Abstract
Telomeric repeat-containing RNA is a new noncoding RNA molecule that performs various biofunctions. Heterogeneous nuclear ribonucleoprotein (hnRNP) A1 is an RNA-binding protein involved in the telomere maintenance machinery. To date, little is known about how hnRNPA1 binds to telomeric RNA. In this study, we investigated the binding affinity and recognition mechanism of telomere RNA with the RNA recognition motif of hnRNPA1. Using the photochemical cross-linking method, we showed that the telomere RNA G-quadruplex with loops is important in the interaction of telomere RNA with hnRNPA1. Using small-molecule probes, we directly visualized the complex formed by the telomere RNA G-quadruplex and hnRNPA1 in vitro and in live cells. The results suggested that the structure-dependent binding of hnRNPA1 to telomere RNA regulates the telomere function. Therefore, our study provides new insights into the interactions between the RNA G-quadruplex and proteins at the telomere.
Graphical abstract
INTRODUCTION
Telomeric RNA molecules containing heterogeneous lengths of UUAGGG repeats were detected in several human and rodent cell lines.1,2 Human telomeric RNA performs various biofunctions, such as the regulation of the telomere length,3–12 heterochromatin formation,13,14 and end protection.15,16 Recent studies suggest that telomeric RNA forms a G-quadruplex that plays an important role in telomere protection and regulation.3,16,17 We and others have shown that human telomeric RNA forms G-quadruplex structures.18–24 Additionally, we have demonstrated that telomeric RNA G-quadruplex structures play an important role in providing a protective structure to the telomere ends.16 This suggests that RNA G-quadruplexes and their related proteins are involved in telomere protection.
The heterogeneous nuclear ribonucleoprotein (hnRNP) family has been shown to regulate both the telomere length and telomerase.25–33 Recently, telomeric RNA was found to promote the binding of POT1 to telomeric single-stranded DNA by removing hnRNPA1.28 This suggests that hnRNPA1, telomere RNA, and POT1 act together to facilitate the telomere capping, which preserves genomic integrity. In this study, we investigated the recognition mechanism and binding affinity of the telomere RNA G-quadruplex with hnRNPA1. We found that hnRNPA1 binds to the human telomere RNA G-quadruplex, and the loops present in the telomere RNA G-quadruplex are required for its binding to hnRNPA1.
We performed a photochemical cross-linking experiment to confirm whether the loops in the telomere RNA G-quadruplex are the recognition site of hnRNPA1.
Furthermore, we developed a small macrocyclic molecule, 7OTD, and its fluorescent derivative, Cy5–7OTD (a Cy5 fluorophore linked to 7OTD), which act as ligands for the telomere RNA G-quadruplex. We found that these ligand molecules interact strongly and selectively with the telomere RNA G-quadruplex. Using Cy5–7OTD as a probe, we directly visualized the complex formed by the telomere RNA G-quadruplex and hnRNPA1 in live cells. The results suggest that telomere RNA and hnRNPA1 regulate the telomere function by binding in a structure-dependent manner, and the loops in the telomere RNA G-quadruplex structure are necessary for the binding of hnRNPA1 to human telomeric RNA.
RESULTS AND DISCUSSION
Structure-Dependent Binding of hnRNPA1 to the Telomere RNA G-Quadruplex
Telomere RNA with different lengths has been reported to form G-quadruplex structures with different topologies: tetramolecular (tetramer), bimolecular (dimer), and intramolecular with or without loops (Figure 1).18–24 We performed NMR experiments to reveal the structural features of different lengths of UUAGGG repeats. For the imino proton, in the 1H NMR spectra of these RNAs with Na+ or K+, the peaks of guanine imino protons in the G-quadruplex structures were observed at approximately 10.5–12.0 ppm (Figure S1, Supporting Information). These peaks were expectedly protected from the D2O exchange, and some peaks were still observed after being dissolved in D2O (Figure S2, Supporting Information). These results are in agreement with previous studies that showed that telomere RNA forms G-quadruplexes by using X-ray crystallography and NMR spectroscopy.18–24 To further reveal the structural features of telomere RNA, we synthesized a guanosine analogue, 7-deazaguanosine (7dG), and incorporated it into telomere RNA sequences (Scheme S1 and Figures S13–S22, Supporting Information). The 7-deazaguanosine-containing RNA does not show the imino signal (Figure S3, Supporting Information), suggesting that it was not able to form a G-quadruplex.34 This confirms that the substitution of N7 with a carbon in 7-deazaguanosine suppresses the hydrogen bonding of base pairs and prevents the formation of the G-quadruplex by the same sequence of RNA.
Figure 1.
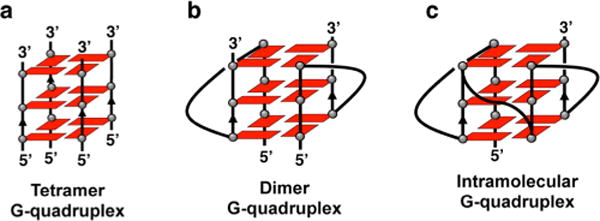
Schematic structures of the human telomere RNA G-quadruplex: (a) tetramolecular G-quadruplex without loops (tetramer), (b) bimolecular RNA G-quadruplex with loops (dimer), and (c) intramolecular RNA G-quadruplex with loops.
RNase T1 footprinting was used to further confirm the formation of the RNA G-quadruplex.34–36 G residues in the G-quadruplex are protected from cleavage, but the same sequence of 7-deazaguanosine-containing RNA was cleaved sufficiently at G residues (Figure S4a, Supporting Information). RNase T1 digestion was carried out in K+ and Li+ buffers to further confirm that these RNAs fold into the G-quadruplex. G residues are protected from the digestion of RNase T1 in K+ buffer relative to Li+ buffer (Figure S4b). The result of the cleavage pattern of G residues suggests that these telomere sequences can form the RNA G-quadruplex.
To investigate the binding of different telomere RNA G-quadruplexes to hnRNPA1, we performed an electrophoretic mobility shift assay using 32P-labeled human telomere RNA sequences with different lengths of UUAGGG repeats. We found that hnRNPA1 binds to the bimolecular and intra-molecular RNA G-quadruplexes with loops, but it does not bind to the tetramolecular RNA G-quadruplex abasic loops and the single-stranded RNA used as a control (Figure 2). To further confirm the structure-specific binding, we performed an electrophoretic mobility shift assay using the 7-deazaguanosine-containing telomere RNA. We found that hnRNPA1 binds to the RNA G-quadruplex formed by telomere RNA, but it does not bind to the same sequence of 7-deazaguanosine-containing RNA (Figure S5, Supporting Information). We performed NMR experiments to characterize the hnRNPA1-RNA complex. The addition of hnRNPA1 into 12-mer RNA (at a 1:1 RNA:protein ratio) resulted in the disappearance of partial 1H NMR signals of guanine imino protons, and new peaks of upfield-shifted broadened imino proton resonances appeared (Figure S6, Supporting Information). The results show the preferential binding of hnRNPA1 to telomere RNA G-quadruplexes bearing loops. We further performed a concentration-dependent experiment to examine the binding ability of hnRNPA1 to the telomere RNA G-quadruplex. At lower concentrations of hnRNPA1, free RNA was observed as the band with a faster mobility (Figure 3a). By increasing the concentration of hnRNPA1, the amount of RNA–protein complex progressively increased and the amount of free RNA decreased. We noted that the experiment with the 24-mer RNA shows two gel-shifted species (Figure 3a), and this will be investigated further to determine whether a multiple binding event or some other phenomenon occurs.
Figure 2.
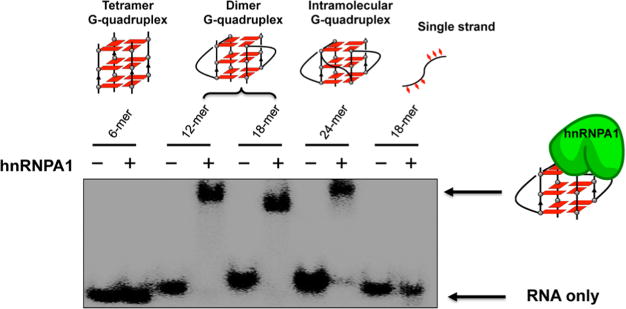
Telomere RNA G-quadruplex binding selectivity of hnRNPA1. The experiment was performed with hnRNPA1 and 32P-labeled 6-mer UUAGGG RNA (tetramer), 12-mer UUAGGGUUAGGG RNA (dimer), 18-mer UUAGGGUUAGGGUUAGGG RNA (dimer), 24-mer UUAGGGUUAGGGUUAGGGUUAGGG RNA (intramolecular G-quadruplex), and single-stranded GAGUAACCCGUAUCGUGA RNA using 8% PAGE in 1× TBE buffer with 20 mM NaCl at 4 °C for 2 h (80 V).
Figure 3.
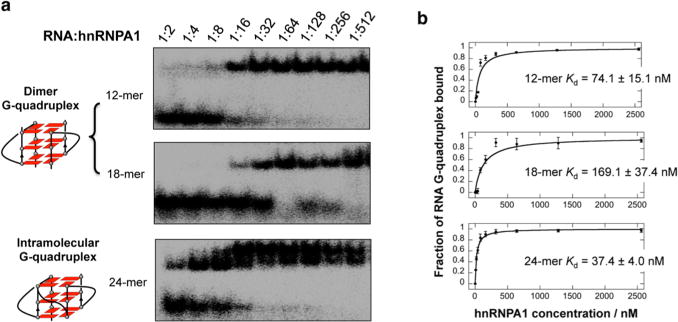
(a) Binding affinity of hnRNPA1 to dimer and intramolecular G-quadruplexes. The ratio of RNA and hnRNPA1 is shown for each lane ([RNA] = 5 nM). (b) Binding curve of hnRNPA1 to RNA from the gel shift assay. The Kd values were obtained by fitting the mobility shift data to the equation given in the Methods. Error bars for each data point represent the range of three independent experiments.
The dissociation constants (Kd) were given by fitting the mobility shift data to the hyperbolic equation. This analysis yielded a Kd of 74.1 or 169.1 nM for the bimolecular G-quadruplex and 37.6 nM for the intramolecular G-quadruplex (Figure 3b). These results suggested the preferential binding of hnRNPA1 to RNA G-quadruplex structures bearing loops in a structure-dependent and sequence-independent manner.
All G-quadruplexes were identified using CD spectroscopy. A negative peak at approximately 240 nm and a positive peak at approximately 265 nm confirm parallel-stranded G-quadruplexes (Figure S7, Supporting Information). The addition of hnRNPA1 to these G-quadruplexes did not significantly change the spectra, which indicates that hnRNPA1 does not affect the G-quadruplex structures (Figure S7).
Photo-Cross-Linking Study To Identify That the Loop of the RNA G-Quadruplex Is the Recognition Site of hnRNP A1
Photo-cross-linking is a useful method to study the interaction points of nucleic acid–protein complexes.37–39 Therefore, we performed a photochemical cross-linking experiment to identify the loop in the telomere RNA G-quadruplex that is recognized by hnRNPA1. Thymine (T) in DNA and uracil (U) in RNA can be replaced by 5-halouracil (IU or BrU) at a single position. Ultraviolet irradiation of the complexes of 5-halouracil-substituted DNA and RNA with protein can produce covalent bonds between the amino acid and nucleobase residues at the interaction site of the nucleic acid–protein complex. The covalent complex generated by photo-cross-linking can be used to identify the contact points in nucleoprotein complexes. Recently, we probed local DNA conformations using the photoreactions of 5-halouracil-substituted DNA.38
In this study, we prepared a 32P-labeled UUAGGGU(BrU)-AGGG (12-mer) telomere RNA by replacing U with BrU at the loop. As shown in Figure 4a, 32P-labeled telomere RNA containing a BrU was photo-cross-linked to hnRNPA1 by irradiation at 302 nm. Samples were then analyzed by denaturing polyacrylamide gel electrophoresis (PAGE). The photo-cross-linked product was found in the presence of hnRNPA1 with irradiation, but it was not found in the absence of hnRNPA1 and without irradiation (Figure 4b). We observed a time-dependent appearance of a lower mobility band compared to the mobility of the free RNA G-quadruplex band (Figure 4c). The intensity of the band with low mobility increases with a longer irradiation time. This increase in band intensity suggests the photo-cross-linking between the telomere RNA G-quadruplex and hnRNPA1. The photo-cross-linking yields were fitted as a time response, and the RNA–protein complex results in a cross-linking yield of approximately 50% (Figure 4d).
Figure 4.
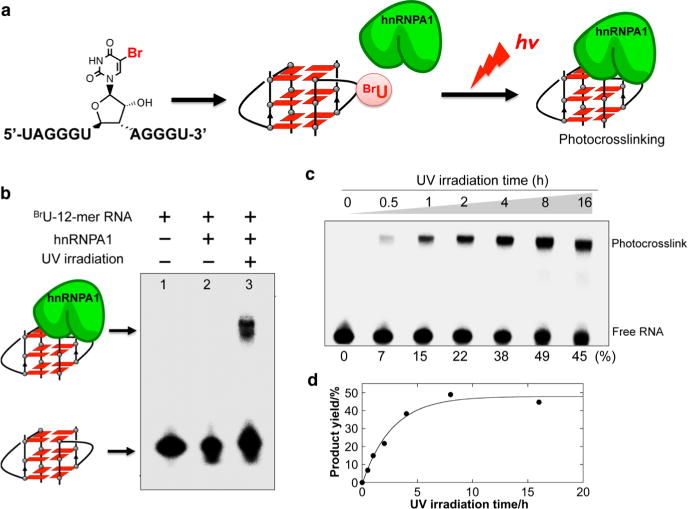
(a) Schematic overview of the photo-cross-linking experiment of hnRNAPA1 and the telomere RNA G-quadruplex containing a 5-bromouracil at the loop. (b) Denaturing PAGE of the reaction mixture containing 32P-labeled 12-mer RNA in the presence or absence of hnRNPA1 with or without irradiation. (c) Photo-cross-linking reaction as a function of time. Conversion yields for each irradiation time are shown at the bottom of the gel. (d) Photo-cross-linking yields of the RNA G-quadruplex to hnRNPA1. The band signals for each cross-linked and un-cross-linked RNA were quantitated, and the cross-linked product in each reaction was plotted versus the UV irradiation time.
The cross-linked RNA–protein complex was then enzymatically digested with trypsin for matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectral characterization. As expected, the peptide with the cross-linked telomere RNA was detected by mass spectrometry (MS) as opposed to the absence of the peptide from the free hnRNP A1 (Figure S8, Supporting Information). The photo-cross-linking study provides evidence that the loop in the RNA G-quadruplex is the recognition site of hnRNPA1; thus, the loop plays a key role in the binding of hnRNPA1 to the RNA G-quadruplex.
Detection of the hnRNPA1 and RNA G-Quadruplex Complex in Vitro and in Live Cells by a Small-Molecule Probe
The properties of telomere RNA G-quadruplexes suggest that they may become the targets for the development of small-molecule ligands. Indeed, a few small molecules have been designed and used to target telomere RNA G-quadruplexes.22,40,41 Recently, we showed that a series of macrocyclic molecules have a high selectivity toward G-quadruplex structures. We also determined the NMR structure of a complex of G-quadruplex and a macrocyclic hexaoxazole (6OTD) molecule.42 These studies provide a platform for the design of compounds that target the topology of the G-quadruplex.
On the basis of the previous studies, we designed and synthesized a macrocyclic heptaoxazole (7OTD) molecule and its fluorescence derivative Cy5–7OTD by introducing an additional oxazole moiety in 6OTD (Figure 5a).42,43 The ligand was expected to efficiently interact with the telomere RNA G-quadruplex and serve as a probe for telomere RNA G-quadruplex visualization.
Figure 5.
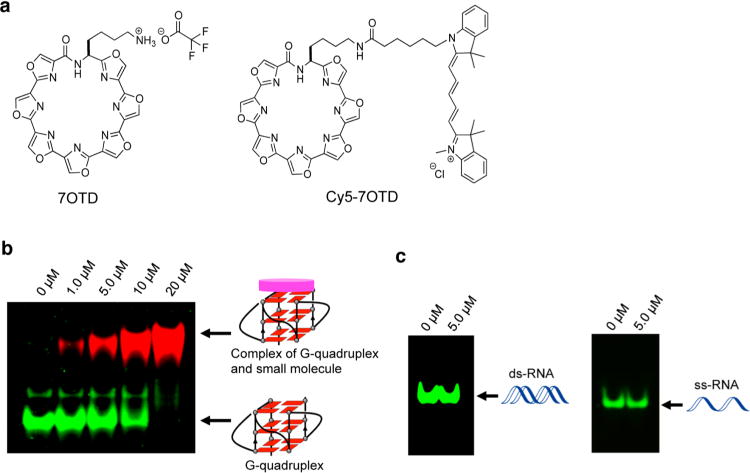
(a) Chemical structure of macrocyclic heptaoxazole (7OTD) and its fluorescence derivative Cy5–7OTD (a Cy5 fluorophore linked to 7OTD). (b) Visualization of the telomere RNA G-quadruplex formed by 24-mer RNA with Cy5–7OTD. RNA bands were stained with GelStar (green). Complexes of the G-quadruplex and Cy5–7OTD were detected by the fluorescence of Cy5–7OTD (red). Concentrations of Cy5–7OTD are shown above the gel. (c) Visualization of ds-RNA and ss-RNA with Cy5–7OTD. RNA bands were stained with GelStar (green). Concentrations of Cy–7OTD are shown above the gel. Conditions: 20% PAGE in 1× TBE buffer with 20 mM NaCl at 4 °C for 8 h (80 V).
The ligand’s ability to bind with the telomere RNA G-quadruplex was evaluated via a circular dichroism (CD) melting experiment. 7OTD attains a significant stabilization by a melting temperature (Tm) higher than 90 °C (Figure S9a, Supporting Information). Indeed, 7OTD gives a higher stabilization with the RNA telomere G-quadruplex structure than with a DNA telomere G-quadruplex structure (Figure S9b), indicating its high RNA G-quadruplex binding affinity.
We further performed an electrophoretic mobility shift assay using Cy5–7OTD to detect the interaction of the ligand with the RNA G-quadruplex structure. As shown in Figure 5b, free RNA was visualized as a green band with a high mobility (the RNA sample was stained with GelStar), whereas red bands were visualized by increasing the Cy5–7OTD concentration, which indicates the formation of a G-quadruplex and Cy5–7OTD complex. The G-quadruplex–ligand complex is visualized as a red band due to the fluorescence of Cy5–7OTD, and the band had a low mobility. With an increase in the concentration of Cy5–7OTD, the intensity of the red band increased concomitantly with a decrease in the intensity of the green band until there was a complete loss of the green band. This observation further confirms that Cy5–7OTD interacts with the G-quadruplex. Consistent with the finding that 7OTD selectively binds to the RNA G-quadruplex, red bands were not visualized when single- and double-stranded RNAs were used as controls for the experiment (Figure 5c). A hairpin RNA was used to further confirm that Cy5–7OTD does not bind to other RNA structures (Figure S10, Supporting Information).
After we confirmed the efficient binding of the ligand to the RNA telomere G-quadruplex, we probed the RNA G-quadruplex and hnRNPA1 complex with Cy5–7OTD (Figure 6). The 12-mer G-quadruplex shows a green band. The interaction of the RNA G-quadruplex with Cy5–7OTD was confirmed by the presence of a red band. The complex formed by the RNA G-quadruplex and hnRNPA1 was directly visualized in the upper portion of the gel because of the large molecular weight of hnRNPA1. Importantly, we observed the red band on the upper portion of the gel only after the addition of Cy5–7OTD to the RNA G-quadruplex and hnRNPA1 complex; thus, this complex can be directly visualized when Cy5–7OTD is used as a probe. We performed a CD spectroscopy experiment to investigate whether Cy5–7OTD influences the structure of the G-quadruplex and hnRNPA1 complex. The results showed that the addition of Cy5–7OTD to the G-quadruplex and hnRNPA1 complex did not affect the structure (Figure S11, Supporting Information).
Figure 6.
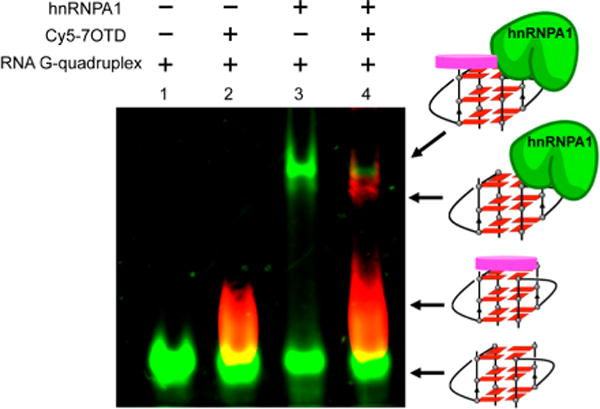
Visualization of the G-quadruplex and hnRNPA1 complex by Cy5–7OTD: lane 1, 12-mer RNA G-quadruplex labeled with fluorescein amidite (FAM) (green); lane 2, the G-quadruplex and Cy5–7OTD complex is indicated as the red color of Cy5–7OTD; lane 3, G-quadruplex and hnRNPA1 (green) complex; lane 4, Cy5–7OTD indicates the G-quadruplex and hnRNPA1 complex by the red fluorescence.
Subsequently, we visualized the RNA telomere G-quadruplex and hnRNPA1 complex in live cells using Cy5–7OTD (Figure 7). HeLa cells were incubated with 1 μM Cy5–7OTD and were visualized using fluorescence microscopy. The telomere RNA G-quadruplex can be visualized as red fluorescence by using Cy5–7OTD. The presence of hnRNPA1 in HeLa cells is shown by the immunofluorescence experiment using anti-hnRNPA1 antibodies that emit green fluorescence. Analysis of fluorescence microscopy images showed that the green fluorescence from hnRNPA1 overlapped with the red fluorescent foci from the RNA G-quadruplex. The signal of the RNA G-quadruplex was greatly reduced after ribonuclease (RNase) treatment, confirming that it originated from the telomere RNA G-quadruplex. We used a control RNA to further confirm that the colocalization resulted from the hnRNPA1–RNA complex and was not a simple consequence of single-stranded RNA recognition. As shown in Figure S12, Supporting Information, this colocalization was not observed when a 7-deazaguanosine-containing telomere RNA probe with a fluorophore (Cy3) at its 3′-terminus was used as a control. These results show that the telomere RNA G-quadruplex and hnRNPA1 form a complex in live cells.
Figure 7.
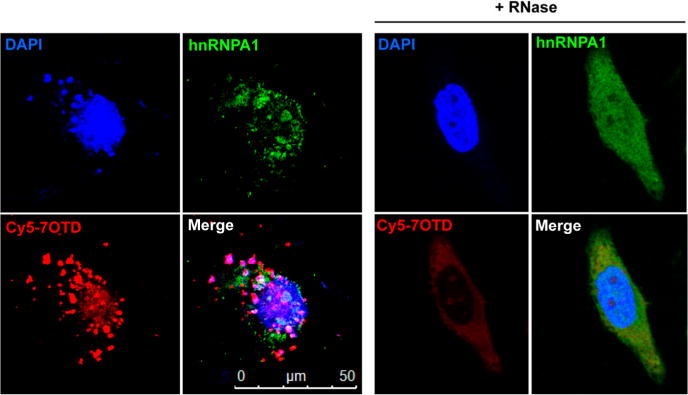
Fluorescence microscopy images of HeLa cells treated with Cy5–7OTD and then immunostained with antibodies to hnRNPA1. G-quadruplex RNA was detected by Cy5–7OTD (red). hnRNPA1 was immunostained using antihuman hnRNPA1 antibodies (green). Blue fluorescence indicates foci derived from DNA stained by DAPI. The merged panel shows a colocalization event. RNase-treated cells show a greatly reduced RNA signal.
In summary, we showed that the loops in the RNA G-quadruplex are important in the interaction between telomere RNA and hnRNPA1. The photo-cross-linking study shows that the loop in the RNA telomere G-quadruplex is the recognition site of hnRNPA1 in its binding to the RNA G-quadruplex. Using a small-molecule probe, we directly visualized the telomere RNA G-quadruplex and hnRNPA1 complex in vitro and in live cells. These results indicate a structure-dependent binding of hnRNPA1 to telomere RNA. Our study provides new insights into the interactions of the RNA G-quadruplex and proteins at the telomere.
METHODS
RNA, Protein, and Ligand
The chemical synthesis of 7-deazaguanosine-containing RNA is shown in the Supporting Information. All RNAs were prepared with an automatic DNA/RNA synthesizer (Nihon Techno Service Co., Ltd.). RNA samples were purified by reversed-phase HPLC (Jasco). RNAs were labeled with [γ-32P]ATP (PerkinElmer) and T4 polynucleotide kinase (2021S, TaKaRa Bio Inc.).
We transformed and expressed pET-15b vector (Novagen) containing the RNA-binding domain of hnRNPA1 from human in the Escherichia coli strain C41(DE3).44 On the basis of the manufacturer’s protocol, the protein was purified using Ni–nitrilotriacetic acid (NTA) affinity resin (Nacalai Tesque) and the bound nucleic acids were removed from E. coli by using Bensonase endonuclease digestion (Sigma-Aldrich), then repurified using Ni–NTA affinity resin, and concentrated by using a Vivaspin filter (GE Healthcare).
7OTD and its fluorescent derivative Cy5–7OTD were prepared by introducing an additional oxazole moiety in 6OTD according to the previous report.42,43
NMR Experiments
1H imino proton NMR was measured on a Bruker AVANCE 400 or 600 MHz spectrometer. Spectra were recorded at 23 °C. We dissolved the RNA samples (0.25 or 0.5 mM) in 150 μL of 90% H2O/10% D2O, 150 mM NaCl/KCl, and 20 mM sodium or potassium phosphate buffer (pH 7.0). For the hydrogen–deuterium exchange (HDX) experiments, NMR samples in aqueous solution were dried. A 150 μL portion of of 99% D2O was then added to dissolve the dried samples. Each sample was kept at room temperature for 2 h before each NMR measurement.
CD Experiments
We performed CD spectra using a J-820 CD spectrophotometer. The CD spectra of RNAs with or without hnRNPA1 were obtained in a buffer, 10 mM Tris–HCl buffer, pH 7.0, 100 mM KCl, 0.1 mg/mL bovine serum albumin (BSA), 5 mM dithiothreitol, and 10% (v/v) glycerol at room temperature for 2 h. The CD spectra of RNAs with or without 7OTD were obtained in 10 mM Tris–HCl, pH 7.0, 100 mM NaCl.
Gel Shift Assay
Binding experiments of 32P-labeled RNAs and hnRNPA1 were performed in a buffer, 10 mM Tris–HCl, pH 7.0, 100 mM NaCl, 5 mM dithiothreitol, 0.1 mg/mL BSA, and 10% (v/v) glycerol. The samples were loaded onto a nondenatured polyacrylamide gel (8%) after incubation for 0.5 h at room temperature. Electrophoresis was performed in 1× TBE (Tris/borate/EDTA) buffer with the addition of 20 mM NaCl at 80 V and 4 °C. The gels were exposed in a phosphorimager cassette and imaged by an FLA-7000 bioimager (GE Healthcare). The bands were analyzed with an ImageQuant TL (GE Healthcare). Kd was obtained from the equation u = [P]/{Kd + [P]} using KaleidaGraph 4.5 (Synergy Software) according to a previous study.45 u is the fraction of free DNA.
The binding reaction of RNAs with 7OTD was carried out in 100 mM NaCl, 10 mM Tris–HCl buffer, pH 7.0, for 2 h. The samples were loaded onto 20% native polyacrylamide gel. The gel was stained with GelStar for 20 min and visualized on a phosphorimager (LAS-3000, Fujifilm). The binding reaction of 7OTD with the RNA G-quadruplex and hnRNPA1 complex was performed in a binding buffer (100 mM NaCl, 10 mM Tris–HCl, pH 7.0, 5 mM dithiothreitol, 0.1 mg/mL BSA, and 10% (v/v) glycerol).
RNases T1 Footprinting
We digested the 32P-labeled RNA with RNase T1 (0.2 units) in 100 mM LiCl or KCl, 10 mM Tris–HCl, pH 7.0, for 5 min. The digestion reaction was stopped with the addition of stop buffer (10 M urea, 0.7 mM EDTA, and 10 mM Tris–HCl, pH 7.0).
Photo-Cross-Linking
After being incubated at room temperature for 2 h, the samples were irradiated with 302 nm light from a Benchtop UV transilluminator (UVP, 6900 μW/cm2) on ice for 8 h. The samples were digested by trypsin (from porcine pancreas, mass spectrometry grade, 202-15951, Wako Chemicals) in 50 mM Tris–HCl, pH 8.5, at 37 °C for 12 h. The digested samples were desalted with a micro-C18 ZipTip (Millipore). To carry out a MALDI-TOF mass spectrometer experiment, 2 μL of sample was mixed with α-cyano-4-hydroxycinnamic acid (Bruker Daltonics) and matrix solution (10 mg/mL in 50% acetonitrile (ACN)/0.1% trifluoroacetic acid (TFA)). The sample–matrix mixture was placed on a MALDI target plate and allowed to air-dry at room temperature.
Immunofluorescence Analysis
HeLa cells were grown on a 3.5 cm dish (glass bottom) and incubated with 1 μM Cy5–7OTD for 18 h. Then the cells were rinsed briefly and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min. The cells were washed three times using PBS and permeabilized with 0.2% Triton X-100 in PBS for 10 min. The cells were incubated with primary antibody (no. 8483, Cell Signaling Technology) at 1:800 dilution in antibody buffer (0.3% Triton X-100 and 1% bovine serum albumin) in PBS at 4 °C overnight. Then the cells were washed with PBS and incubated with secondary antibody that conjugated with Alexa 488 (no. 4412, Cell Signaling Technology) at 1:1000 dilution in antibody buffer for 1 h. The cells were incubated with 5 μg/mL Hoechst for 20 min after being washed with PBS. To perform the enzymatic experiment, we treated the cells with 200 μg/mL RNase A for 30 min at 37 °C. Digital images were captured using a TCS SP8 confocal microscope. Transfection of the Cy3-labeled 7-deazaguano-sine-containing probe RNA 5′-UUA7dG7dGGUUA7dG7dGG-Cy3-3′ (0.4 μM) was performed according to a previous study.16
Supplementary Material
Acknowledgments
X.L. is supported by the Kohnan Asia Scholarship Foundation. H.-L.B. is supported by the Nakatani Foundation Scholarship. T.I. is supported by JSPS KAKENHI (Grant 16K17938). This work was funded by JSPS KAKENHI (Grant 26288083 to Y.X.). Support from the Takeda Science Foundation is also acknowledged.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b01599.
Complete methods, sample preparation, NMR spectroscopy, and other additional results (PDF)
ORCID
Kazuo Nagasawa: 0000-0002-0437-948X
Yan Xu: 0000-0003-0379-8866
Notes
The authors declare no competing financial interest.
References
- 1.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 2.Schoeftner S, Blasco MA. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 3.Takahama K, Takada A, Tada S, Shimizu M, Sayama K, Kurokawa R, Oyoshi T. Chem Biol. 2013;20:341–350. doi: 10.1016/j.chembiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Zhao L, Lu S. Int J Biol Sci. 2015;11:316–323. doi: 10.7150/ijbs.10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caslini C, Connelly JA, Serna A, Broccoli D, Hess JL. Mol Cell Biol. 2009;29:4519–4526. doi: 10.1128/MCB.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarthy J, Bae NS, Scrafford J, Baumann P. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rippe K, Luke B. Nat Struct Mol Biol. 2015;22:853–858. doi: 10.1038/nsmb.3078. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Deng Z, Dahmane N, Tsai K, Wang P, Williams DR, Kossenkov AV, Showe LC, Zhang R, Huang Q, Conejo-Garcia JR, Lieberman PM. Proc Natl Acad Sci U S A. 2015;112:E6293–E6300. doi: 10.1073/pnas.1505962112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Nat Struct Mol Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 11.Azzalin CM, Lingner J. Trends Cell Biol. 2015;25:29–36. doi: 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Nat Commun. 2014;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. Mol Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Z, Campbell AE, Lieberman PM. Cell Cycle. 2010;9:69–74. doi: 10.4161/cc.9.1.10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. FEBS Lett. 2010;584:3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Ishizuka T, Yang J, Ito K, Katada H, Komiyama M, Hayashi T. J Biol Chem. 2012;287:41787–41796. doi: 10.1074/jbc.M112.342030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biffi G, Tannahill D, Balasubramanian S. J Am Chem Soc. 2012;134:11974–11976. doi: 10.1021/ja305734x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Kaminaga K, Komiyama M. J Am Chem Soc. 2008;130:11179–11184. doi: 10.1021/ja8031532. [DOI] [PubMed] [Google Scholar]
- 19.Martadinata H, Phan AT. J Am Chem Soc. 2009;131:2570–2578. doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Suzuki Y, Ito K, Komiyama M. Proc Natl Acad Sci U S A. 2010;107:14579–14584. doi: 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collie GW, Parkinson GN, Neidle S, Rosu F, De Pauw E, Gabelica V. J Am Chem Soc. 2010;132:9328–9334. doi: 10.1021/ja100345z. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y. Chem Soc Rev. 2011;40:2719–2740. doi: 10.1039/c0cs00134a. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Ishizuka T, Kimura T, Komiyama M. J Am Chem Soc. 2010;132:7231–7233. doi: 10.1021/ja909708a. [DOI] [PubMed] [Google Scholar]
- 24.Collie GW, Haider SM, Neidle S, Parkinson GN. Nucleic Acids Res. 2010;38:5569–5580. doi: 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 26.Lopez de Silanes I, Stagno d’Alcontres M, Blasco MA. Nat Commun. 2010;1:33. doi: 10.1038/ncomms1032. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q-S, Manche L, Xu R-M, Krainer AR. RNA. 2006;12:1116–1128. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redon S, Zemp I, Lingner J. Nucleic Acids Res. 2013;41:9117–9128. doi: 10.1093/nar/gkt695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeiffer V, Lingner J. PLoS Genet. 2012;8:e1002747. doi: 10.1371/journal.pgen.1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suva ML, Benes CH, Haber DA, Boussin FD, Zou L. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishikawa F, Matunis MJ, Dreyfuss G, Cech TR. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weldon C, Behm-Ansmant I, Hurley LH, Burley GA, Branlant C, Eperon IC, Dominguez C. Nat Chem Biol. 2017;13:18–20. doi: 10.1038/nchembio.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 36.Morris MJ, Basu S. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 37.Willis MC, Hicke BJ, Uhlenbeck OC, Cech TR, Koch TH. Science. 1993;262:1255–1257. doi: 10.1126/science.7694369. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Tashiro R, Sugiyama H. Nat Protoc. 2007;2:78–87. doi: 10.1038/nprot.2006.467. [DOI] [PubMed] [Google Scholar]
- 39.Tashiro R, Wang AH, Sugiyama H. Proc Natl Acad Sci U S A. 2006;103:16655–16659. doi: 10.1073/pnas.0603484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collie G, Reszka AP, Haider SM, Gabelica V, Parkinson GN, Neidle S. Chem Commun. 2009:7482–7484. doi: 10.1039/b901889a. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y. Curr Pharm Des. 2012;18:2096–2101. doi: 10.2174/138161212799958378. [DOI] [PubMed] [Google Scholar]
- 42.Chung WJ, Heddi B, Tera M, Iida K, Nagasawa K, Phan AT. J Am Chem Soc. 2013;135:13495–13501. doi: 10.1021/ja405843r. [DOI] [PubMed] [Google Scholar]
- 43.Iida K, Nagasawa K. Chem Rec. 2013;13:539–548. doi: 10.1002/tcr.201300015. [DOI] [PubMed] [Google Scholar]
- 44.Guo L, Shorter J. Mol Cell. 2015;60:189–192. doi: 10.1016/j.molcel.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahama K, Kino K, Arai S, Kurokawa R, Oyoshi T. FEBS J. 2011;278:988–998. doi: 10.1111/j.1742-4658.2011.08020.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


