellular traction on the scaffold.9,10 In this work, we focus on the change in properties prior to motility because previous work determined that migration occurs in two steps: (1) the cell creates a degradation profile in the scaffold that enables attachment and spreading and is not motile and (2) the cell is motile and has degraded the entire scaffold on the length scale of our measurements (1 μm).9 hMSC motility is of particular importance in wound healing and tissue regeneration. During wound healing, hMSCs are called to the wound by chemical cues in the environment. In response, they migrate out of their niche and traverse mechanically distinct microenvironments to reach the wound and regulate healing.10 hMSCs secrete matrix metalloproteinases (MMPs) to degrade ECM proteins during migration. The main MMPs secreted by hMSCs are MMP-1, -2, -9, and -13.10,11 In our synthetic hydrogel scaffold, cell-secreted MMPs degrade the peptide cross-linker allowing hMSCs to create paths for facile migration.9
The synthetic hydrogel scaffold used for 3D hMSC encapsulation is composed of four-arm star PEG end-functionalized with norbornene (Mn = 20 000 g mol−1, 3 mM) cross-linked with an MMP degradable peptide sequence, KCGPQG↓IWGQCK (Mn = 1305 g mol−1, 3.9 mM).12 These molecules undergo a step-growth photopolymerization reaction to form the hydrogel scaffold initiated by lithium phenyl-2,4,6- trimethylbenzoylphosphinate (LAP).13 The MMP degradable peptide cross-linker is chosen for its fast degradation kinetics, enabling facile cell-mediated degradation.2 An adhesion ligand, CRGDS (Mn = 594 g mol−1, 1 mM) is tethered to the scaffold to enable cellular attachment. hMSCs are encapsulated in the scaffold by addition of a concentrated cell suspension in 1× phosphate buffered saline to the precursor solution. The final concentration of hMSCs is 2 × 105 cells mL−1. This low concentration is chosen to enable measurements of the rheological changes in the pericellular region due to hMSC-mediated degradation from a single cell.9
Multiple particle tracking microrheology (MPT) is used to measure dynamic changes in hydrogel properties around each hMSC. In MPT, fluorescent probe particles are embedded in the material and the Brownian motion of the particles is measured.5,14–16 This motion is related to the rheological properties of the material using the Generalized Stokes–Einstein Relation: , where a is the particle radius, kBT is the thermal energy, J(τ) is the creep compliance, 〈Δr2(τ)〉 is the measured ensemble averaged mean-squared displacement (MSD) and τ is the lag time. The logarithmic slope of the MSD, , determines the state of the material. α → 0 indicates that particles are arrested in a gel scaffold. α = 1 indicates particles are freely diffusing in a liquid. 0 < α < 1 is indicative of particles in a viscoelastic fluid or solid.14,15,17 The state of the material is quantitatively defined by comparison of α to the critical relaxation exponent, n, determined using time-cure superposition. n is the value of α where, during degradation, the last sample-spanning network cluster breaks and the material transitions from a gel to a sol.12,18,19 When n < α the material is a viscoelastic liquid and when n > α the material is a viscoelastic solid.12,20,21 n is a material property and has been previously reported as n = 0.25 ± 0.16 for this hydrogel scaffold.12
Particle image velocimetry (PIV) is also used to quantify the displacement of the particles on long time scales, over several minutes.9 This analysis determines the impact of cytoskeletal tension on the hydrogel scaffold by quantifying particle displacement between two images taken 4–6 min apart. This analysis determines the direction and velocity of particles due to the cell pulling on the scaffold. Unlike traction force microscopy (TFM), we are not measuring the traction a cell has on the scaffold. Instead, we are determining the displacement of the scaffold structure due to cellular traction. It should be noted that TFM cannot be used for this hydrogel scaffold because a main assumption of TFM is that the properties of the material are not changing.22 This assumption is violated in this material which is designed for facile cell-mediated degradation. PIV analyzes bright-field images taken prior to MPT data collection of the same field of view using ImageJ.23
Data are taken 18–48 h after encapsulation in two hydrogels per stock solution with 3–5 different cells measured per gel. Three biological replicates are also measured for each experimental condition. Details of the methods of cell encapsulation, hMSC treatment, data acquisition and results of control experiments are presented in the Supporting Information. We measure the change in the rheological properties of the pericellular region around hMSCs that are untreated, treated with a myosin II inhibitor to prevent cytoskeletal tension on the network and treated with an MMP inhibitor to prevent enzymatic degradation of the hydrogel scaffold. Myosin IIs are adenosine triphosphate-driven molecular motors found in eukaryotic cells that have many diverse functions, including muscle contraction and cortical tension.24 hMSC cytoskeletal tension on the hydrogel network is due to myosin II activity. MMP inhibited hMSCs do not degrade the scaffold over the data acquisition window. This work is detailed in the Supporting Information. Because cells that cannot secrete MMPs are unable to degrade the scaffold, we conclude that cytoskeletal tension does not have a major contribution in degradation of the hydrogel scaffold in the pericellular region. Therefore, we will focus our discussion on untreated hMSCs and myosin II inhibited hMSCs.
MPT characterizes changes in the pericellular region of hMSCs over time. Figure 1a shows the changes of the logarithmic slope of the MSD, α, through time for six untreated hMSCs in a single hydrogel. Measurements capture hMSCs at three time points during motility. In the first time point hMSCs are encapsulated in the gel network and do not degrade the scaffold during the measurement time period. The second time point shows hMSCs initially in a gel scaffold, α = 0, and through time they degrade the hydrogel past the gel–sol transition, α = n = 0.25, to the liquid phase, α → 1. The third time point shows that at the time that data acquisition of the pericellular region is begun the cell has already degraded the material (α = 1) and motility is observed. This degradation is a local phenomenon, which has an impact on the bulk rheological properties. With this low concentration of hMSCs, each cell will carve a trail through the scaffold. This will change the bulk modulus of the scaffold but the bulk hydrogel does not undergo a gel–sol transition over the measurement window.
Figure 1.
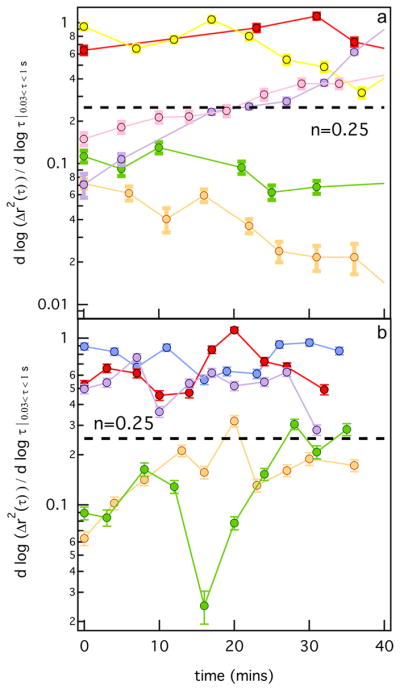
Logarithmic slope of mean-squared displacement, α, in the pericellular region through time for (a) untreated and (b) myosin II inhibited hMSCs. The dashed line represents the gel–sol transition determined previously by Schultz et al. using time-cure super-position. 12
Figure 1b shows the changes of the α value through time around encapsulated hMSCs after inhibition of myosin II. Again, the pericellular region is characterized at the same time points during scaffold degradation as were measured for the untreated hMSCs. We measure hMSCs that are degrading the hydrogel past the gel–sol transition and those that have already degraded the scaffold. Untreated and myosin II inhibited hMSCs differ in the time scale of degradation from a gel to a sol, which is slower around myosin II inhibited cells. Both sets of data show similar trends in degradation, confirming that the inhibition of cytoskeletal tension on the network is not changing cell-mediated scaffold degradation.
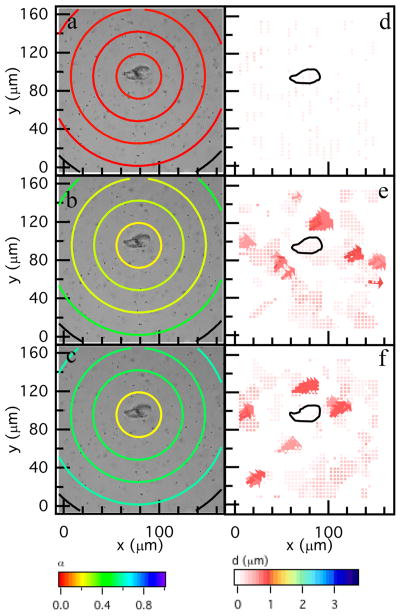
The spatial hydrogel microenvironment during hMSC-mediated degradation prior to motility is characterized using MPT and PIV, Figure 2. These data are a biological and material replicate of data previously published in Schultz et al. using hMSCs isolated from a different donor.9 It is presented here for comparison with myosin II inhibited hMSCs and shows the repeatability of the measured degradation profile in the pericellular region. For all experiments, t = 0 is the time that the cell is identified during the acquisition window, which is 18–48 h after encapsulation. MPT quantitatively characterizes the temporal changes in material properties as a function of the distance from the cell center, Figure 2a–c. In this figure, α is calculated for particles in a given area, which is represented by the colored ring on the graph. The first circle has a radius of 23 μm and this radius is increased by 23 μm to the edge of the field of view. Only the particles within the bounding ring are used to calculate α.9 The color of the ring is the value of α. Warm colors indicate small movement of probes in the gel phase; while, cool colors indicate probes are diffusing in a viscoelastic liquid. The expectation for the degradation profile was that the largest degradation would occur directly around the cell, the source of the enzymes that degrade the scaffold, with an increasing cross-link density moving further from the cell center. Surprisingly, measurements show that the highest cross-link density is directly around the cell with the scaffold remaining in the gel phase during the 60 min measurement period. The cross-link density decreases as a function of the distance away from the cell center, Figure 2b, c. The cell is creating a microenvironment that enables spreading and attachment to the scaffold prior to migration.9
Figure 2.
Spatial degradation profiles around an encapsulated hMSC prior to motility. The left column are MPT measurements and the right column are PIV measurements. MPT data are collected through time after identification of the hMSC at (a) 0, (b) 36 and (c) 60 min. quantifies the material microenvironment as a function of the distance from the cell center. The state of the material is determined by comparing α to the critical relaxation exponent, n. When α > n the material is a sol and when α < n the material is a gel. PIV measures the distance probes travel over larger time scales, namely between (d) 0–6, (e) 30–36 and (f) 54–60 min. PIV measurements indicate directed motion of probes which is a result of cytoskeletal tension on the network. The black outline is the outline of the hMSC.
Figure 2d–f are PIV measurements. The color and size of the arrows are particle displacement between images taken several minutes apart. Warm colors represent small displacements and cool colors represent large displacements. The cell is outlined in Figure 2d–f. Lack of arrows in Figure 2d is undetectable particle displacement indicating no cytoskeletal tension in the pericellular region. In Figure 2e, f, arrows near the cell center indicate cytoskeletal tension on the network. There are small displacement of particles at the edge of the field of view that are caused by particle imaging distortion.
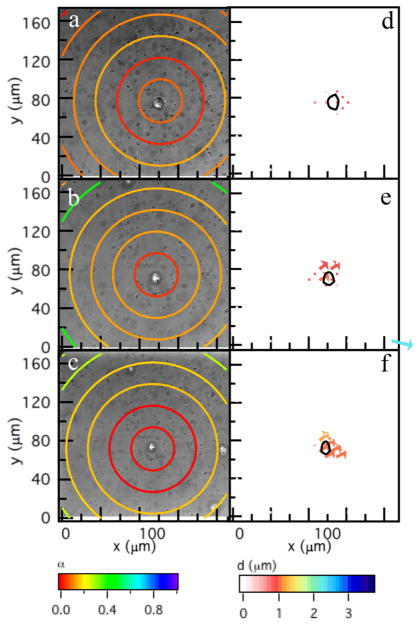
To separate the role of cell-secreted enzymes and cytoskeletal tension in material degradation, cytoskeletal tension is inhibited using blebbistatin, a myosin II inhibitor. MPT data of the microenvironment around myosin II inhibited hMSCs shows the same degradation profile as untreated hMSCs, Figure 3a–c. PIV shows minimal cytoskeletal tension directly under the cell, Figure 3d–f. These results demonstrate that matrix degradation is due to cell-secreted MMPs and cytoskeletal tension has a negligible influence on scaffold degradation.
Figure 3.
Dynamic microrheological characterization around an encapsulated hMSC after myosin II inhibition. MPT measurements (left column) at (a) 0, (b) 24, and (c) 35 min. The colored rings represent the logarithmic slope of the MSD, . Warm colors are a low value of α representing a gel while cool colors represent a liquid. PIV measurements (right column) are between (d) 0–5, (e) 24–30, and (f) 30–35 min and measure cytoskeletal tension on the scaffold. The hMSC is outlined in d–f. The area of this plot has been reduced to match Figure 2, the complete field of view measurements are in the Supporting Information.
These experiments together suggest that there is a chemical mechanism that is creating the measured degradation profile around untreated hMSCs. hMSCs can also secrete a second set of enzymes, tissue inhibitor of metalloproteinases (TIMPs). TIMPs bind to MMPs and inhibit their activity, making them unable to degrade the hydrogel scaffold.11,25 TIMPs also slowly unbind from MMPs, making MMPs active once they have diffused away from the cell.25 Four TIMPs have been identified (TIMP-1–4) and hMSCs mainly secrete TIMP-1 and -2.11,25 We hypothesize that TIMPs are inhibiting MMP activity close to the cell enabling spreading and attachment prior to migration.
To understand the role of TIMPs in scaffold degradation we model this mechanism using Michaelis–Menten competitive inhibition kinetics. To describe this mechanism, MMPs are the enzymes, E, which are inhibited by TIMPs, I. The derivation and complete set of equations are available in the Supporting Information. The kinetic parameters are measured in previous work by Olson et al. that investigated MMP–TIMP binding and unbinding in solution experiments.25 The rate of MMP–TIMP unbinding, R, as a function of distance from the cell, r, is calculated from the following equation
| (1) |
where [] indicates concentration, [S0] and [I0] are the initial concentrations of the substrate (cross-linker) and inhibitor (TIMP), kon and koff are the rate of binding and unbinding of TIMPs to MMPs, kdes and kads are the desorption and adsorption rate, r0 is the radius of the cell, and ϕ is the Thiele modulus. The Thiele modulus is defined as , where D is diffusivity and kcat is the catalytic rate constant. Figure 4 shows the unbinding of TIMP-1 from MMP-2. This model predicts that MMP–TIMP binding will inhibit MMP activity and degradation approximately 50 μm from the center of the cell. When compared to MPT measurements, the area where degradation first occurs is approximately 50 μm from the cell center, shown in Figures 2 and 3, which agrees with the model. For comparison, we determine the distance away from the cell where degradation begins using data from MPT measurements. The average distance where degradation first occurs around a cell (using three biological replicates, and in each replicate data is taken over 2 days in two hydrogels per stock solution) for untreated hMSCs is 49.3 ± 25.3 μm and for myosin II inhibited hMSCs is 54.05 ± 28.19 μm. This model is predicting activity due to MMP–TIMP unbinding. Therefore, in the area past the maximum reaction rate (over 70 μm from the cell center) the model predicts a decrease in MMP–TIMP unbinding, which is expected since the maximum unbinding rate has already been achieved (Figure S10).
Figure 4.
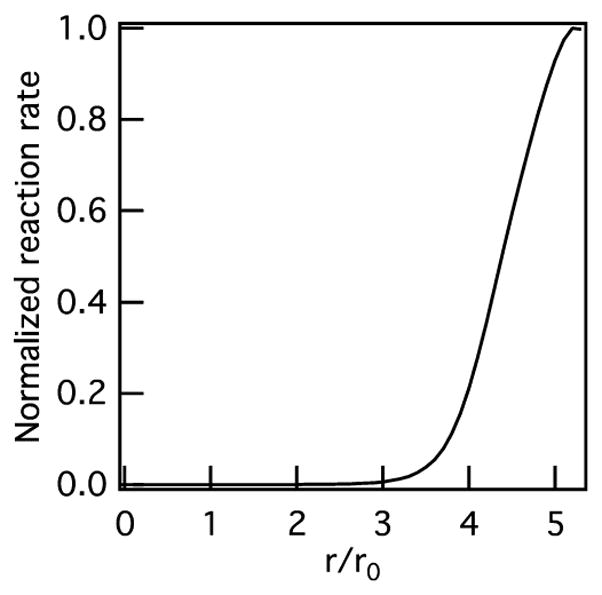
Reaction rate of unbinding of TIMP–1 from MMP–2 as a function of normalized distance from the cell calculated using Michaelis–Menten competitive inhibition kinetics.
In summary, multiple particle tracking microrheology is used to measure the degradation profile in the pericellular region prior to 3D hMSC migration. We measure a degradation profile where scaffold degradation does not occur directly around the cell and the cross-link density decreases as the distance away from the cell is increased. We then determine the contributions of two cell-mediated degradation mechanisms to the measured degradation profile: enzymatic degradation by cell-secreted MMPs and cytoskeletal tension. To decouple their impact on material degradation, we inhibit both MMPs using an MMP inhibitor and cytoskeletal tension by inhibiting myosin II activity using blebbistatin. MMP inhibition results in no degradation of the hydrogel scaffold. Measurements of scaffold degradation in the pericellular region of untreated and myosin II inhibited hMSCs show the degradation profiles are the same. In the long time scale, t ≈ 4–6 min, PIV of untreated cells show that hMSCs adhere and pull on the scaffold prior to motility. In myosin II inhibited hMSCs, no cytoskeletal tension on the scaffold is measured. Taken together, cytoskeletal tension has a minimum effect on hydrogel degradation. The measured degradation profile is due to chemical degradation by hMSC-secreted enzymes. We hypothesize that TIMPs are inhibiting MMP activity close to the cell creating this unique degradation profile and an environment for the cell to spread and attach. This mechanism is modeled using Michaelis–Menten competitive inhibition kinetics and agrees well with MPT measurements. The role of TIMPs in matrix degradation has not been well studied and is the subject of our future investigations. Understanding these fundamental phenomena will improve the design and engineering of biomaterials that can direct cellular processes to enhance biological applications, such as tissue regeneration and wound healing.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number R15GM119065. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors acknowledge Professor Kristi S. Anseth for helpful discussions.
ABBREVIATIONS
- ECM
extracellular matrix
- MMP
matrix metalloproteinase
- TIMP
tissue inhibitor of metalloproteinase
- MPT
multiple particle tracking microrheology
- PIV
particle image velocimetry
- MSD
mean-squared displacement
Footnotes
Author Contributions
M.D and K.M.S designed the project. M.D. and K.M.S. performed the experiments and data analysis and wrote the manuscript. M.D., H.S.C., and K.M.S. developed the model.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsbiomaterials. 7b01005.
Experimental details for materials, device fabrication, methods, control experiments, and additional figures (PDF)
References
- 1.Ferreira LS, Gerecht S, Fuller J, Shieh HF, Vunjak-Novakovic G, Langer R. Bioactive hydrogel scaffolds for controllable vascular differentiation of human embryonic stem cells Bioactive hydrogel scaffolds for controllable vascular differentiation of human embryonic stem cells. Biomaterials. 2007;28:2706–2717. doi: 10.1016/j.biomaterials.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patterson J, Hubbell J. Enhanced proteolytic degradation of molecularly engineered PEG hydrogels in response to MMP-1 and MMP-2. Biomaterials. 2010;31:7836–7845. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 3.Peppas BNA, Hilt JZ, Khademhosseini A, Langer R. Hydrogels in Biology and Medicine: From Molecular Principles to Bionanotechnology. Adv Mater. 2006;18:1345–1360. [Google Scholar]
- 4.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677– 689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 5.Bloom RJ, George JP, Celedon A, Sun SX, Wirtz D. Mapping local matrix remodeling induced by a migrating tumor cell using three-dimensional multiple-particle tracking. Biophys J. 2008;95:4077–4088. doi: 10.1529/biophysj.108.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz MP, Fairbanks BD, Rogers RE, Rangarajan R, Zaman MH, Anseth KS. A synthetic strategy for mimicking the extracellular matrix provides insight about tumor cell migration. Integr Biol. 2010;2:32–40. doi: 10.1039/b912438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization. Adv Mater. 2009;21:5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peyton SR, Raub CB, Keschrumrus VP, Putnam AJ. The use of poly(ethylene glycol) hydrogels to investigate the impact of ECM chemistry and mechanics on smooth muscle cells. Biomaterials. 2006;27:4881–4893. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Schultz KM, Kyburz KA, Anseth KS. Measuring dynamic cell-material interactions and remodeling during 3D human mesenchymal stem cell migration in hydrogels. Proc Natl Acad Sci U S A. 2015;112:E3757–E3764. doi: 10.1073/pnas.1511304112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ries C, Egea V, Karow M, Kolb H, Jochum M, Neth P. MMP-2, MT1-MMP, and TIMP-2 are essential for the invasive capacity of human mesenchymal stem cells: differential regulation by inflammatory cytokines. Blood. 2007;109:4055–4063. doi: 10.1182/blood-2006-10-051060. [DOI] [PubMed] [Google Scholar]
- 11.Lozito TP, Jackson WM, Nesti LJN, Tuan RS. Human mesenchymal stem cells generate a distinct pericellular zone of MMP activities via binding of MMPs and secretion of high levels of TIMPs. Matrix Biol. 2014;34:132–143. doi: 10.1016/j.matbio.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Schultz KM, Anseth KS. Monitoring degradation of matrix metalloproteinases-cleavable PEG hydrogels via multiple particle tracking microrheology. Soft Matter. 2013;9:1570–1579. [Google Scholar]
- 13.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl- 2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials. 2009;30:6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crocker JC, Grier DG. Methods of Digital Video Microscopy for Colloidal Studies. J Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 15.Mason TG, Weitz DA. Optical Measurements of Frequency-Dependent Linear Viscoelastic Moduli of Complex Fluids. Phys Rev Lett. 1995;74:1250. doi: 10.1103/PhysRevLett.74.1250. [DOI] [PubMed] [Google Scholar]
- 16.Savin T, Doyle PS. Static and dynamic errors in particle tracking microrheology. Biophys J. 2005;88:623–638. doi: 10.1529/biophysj.104.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz KM, Furst EM. Microrheology of biomaterial hydrogelators. Soft Matter. 2012;8:6198–6205. [Google Scholar]
- 18.Adolf D, Martin JE. Time-cure superposition during crosslinking. Macromolecules. 1990;23:3700–3704. [Google Scholar]
- 19.Muthukumar M, Winter HH. Fractal dimension of a crosslinking polymer at the gel point. Macromolecules. 1986;19:1284– 1285. [Google Scholar]
- 20.Schultz KM, Baldwin AD, Kiick KL, Furst EM. Measuring the Modulus and Reverse Percolation Transition of a Degrading Hydrogel. ACS Macro Lett. 2012;1:706–708. doi: 10.1021/mz300106y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen TH, Furst EM. Microrheology of the liquid-solid transition during gelation. Phys Rev Lett. 2008;100:146001–146004. doi: 10.1103/PhysRevLett.100.146001. [DOI] [PubMed] [Google Scholar]
- 22.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods. 2010;7:969–973. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng Q. PIV (Particle Image Velocimetry) – ImageJ plugin. 2014 https://sites.google.com/site/qingzongtseng/piv.
- 24.Kovacs M, Toth J, Hetenyi C, Malnasi-Csizmadia A, Sellers JR. Mechanism of Blebbistatin Inhibition of Myosin II. J Biol Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 25.Olson MW, Gervasi DC, Mobashery S, Fridman R. Kinetic Analysis of the Binding of Human Matrix Metalloproteinase-2 and –9 to Tissue Inhibitor of Metalloproteinase (TIMP)-1 and TIMP-2. J Biol Chem. 1997;272:29975–29983. doi: 10.1074/jbc.272.47.29975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.