Abstract
CD26 is a T cell activation antigen that contains dipeptidyl peptidase IV activity and is known to bind adenosine deaminase. The mechanism by which CD26 costimulation potentiates T cell receptor-mediated T cell activation, leading to subsequent exertion of T cell effector function, is still not clearly defined. In this article, we demonstrate that CD26 localizes into lipid rafts, and targeting of CD26 to rafts is necessary for signaling events through CD26. Importantly, aggregation of CD26 by anti-CD26 mAb crosslinking also causes coaggregation of CD45 into rafts. Moreover, we show that CD26 directly binds to the cytoplasmic domain of CD45. Our results therefore indicate a mechanism whereby CD26 engagement promotes aggregation of lipid rafts and facilitates colocalization of CD45 to T cell receptor signaling molecules p56Lck, ZAP-70, and TCRζ, thereby enhancing protein tyrosine phosphorylation of various signaling molecules and subsequent interleukin-2 production.
The CD26 is a 110-kDa cell-surface glycoprotein that possesses dipeptidyl peptidase IV (EC 3.4.14.5) activity in its extracellular domain (1, 2) and has an important role in T cell activation (3). Although constitutively expressed in the liver, intestine, and kidney, CD26 expression level is tightly regulated on T cells, and its density is markedly enhanced after T cell activation (4). In the resting state of human T lymphocytes, CD26 is expressed on a subset of CD4+ memory T cells, and this CD4+CD26high T cell population has been shown to respond maximally to recall antigens (5). In fact, CD26 itself is involved in the processes of T cell signal transduction.
Activation of resting T cells requires two independent signals, the first stemming from recognition by the T cell receptor (TCR) complex of processed antigen peptides plus major histocompatibility complex molecules on antigen-presenting cells, with the second signal being provided by other receptor–ligand interactions between T cells and antigen-presenting cells (6, 7). Intensive work over the past several years has characterized the T cell surface molecule CD26 as a receptor capable of generating T cell costimulatory signals (8, 9). Crosslinking of CD26 and CD3 with immobilized mAbs induced T cell activation and IL-2 production (5). Moreover, anti-CD26 antibody treatment of T cells led to decreased surface expression of CD26 via its internalization, and this modulation of CD26 resulted in an enhanced proliferative response to anti-CD3 or anti-CD2 stimulation (9). We also demonstrated previously that ligation of CD26 by the anti-CD26 mAb 1F7 induced increased tyrosine phosphorylation of signaling molecules such as c-Cbl, mitogen-activated protein kinase [MAP kinase, or extracellular-regulated kinase (ERK1/2)], Zap70, p56Lck, and CD3ζ (10). However, the exact mechanism responsible for CD26-mediated tyrosine phosphorylation and costimulation is unclear, particularly in view of the fact that CD26 has a short conserved cytoplasmic domain consisting of only six amino acid residues, without intrinsic protein tyrosine kinase function or known binding motifs for tyrosine kinases (5). Antibody-induced modulation of CD26 on T cells resulted in a concurrent decrease in CD45 expression, enhanced phosphorylation of the TCR-associated zeta chain, and increased p56Lck src-kinase activity (11). CD26 is therefore involved in essential T cell signaling events through its physical and functional association with structures with key roles in T cell activation.
Recent studies demonstrated that T cell activation induced by anti-CD3 stimulation led to a redistribution of the detergent-insoluble glycolipid-enriched domains (DIGs) (also called lipid rafts, membrane rafts, or rafts) (12–15). The raft redistribution resulted in the association of TCR/CD3 and raft-associated signal-transducing molecules, indicating the role of such membrane compartmentation in efficient T cell activation (13, 14, 16). Most recently, CD2, CD5, CD9, and CD44 costimulatory molecules were shown to be present in rafts (17). These studies showed that the raft-resident costimulatory molecules strengthened TCR-mediated signaling when coligated with CD3 by inducing aggregation of rafts as well as enhancing association of TCR/CD3 and rafts.
In this article, we show that CD26 costimulatory molecules are present in rafts and that ligation of CD26 by the anti-CD26 mAb 1F7 increases the recruitment of CD26 molecules to rafts, thereby inducing increased tyrosine phosphorylation of signaling molecules. Moreover, crosslinking of CD26 with anti-CD26 mAb induces the aggregation of lipid rafts, colocalizing CD26 with the protein tyrosine phosphatase (PTP) CD45. Our work also demonstrates that CD26 binds to the cytoplasmic domain of CD45. Consistent with the notion that modulation of CD26 on T cells results in enhanced phosphorylation of c-Cbl, Zap70, Erk1/2, p56Lck, and CD3ζ to anti-CD3 stimulation (11), our findings provide a mechanism by which CD26 molecules exhibit their costimulatory ability through anti-CD26 mAb-induced internalization.
Materials and Methods
Cells.
The properties and maintenance of the human T-cell line Jurkat transfected with a CD26 cDNA (J. CD26) have been described previously (2). Human peripheral blood was obtained from the transfusion service of the institute. Mononuclear cells (human peripheral blood mononuclear cells) were isolated by density gradient centrifugation using Ficoll/Paque (Amersham Pharmacia), and then T cells were positively selected using sheep red blood cells.
Antibodies and Reagents.
Anti-CD26 mAb 1F7, anti-CD3 mAb OKT3, and anti-CD45RO mAb UCHL1 were previously described (1, 11). The following antibodies were purchased: mouse monoclonal anti-Lck (PharMingen), mouse monoclonal anti-phosphotyrosine (4G10) (Sigma), mouse monoclonal anti-TCRζ (Beckman Coulter), rabbit polyclonal anti-human c-Cbl, rabbit polyclonal anti-human ZAP70 (Santa Cruz Biotechnology), mouse monoclonal anti-Erk1 (Transduction Laboratories, Lexington, KY), goat anti-mouse IgG (H+L) (Jackson ImmunoResearch), and horseradish peroxidase-labeled anti-mouse and anti-rabbit Ig (Amersham Pharmacia). FITC-conjugated anti-CD26 mAb (Beckman Coulter), Cholera toxin B (CTB), FITC-conjugated CTB, Texas red-conjugated streptavidin, mouse monoclonal anti-human β-actin antibody, and cytochalasin D were purchased from Sigma. Sulfosuccinimidyl-6-biotinamido hexanoate was purchased from Pierce, and enhanced chemiluminescence reagents were obtained from NEN.
Sucrose Gradient Centrifugation.
To obtain the DIGs/rafts membrane fraction, J. CD26 cells and purified T cells (1 × 108) were lysed on ice with 1 ml of 1% Triton X-100 and 1 mM PMSF in MNE buffer (25 mM MES, pH 6.5/150 mM NaCl/5 mM EDTA). Following a 30-min incubation at 4°C with gentle agitation, the lysates were homogenized with 20 strokes of a loose-fitting Dounce homogenizer, gently mixed with an equal volume of 80% sucrose (wt/vol) in MNE buffer, and placed in the bottom of a SW41Ti centrifuge tube (Beckman Coulter). The sample was then overlaid with 6.5 ml of 30% sucrose and 3.5 ml of 5% sucrose in MNE buffer and spun for 16–20 h at 410,000 rpm at 4°C in a Beckman SW41Ti swing rotor using an XL-90 Ultracentrifuge (Beckman). One-milliliter fractions were harvested serially from the top of the gradient. The DIGs/rafts fraction was obtained in fractions 4 and 5.
Stimulation of Cells and Preparation of Lysates from Stimulated Cells.
T cells (1.0 × 106 per sample) were treated with cytochalasin D (20 mM stock solution in DMSO) at indicated concentrations for 20 min at 37°C. These samples were then used to analyze protein phosphorylation in tyrosine residues as described previously (11), following incubation with medium alone, 0.05 μg/ml OKT3, or 10 μg/ml 1F7 mAb for 15 min at 4°C. Cells were washed once with cold medium and then incubated with 200 μl of medium containing goat anti-mouse IgG (H+L, 5 μg/ml) at 37°C for 15 min. The reactions were terminated by the addition of 1 ml of ice-cold PBS containing 5 mM EDTA/10 mM NaF/10 mM Na pyrophosphate/0.4 mM Na3VO4. Cells were centrifuged and then solubilized in lysis buffer [1% (vol/vol) Nonidet P-40/150 mM NaCl/50 mM Tris⋅HCl, pH 8.0/5 mM EDTA/1 mM PMSF/10 mM iodacetamide/10 mM NaF/10 mM Na pyrophosphate/0.4 mM Na3VO4] for 15 min on ice, and then insoluble material was removed by centrifugation at 12,000 rpm for 5 min. Lysates were also immunoprecipitated by adding 2 μg/ml of indicated mAbs plus protein Sepharose-A beads (Amersham Pharmacia). After a 2-h incubation period at 4°C under constant agitation, the beads were washed four times in lysis buffer.
Western Blotting.
Proteins were separated by SDS/PAGE in reducing condition and then transferred to a polyvinylidene difluoride membrane (Millipore) in 25 mM Tris/192 mM glycine/20% methanol, and the membrane was blocked for 1 h at room temperature in PBS with 0.05% Tween 20 (PBS-T) containing 3% BSA or 5% nonfat milk. Specific antigens and phosphorylated tyrosine were probed by the corresponding mAbs and antiphosphotyrosine mAb 4G10, respectively, followed by an anti-mouse or anti-rabbit horseradish peroxidase-Ig. The proteins were revealed by enhanced chemiluminescence.
Capping and Immunofluorescence Confocal Laser Microscopy.
All procedures were performed at 4°C unless otherwise described. For observation of CD26 in rafts, J. CD26 cells were suspended at 1 × 105 per ml in PBS and, after blocking in ice-cold 3% BSA/PBS, incubated with 5 μg/ml of FITC-conjugated anti-CD26 mAb (Ta1) and biotinylated CTB for 30 min. Following washing in ice-cold 3% BSA/PBS, cells were then incubated with Texas red-conjugated streptavidin at 37°C for 30 min to induce capping. Cells (1.0 × 105 cells per slip) were then attached to poly-L-lysine-coated coverslips (Matsunami Glass, Tokyo) and processed for confocal laser microscopy. For observation of colocalization of CD26 and CD45 in rafts, T cells after being blocked with 3% BSA/PBS were incubated with 5 μg/ml of the following primary antibodies in 3% BSA/PBS: FITC-conjugated anti-CD45 (UCHL1), biotinylated anti-CD26 (5F8), and biotinylated CTB or FITC-conjugated CTB. Cells were then incubated either in media alone or with 10 μg/ml anti-CD26 mAb 1F7 at 37°C for 1 h, and then washed twice with ice-cold PBS. As previously reported, 5F8 and 1F7 mAbs defined distinct epitopes of CD26 (18). After being washed in ice-cold 3% BSA/PBS, cells were stained with Texas red-conjugated streptavidin for 15 min and then for 30 min at 37°C to induce lipid raft patching. After being attached to poly-L-lysine-coated coverslips (1.0 × 105 cells/slip), cells were fixed with 3% paraformaldehyde in PBS for 15 min at room temperature. Confocal laser microscopy was performed with an Olympus IX70 confocal microscope with 60 objective lenses (Olympus, New Hyde Park, NY), using laser excitation at 488 and 568 nm. The widths of FITC and Texas red emission channels were set to maximize specificity.
Coimmunoprecipitation.
The soluble CD26 (sCD26) molecules generated in Chinese hamster ovary cells were labeled with Oregon green (OG) (Molecular Probes) according to the manufacturer's protocol. cDNA for glutathione S-transferase (GST)-tagged CD45 and CD45/LAR (leukocyte common antigen-related molecule) swap mutants were generated as described previously (19). Briefly, the wild-type CD45 RO cDNA was inserted into the BamHI site of pGEX-2T. The resultant plasmid was named pGST-CD45 WT. Amplification of the wild-type CD45 extracellular domain sequences (amino acid positions 1–582) was performed between synthesized oligonucleotide primers, GGCTTCCAGATATGACCATG and GCCAGAAATGCTATCAGTGC. The amplified PCR product was subsequently cloned into the BamHI site of pGEX-2T. The resultant plasmid was named pGST-extracellular CD45 WT. The plasmid encoding the wild-type CD45 cytoplasmic domain, pGST-cytoplasmic CD45WT (positions 584-1281), was kindly provided by Dr. Haruo Saito (Dana-Farber Cancer Institute, Boston). The plasmids encoding CD45 cytoplasmic domain (position 584-1281), cytoplasmic domain (position 1275–1881), or chimerical PTPases (position 584–895 of CD45 and 1590–1881 of LAR or 1275–1590 of LAR and 897-1281 of CD45) containing Cys to Ser (CS) point mutations, which were pGST-CD45 CS (position 828), pGST-LAR CS (position 1522), pGST-CD45/LAR CS (position 828), and pGST-LAR/CD45 CS (position 1522), were also gifts from Dr. Haruo Saito; 10 μg/ml of sCD26-OG was then incubated with recombinant GST-tagged proteins at 4°C for 60 min and isolated with glutathione-conjugated agarose beads. After washing for four times in PBS, the beads were separated by SDS/PAGE under reducing conditions, and sCD26-OG was detected by fluoroimager (STORM, Molecular Dynamics).
Results
CD26 Is Present in Membrane Rafts.
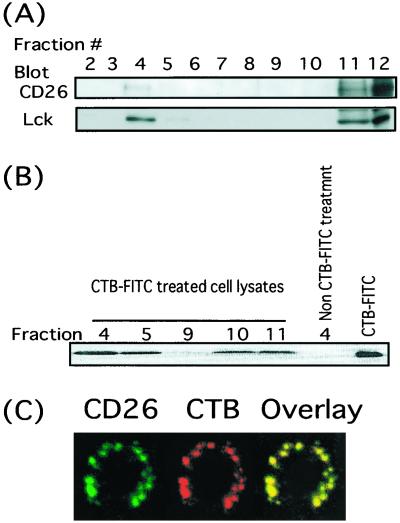
Recent studies have shown that lipid rafts have an important role in the generation of TCR-related signaling along with various types of costimulatory molecules (e.g., CD2, CD5, CD9, and CD44) (17). In view of the ability of CD26 to serve as a costimulatory molecule for the CD3/TCR pathway of activation, we first searched for the presence of CD26 molecules inside the raft fraction. As shown Fig. 1A, the localization of p56Lck to the raft fraction is confirmed, and importantly, we find that CD26 molecules (1F7) also exist in the raft fraction, but they are present in the cytosol and heavy membrane fractions as well. Consistent with previous studies, the raft fractions are enriched for glycosphingolipids as determined by reactivity with CTB, which recognizes the ganglioside GM1 (Fig. 1B). To confirm the association of CD26 with the rafts, we used a nondestructive approach to investigate the localization of CD26 within intact membrane rafts. Membrane rafts were stained with biotinylated CTB, which binds to the plasma membrane ganglioside GM1, followed by capping with Texas red-conjugated streptavidin. Lateral crosslinking of GM1 can therefore manipulate membrane rafts, forming distinct patches (Fig. 1C). CD26 molecules were also stained with FITC-conjugated Ta1 (anti-CD26 mAb). The subsequent appearance of the yellow dots observed in Fig. 1C thus indicates that CTB (red) and CD26 (green) are present in close physical proximity. Consistent with our biochemical analyses, CD26 is substantially localized in GM1 patches (Fig. 1A).
Figure 1.
Presence of CD26 in the DIGs/rafts fraction. (A) Lysates from CD26-transfected Jurkat cells were fractionated on a sucrose density gradient as described in Materials and Methods. Protein contents of each fraction of the sucrose gradient were resolved by SDS/PAGE, and protein patterns were analyzed on immunoblots stained with the corresponding Abs. (B) Cells were incubated with FITC-conjugated CTB, and the lysates were fractionated on a sucrose density gradient. Proteins and FITC-conjugated CTB reactive with GM1 contents of each fraction of the sucrose gradient were resolved by SDS/PAGE and analyzed with fluoroimager (STORM). (C) Patched T cells were stained with GM1-reactive biotinylated CTB followed by streptavidin-conjugated Texas red and FITC-Ta1 against CD26. Stained cells were examined on a confocal microscopy, and single confocal sections were taken.
Internalization of CD26 Triggers Its Recruitment to Membrane Rafts.
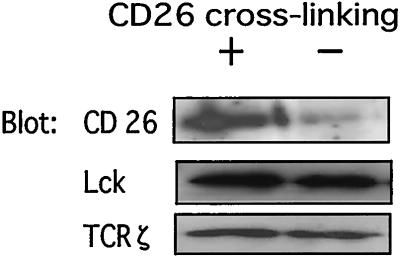
Our previous studies showed that anti-CD26 antibody treatment of T cells led to a decrease in the surface expression on CD26 via its internalization, and such modulation resulted in an enhanced proliferative response to anti-CD3 stimulation, as well as enhanced tyrosine phosphorylation of signaling molecules (5, 9, 10). Because Fig. 1 demonstrates that CD26 molecules are present in rafts, we next examined the relationship between antibody-mediated internalization of CD26 and its localization to rafts. Resting T cells isolated from peripheral blood mononuclear cells were used to analyze the relative amount of CD26 molecules present in the raft fractions in control cells as well as in cells treated with anti-CD26 mAb (1F7) to induce its modulation. Fig. 2 shows that treatment with 1F7 triggers the recruitment of CD26 molecules to membrane rafts. Furthermore, recruitment of p56Lck and TCRζ molecules to rafts is not affected by 1F7 treatment. Because CD26 modulation induced by 1F7 plays an important role in CD26-mediated T cell costimulation (9), these results suggest that T cell activation induced by CD26 is essentially mediated by the localization of CD26 in membrane rafts.
Figure 2.
Recruitment of CD26 to lipid rafts after crosslinking of anti-CD26 mAb. T cells were treated with anti-CD26 antibody or media alone. After modulation of CD26, lysates of the cells were prepared as described in Materials and Methods. Protein contents of each fraction of the sucrose gradient were resolved by SDS/PAGE, and protein patterns were analyzed on immunoblots stained with the corresponding Abs.
Disrupting the Structure of Membrane Rafts Impairs Tyrosine Phosphorylation Induced by Crosslinking of CD26.
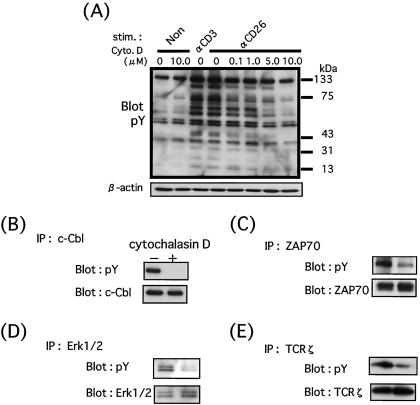
We next examined whether the presence of CD26 in rafts is necessary for the tyrosine phosphorylation induced by crosslinking of CD26. To analyze the function of rafts on CD26 signaling, resting T cells isolated from peripheral blood mononuclear cells were first treated with either medium alone or cytochalasin D, which is known to disturb the structure of DIGs/rafts by sequestrating membrane cholesterol through depolymerization of actin skeleton (13, 20). As shown in Fig. 3A, after 15 min of CD26 stimulation, cells incubated in medium alone display tyrosine phosphorylation of several cellular proteins, especially the 120-kDa, 70-kDa, 40-kDa, and 16-kDa proteins as previously reported (11). On the other hand, cytochalasin D pretreatment markedly prevents the tyrosine phosphorylation of T cells induced by crosslinking of CD26. Importantly, increasing inhibition of tyrosine phosphorylation is observed at increasing doses of cytochalasin D. Of note is the fact that cell viability remains unchanged by the increasing concentrations of cytochalasin D (data not shown). To define the tyrosine-phosphorylated proteins inhibited by the disruption of rafts, we conducted immunoprecipitation with the indicated antibodies and immunoblotting experiments. Fig. 3 B–E shows that c-Cbl, ZAP70, Erk1/2, and TCRζ proteins are phosphorylated following stimulation by anti-CD26 mAb and that cytochalasin D pretreatment substantially reduces the tyrosine phosphorylation of these molecules. To further determine the relationship between CD26 and membrane rafts, we used nystatin in addition to cytochalasin D to disrupt membrane rafts. Nystatin is a fungal metabolite that binds to membrane cholesterol and directly perturbs raft integrity without causing cell destruction. Similar results were found with nystatin as those obtained with cytochalasin D (data not shown). These findings support the notion that ligation of CD26 molecules induces tyrosine phosphorylation of various signaling proteins and that inhibition of the localization of CD26 molecules in membrane rafts results in the impairment of tyrosine phosphorylation of these cellular proteins, thus leading to disruption of the signaling cascade.
Figure 3.
Disrupting the structure of DIGs/rafts impairs tyrosine phosphorylation induced by crosslinking of CD26. (A) T cells were treated with cytochalasin D as indicated and then incubated with OKT3 or 1F7. After being washed, cells were incubated in goat anti-mouse IgG (H+L) containing medium. Total lysates were prepared, resolved by SDS/PAGE, and analyzed by Western blotting as described in Materials and Methods. The inhibitory effect of raft depletion by cytochalasin D on CD26-mediated tyrosine phosphorylation was observed in a dose-dependent manner. As indication that a similar amount of proteins was applied to each lane, the membrane was reprobed with anti-human β-actin antibody (B–E). T cells were treated with cytochalasin D at a concentration of 10 μg/ml and then incubated with 1F7. After being washed, cells were incubated in goat anti-mouse IgG (H+L) containing medium. Total lysates were prepared as described in Materials and Methods, immunoprecipitated with anti-c-Cbl (B), anti-ZAP70 (C), anti-Erk1/2 (D), and anti-TCRζ (E). After analysis using anti-phosphotyrosine Ab (4G10), the membranes were reprobed with specific antibodies. Raft disruption by cytochalasin D inhibits phosphorylation of specific cellular proteins induced by CD26 ligation.
CD26 Binds to the Cytoplasmic Region of CD45.
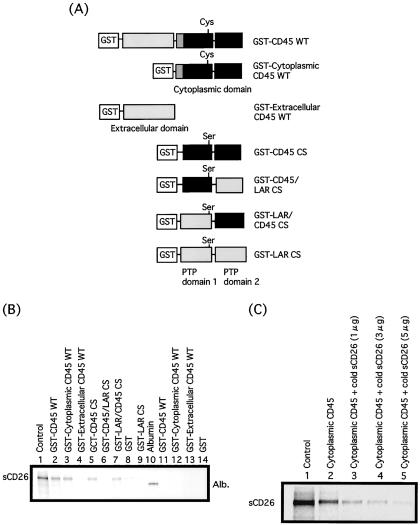
Because of its short cytoplasmic tail, CD26 is most likely unable to transmit signals by itself, relying instead on its physical and functional association with other T cell signaling molecules (5), including CD45. We have shown previously that CD26 comodulated on the T cell surface with CD45RO, a known membrane-linked protein tyrosine phosphatase, following the ligation of CD26 by anti-CD26 mAb. In addition, anti-CD26 mAb coimmunoprecipitated CD45 with CD26 (11). We therefore hypothesized that following antibody-mediated internalization of CD26, the internalized CD26 molecules would associate with the CD45 phosphatase in rafts. To test this hypothesis, we analyzed the binding between sCD26 produced by Chinese hamster ovary cells and GST-tagged extracellular and cytoplasmic regions of CD45. To further study the binding of CD26 and the cytoplasmic region of CD45, we also established the swap mutants of CD45 and LAR (Fig. 4A). sCD26-OG is incubated with the GST-tagged cytoplasmic or extracellular domain of CD45 or with the swap mutants of the cytoplasmic region of CD45 and LAR. As shown in Fig. 4B, sCD26-OG is coprecipitated with cytoplasmic CD45 GST-agarose beads but not with GST beads or extracellular CD45 GST beads. Lanes 5–8 in Fig. 4B also show that the binding domain of CD45 to CD26 is the PTP domain 2 in the cytoplasmic region of CD45. To define whether the CD45 phosphatase activity plays a role in this interaction, we used a mutant CD45 phosphatase domain in which the amino acid residue Cys-828 has been changed to serine. Because Cys-828 is the catalytic center of the CD45 phosphatase, the CS mutant has no phosphatase activity. Nonetheless, the CS mutant of the PTP domain 2 still retains binding activity to CD26. As a control, Albumin-OG is not coprecipitated with cytoplasmic CD45 GST. Moreover, excess nonlabeled sCD26 completely inhibits the binding between sCD26-OG and cytoplasmic CD45 (Fig. 4C). We also confirmed that sCD26 cannot bind to the individual isoform of CD45-transfected 300-19 murine B cell lines previously established (data not shown) (21), indicating that CD26 cannot bind to the extracellular domain of CD45. These data thus indicate that CD26 binds directly to the cytoplasmic region (PTP domain 2) but not the extracellular domain of CD45.
Figure 4.
Direct association between CD26 and the cytoplasmic legion of CD45. (A) Structure of the recombinant CD45 and CD45/LAR swap mutants used in this study. The black bar represents the phosphatase domain of CD45. sCD26 was labeled with Oregon Green (sCD26-OG) and incubated with GST-tagged cytoplasmic or extracellular domain of CD45 or CD45/LAR swap mutants. The precipitates were isolated with glutathione-conjugated agarose beads. The precipitates were washed and separated by SDS/PAGE under reducing condition and detected by fluoroimager (STORM). (B) sCD26-OG and albumin-OG were incubated with cytoplasmic CD45 or extracellular CD45 or CD45/LAR swap mutants. (C) Nonlabeled sCD26 at varying concentrations was added in the binding reaction between sCD26-OG and cytoplasmic CD45.
CD45 Is Colocalized in Membrane Rafts after Crosslinking of CD26.
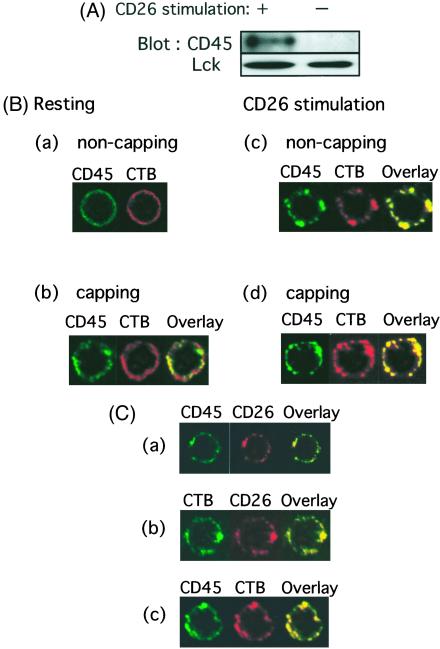
Because our results indicate that CD26 is aggregated in the rafts after antibody-induced down-modulation and internalized CD26 can bind to the cytoplasmic region of CD45, it is possible that CD45 is coaggregated in rafts after crosslinking of CD26. To address this issue, unstimulated or 1F7-stimulated T cells were subjected to sucrose gradient centrifugation to separate lipid raft fractions from Triton X-100 soluble fractions. As previously reported for unstimulated T cells (12, 13, 22), CD45 molecules do not copurify with lipid rafts isolated biochemically by virtue of their Triton X-100 insolubility and low density (Fig. 5A). However, antibody-mediated crosslinking of CD26 results in enriched populations of not only CD26 (Fig. 2) but also CD45 (Fig. 5A) in lipid rafts. On the other hand, there is only a small increase in the levels of p56Lck and TCRζ chain in the lipid raft fractions after stimulation with anti-CD26 mAb. To exclude the possibility that our observations were because of a stabilization of the association of CD45 molecules with membrane rafts by CD26-mAb crosslinking rather than movement into rafts, we treated T cells with anti-CD3 mAb and performed the same analyses as described for Fig. 5A. Similar to previous reports (14), CD45 was not detected in the detergent-insoluble fractions (data not shown).
Figure 5.
Colocalization of CD26 and CD45 in lipid rafts after modulation of CD26. (A) Unstimulated or 1F7-stimulated T cells were subjected to sucrose gradient centrifugation to separate lipid raft fractions from Triton-soluble fractions. Protein contents of fraction 4 of the sucrose gradient were resolved by SDS/PAGE, and protein patterns were analyzed on immunoblots stained with the corresponding Abs. CD45 (UCHL1) was enriched in rafts after CD26 stimulation. (B) Following staining with anti-CD45 (FITC-conjugated UCHL1) and CTB (biotinylated), cells were then analyzed for potential colocalization of CD45 and CTB patches. After staining of these molecules on the cell surface with (b and d) or without (a and c) capping, CD26 was crosslinked and internalized with 1F7 mAb (c and d). (C) Cells were stained by the biotinylated anti-CD26 antibody 5F8 followed by Texas red-conjugated streptavidin and the FITC-conjugated anti-CD45RO antibody UCHL1 (a), FITC-conjugated CTB and biotinylated 5F8 (b), or FITC-conjugated UCHL1 and biotinylated CTB (c). After staining of these molecules on the cell surface, CD26 was crosslinked and internalized with 1F7 mAb.
Expanding on work described above, T cells were next incubated either in media alone or with anti-CD26 mAb (1F7). Following patching with anti-CD45 and CTB, cells were then analyzed for potential colocalization of CD45 and CTB patches. Membrane rafts were stained with biotinylated CTB followed by capping with Texas red-conjugated streptavidin. CD45 were also stained with FITC-labeled UCHL1 (anti-CD45 RO mAb) (Fig. 5Ba). Again, the appearance of the yellow dots would indicate a close physical proximity of CTB (red) and CD45 (green). As shown in Fig. 5Bb, although a relatively small portion of the CD45 molecules colocalizes in the CTB patches, CD45 remains almost uniformly distributed after CTB patching in cells incubated in media alone. On the other hand, CD45 is colocalized with CTB patches in cells treated with the anti-CD26 mAb 1F7 [Fig. 5B (c and d)]. It should be noted that in Fig. 5B (d), the cells underwent crosslinking of CD26 and induction of patching of GM1/rafts by CTB crosslinking to confirm the colocalization of the molecules on GM1/rafts. In Fig. 5Bc, cells underwent crosslinking of CD26 without induction of patching by CTB, and as shown in this figure, CD26 and CTB were aggregated. Therefore, the difference in capping in Fig. 5B (between c and d) was not as apparent as that seen in Fig. 5B (a and b).
We next determined whether the colocalization of CD26 and CD45 is observed in membrane rafts after antibody-mediated internalization of CD26. As shown in Fig. 5C, colocalization (yellow) between CD26 and CD45RO is clearly observed in rafts after internalization of CD26 with 1F7. Therefore, CD26 and CD45RO are directly associated in membrane rafts after ligation of CD26 by its specific mAb.
Discussion
Although the ability of CD26 to costimulate human T cells in conjunction with the CD3/TCR complex is well established, the precise mechanisms of signal transduction following CD26 engagement remain poorly understood. A number of molecules with key roles in T cell function and activation, including CD5, CD2, CD9, CD4, CD43, and CD11a, have also been described to transmit costimulatory signals (23, 24). Recently, it has been shown that these molecules exist in the raft fraction of T cell plasma membranes and that ligation of such raft-resident costimulatory molecules induces the mobilization of rafts (17). In this article, we show that CD26 costimulatory molecules are also present in membrane rafts. Importantly, binding of the CD26 molecules by the anti-CD26 mAb 1F7 leads to the recruitment of additional CD26 molecules to the rafts and subsequent interaction with the CD45 tyrosine phosphatase in the raft fraction.
We have previously shown that CD45, particularly CD45RO, was comodulated with CD26 after CD26 ligation and that anti-CD26 was capable of coprecipitating lower isoforms of CD45 from T cell lysates (11). Moreover, we showed that antibody-mediated modulation of CD26 on the T cell surface resulted in an enhanced proliferative response to anti-CD3 or anti-CD2 stimulation and that ligation of CD26 by 1F7 induced increased tyrosine phosphorylation of such T cell signaling molecules as c-Cbl, ZAP70, Erk1/2, p56Lck, and CD3ζ (9, 11). We also recently demonstrated that internalization of CD26 was mediated by mannose 6-phosphate/insulin-like growth factor II receptor, with its interaction with CD26 playing an important role in CD26-mediated T cell costimulation (25). A puzzling aspect of CD26-mediated signaling involves the fact that CD26 cytoplasmic domain contains only six amino acid residues, with no intrinsic protein tyrosine kinase function nor known binding motif for tyrosine kinases. Addressing this issue, our present work indicates that membrane rafts provide a cellular location for CD26 signaling in T cells to occur through its interaction with CD45. In fact, our study strongly suggests that CD26-mediated tyrosine phosphorylation in T cells occurs within membrane rafts, a process that requires antibody-induced internalization and the subsequent recruitment of CD26 molecules to rafts. The following lines of evidence described in this article support these conclusions: (i) the relative levels of CD26 molecules in rafts are increased following anti-CD26 (1F7)-mediated modulation of CD26, and (ii) perturbation of the structural integrity of the membrane rafts by the actin-depolymerizing agent, cytochalasin D, profoundly affects CD26-mediated tyrosine phosphorylation (especially c-Cbl, ZAP70, Erk1/2, and CD3ζ) in a dose-dependent fashion.
We also demonstrate that the tyrosine phosphatase CD45 aggregates and interacts with CD26 in lipid rafts after anti-CD26 mAb-induced modulation of CD26. Our biochemical and colocalization studies clearly demonstrate the direct association of CD26 and CD45, particularly the cytoplasmic domain 2 of CD45. Moreover, the CS mutant without phosphatase activity still retains the ability to bind to CD26, indicating that the enzymatic activity of CD45 is not directly involved in this interaction. It has been proposed that CD45 is excluded from the contact sites between T cells and antigen-presenting cells (26). Indeed, previous studies have indicated that CD45 is absent from membrane rafts and that TCR-mediated aggregation of rafts facilitates colocalization of p56Lck, LAT (linker for activation of T cells), and TCR while excluding CD45 (14, 26). However, other reports have suggested that CD45 can be also found at the T cell–antigen-presenting cells interaction site, as well as at subsequent immune synapse formation (27, 28). In human leukemic cell lines and normal granulocytes, CD45 can be found in plasma membrane rafts (29). It is therefore conceivable that under some physiological circumstances, CD45 molecules can have access to membrane rafts. The present study clearly demonstrates that CD26 and CD45 are associated following antibody-induced CD26 modulation. Our results thus strongly suggest that following CD26 modulation, the CD26 molecules migrate to membrane rafts where they can physically and functionally associate with CD45, particularly CD45RO. Our findings also suggest that CD26-related signals are transduced within membrane rafts, the site of aggregation of a number of T cell costimulatory molecules. The expression of lipid rafts is a dynamic process in which preassembled intracellular rafts are released from storage components and recycled from the cell surface to the cytoplasm. The ability of the anti-CD26 mAb to increase the association with receptors could occur at any stage of this process and not just act to promote adjacent entry into rafts. However, in Fig. 5 B and C, the level of CTB staining does not seem to increase with CD26 crosslinking. Therefore, anti-CD26 seems to have a direct effect as a chaperon in bringing CD45 into preexisting membrane rafts.
An important regulator of the Src-family kinases that mediates proximal biochemical signaling events, CD45 continuously controls Src-family member kinases by opposing C-terminal Src kinase (Csk) action and dephosphorylating the C-terminal tyrosine in T cells (30–33). Because Csk is a constitutively active enzyme, inhibition of kinase activity is achieved by phosphorylation of this site (32, 34). Several reports show that CD45 is required for antigen receptor signal transduction (35–37). However, localization of certain membrane proteins, but not CD45, to the immunological synapse after antigen activation allows for the recruitment of Src-family kinases to the antigen receptor (12, 13, 21, 38). Recent work also revealed that CD45 fragment forms a symmetrical dimer in which the catalytic site of one molecule is blocked by specific contacts with a structural wedge from the membrane-proximal region of the other and that this dimerization process inhibits phosphatase activity (39). Moreover, the CD45 mutation that inactivates the inhibitory wedge causes polyclonal lymphocyte activation, leading to lymphoproliferation and severe autoimmune nephritis with autoantibody production (40). The present results suggest that CD26 may potentially regulate CD45 phosphatase activity by binding to the CD45 molecule and inhibiting dimerization to regulate TCR signaling. We showed previously that T cells expressing higher levels of CD26 had a strong migratory capacity, as assayed by the use of endothelial cell monolayer on collagen gels (41). Furthermore, in vivo studies revealed that a large number of CD26 positive cells was found in inflamed tissues of patients with multiple sclerosis and rheumatoid arthritis (42, 43), suggesting that CD26 positive T cells functioned as effector T cells. In this regard, it is hypothesized that CD26 expression on T cells is a marker for Th1-type cells (44). In view of these findings, the fact that CD26 binds to the cytoplasmic domain 2 of CD45 tyrosine phosphatase in membrane rafts strongly suggests that steric interference from this interaction may inhibit the activation and function of CD26 positive T cells. Furthermore, the interaction between CD26 and CD45 may regulate the inflammation observed in various immune-mediated disorders, such as autoimmune diseases and graft-versus-host disease following allogeneic organ transplantation. Future studies of the mechanisms by which CD26 and CD45 interact may therefore provide a clue to the pathogenic role of CD26 in various immune-mediated diseases and may lead to the eventual development of novel therapeutic approaches based on our knowledge of CD26 biology.
In conclusion, we show that CD26 costimulatory molecules are present in lipid rafts and that ligation of the CD26 molecules by the anti-CD26 mAb 1F7 increases the recruitment of CD26 molecules with CD45 to rafts, resulting in increased tyrosine phosphorylation of signaling molecules.
Acknowledgments
We thank Dr. Masaya Higuchi for assistance with raft preparation technique and Dr. Haruo Saito for providing human CD45 cDNAs. This work was supported by National Institutes of Health Grant AR33713 and by grants-in-aid from the Ministry of Education, Science, Sports, and Culture and the Ministry of Health, Labor, and Welfare, Japan (to C.M.). N.H.D. is the recipient of a grant from the MD Anderson Cancer Center Physician–Scientist Program and the V Foundation.
Abbreviations
- CTB
cholera toxin B
- DIGs
detergent-insoluble glycolipid-enriched domains
- PTP
protein tyrosine phosphatase
- TCR
T cell receptor
- OG
Oregon green
- GST
glutathione S-transferase
References
- 1.Morimoto C, Torimoto Y, Levinson G, Rudd C E, Schrieber M, Dang N H, Letvin N L, Schlossman S F. J Immunol. 1989;143:3430–3439. [PubMed] [Google Scholar]
- 2.Tanaka T, Camerini D, Seed B, Torimoto Y, Dang N H, Kameoka J, Dahlberg H N, Schlossman S F, Morimoto C. J Immunol. 1992;149:481–486. [PubMed] [Google Scholar]
- 3.Dang N H, Torimoto Y, Deusch K, Schlossman S F, Morimoto C. J Immunol. 1990;144:4092–4100. [PubMed] [Google Scholar]
- 4.Fleischer B. J Immunol. 1987;138:1346–1350. [PubMed] [Google Scholar]
- 5.Morimoto C, Schlossman S F. Immunol Rev. 1998;161:55–70. doi: 10.1111/j.1600-065x.1998.tb01571.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Linsley P S. Curr Opin Immunol. 1992;4:265–270. doi: 10.1016/0952-7915(92)90075-p. [DOI] [PubMed] [Google Scholar]
- 7.Harding F A, McArthur J G, Gross J A, Raulet D H, Allison J P. Nature (London) 1992;356:607–609. doi: 10.1038/356607a0. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, Kameoka J, Yaron A, Schlossman S F, Morimoto C. Proc Natl Acad Sci USA. 1993;90:4586–4590. doi: 10.1073/pnas.90.10.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang N H, Torimoto Y, Sugita K, Daley J F, Schow P, Prado C, Schlossman S F, Morimoto C. J Immunol. 1990;145:3963–3971. [PubMed] [Google Scholar]
- 10.Hegen M, Kameoka J, Dong R P, Schlossman S F, Morimoto C. Immunology. 1997;90:257–264. doi: 10.1046/j.1365-2567.1997.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torimoto Y, Dang N H, Vivier E, Tanaka T, Schlossman S F, Morimoto C. J Immunol. 1991;147:2514–2517. [PubMed] [Google Scholar]
- 12.Montixi C, Langlet C, Bernard A M, Thimonier J, Dubois C, Wurbel M A, Chauvin J P, Pierres M, He H T. EMBO J. 1998;17:5334–5348. doi: 10.1093/emboj/17.18.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xavier R, Brennan T, Li Q, McCormack C, Seed B. Immunity. 1998;8:723–732. doi: 10.1016/s1074-7613(00)80577-4. [DOI] [PubMed] [Google Scholar]
- 14.Janes P W, Ley S C, Magee A I. J Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horejsi V, Drbal K, Cebecauer M, Cerny J, Brdicka T, Angelisova P, Stockinger H. Immunol Today. 1999;20:356–361. doi: 10.1016/s0167-5699(99)01489-9. [DOI] [PubMed] [Google Scholar]
- 16.Anderson H A, Hiltbold E M, Roche P A. Nat Immunol. 2000;1:156–162. doi: 10.1038/77842. [DOI] [PubMed] [Google Scholar]
- 17.Yashiro-Ohtani Y, Zhou X Y, Toyo-Oka K, Tai X G, Park C S, Hamaoka T, Abe R, Miyake K, Fujiwara H. J Immunol. 2000;164:1251–1259. doi: 10.4049/jimmunol.164.3.1251. [DOI] [PubMed] [Google Scholar]
- 18.Dong R P, Tachibana K, Hegen M, Scharpe S, Cho D, Schlossman S F. Mol Immunol. 1998;35:13–21. doi: 10.1016/s0161-5890(98)00015-7. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa T, Itoh M, Kruger N X, Streuli M, Saito H. Proc Natl Acad Sci USA. 1994;91:10928–10932. doi: 10.1073/pnas.91.23.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer K D, Kong Y Y, Nishina H, Tedford K, Marengere L E, Kozieradzki I, Sasaki T, Starr M, Chan G, Gardner S, et al. Curr Biol. 1988;8:544–562. doi: 10.1016/s0960-9822(98)70224-6. [DOI] [PubMed] [Google Scholar]
- 21.Streuli M, Morimoto C, Schrieben M, Schlossman S F, Saito H. J Immunol. 1988;141:3910–3914. [PubMed] [Google Scholar]
- 22.Rodgers W, Rose J K. J Cell Biol. 1996;135:1515–1523. doi: 10.1083/jcb.135.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ledbetter J A, Martin P J, Spooner C E, Wofsy D, Tsu T T, Beatty P G, Gladstone P. J Immunol. 1985;135:2331–2236. [PubMed] [Google Scholar]
- 24.Bierer B E, Peterson A, Gorga J C, Herrmann S H, Burakoff S J. J Exp Med. 1988;168:1145–1156. doi: 10.1084/jem.168.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikushima H, Munakata Y, Ishii T, Iwata S, Terashima M, Tanaka H, Schlossman S F, Morimoto C. Proc Natl Acad Sci USA. 2000;97:8439–8444. doi: 10.1073/pnas.97.15.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penninger J M, Irie-Sasaki J, Sasaki T, Oliveira-dos-Santos A J. Nat Immun. 2001;2:389–396. doi: 10.1038/87687. [DOI] [PubMed] [Google Scholar]
- 27.Johnson K G, Bromley S K, Dustin M L, Thomas M L. Proc Natl Acad Sci USA. 2000;29:10138–10143. doi: 10.1073/pnas.97.18.10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperling A I, Sedy J R, Majunath N, Kupfer A, Ardman B, Burkhardt J K. J Immunol. 1998;161:6459–6462. [PubMed] [Google Scholar]
- 29.Parolini I, Sargiacomo M, Lisanti M P, Peschle C. Blood. 1996;87:3783–3794. [PubMed] [Google Scholar]
- 30.Thomas M L, Brown E J. Immunol Today. 1999;20:406–411. doi: 10.1016/s0167-5699(99)01506-6. [DOI] [PubMed] [Google Scholar]
- 31.Ashwell J D, D'Oro U. Immunol Today. 1999;20:412–416. doi: 10.1016/s0167-5699(99)01505-4. [DOI] [PubMed] [Google Scholar]
- 32.Cho H, Ramer S E, Itoh M, Kitas E, Bannwarth W, Burn P, Saito H, Walsh C T. Biochemistry. 1992;31:133–138. doi: 10.1021/bi00116a019. [DOI] [PubMed] [Google Scholar]
- 33.Cheng S H, Espino P C, Marshall J, Harvey R, Merrill J, Smith A E. J Virol. 1991;65:170–179. doi: 10.1128/jvi.65.1.170-179.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veillette A, Fournel M. Oncogene. 1990;5:1455–1462. [PubMed] [Google Scholar]
- 35.Koretzky G A, Picus J, Schultz T, Weiss A. Proc Natl Acad Sci USA. 1991;88:2037–2041. doi: 10.1073/pnas.88.6.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pingel J T, Thomas M L. Cell. 1989;58:1055–1065. doi: 10.1016/0092-8674(89)90504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volarevic S, Niklinska B B, Burns C M, Yamada H, June C H, Dumont F J, Ashwell J D. J Exp Med. 1992;176:835–844. doi: 10.1084/jem.176.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujimaki W, Iwashima M, Yagi J, Zhang H, Yagi H, Seo K, Imai Y, Imanishi K, Uchiyama T. J Biol Chem. 2001;276:17455–17460. doi: 10.1074/jbc.M101072200. [DOI] [PubMed] [Google Scholar]
- 39.Desai D M, Sap J, Schlessinger J, Weiss A. Cell. 1993;73:541–554. doi: 10.1016/0092-8674(93)90141-c. [DOI] [PubMed] [Google Scholar]
- 40.Majeti R, Xu Z, Parslow T G, Olson J L, Daikh D I, Killeen N, Weiss A. Cell. 2000;103:1059–1070. doi: 10.1016/s0092-8674(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 41.Masuyama J, Berman J S, Cruikshank W W, Morimoto C, Center M D. J Immunol. 1992;148:1367–1374. [PubMed] [Google Scholar]
- 42.Hafler D A, Fox D A, Manning M E, Schlossman S F, Reinherz E L, Weiner H L. N Engl J Med. 1985;312:1405–1411. doi: 10.1056/NEJM198505303122201. [DOI] [PubMed] [Google Scholar]
- 43.Mizokami A, Eguchi K, Kawakami A, Ida H, Kawabe Y, Tsukada T, Aoyagi T, Maeda K, Morimoto C, Nagataki S. J Rheumatol. 1996;23:2022–2026. [PubMed] [Google Scholar]
- 44.Cardero O, Salgado F, Vinulea J, Nogueria M. Immunobiology. 1997;197:522–533. doi: 10.1016/s0171-2985(97)80084-8. [DOI] [PubMed] [Google Scholar]