Abstract
Equilibrium dialysis is a simple and effective technique used for investigating the binding of small molecules and ions to proteins. A three-dimensional (3D) printer was used to create a device capable of measuring binding constants between a protein and a small ion based on equilibrium dialysis. Specifically, the technology described here enables the user to customize an equilibrium dialysis device to fit their own experiments by choosing membranes of various material and molecular-weight cutoff values. The device has dimensions similar to that of a standard 96-well plate, thus being amenable to automated sample handlers and multichannel pipettes. The device consists of a printed base that hosts multiple windows containing a porous regenerated-cellulose membrane with a molecular-weight cutoff of ~3500 Da. A key step in the fabrication process is a print-pause-print approach for integrating membranes directly into the windows subsequently inserted into the base. The integrated membranes display no leaking upon placement into the base. After characterizing the system’s requirements for reaching equilibrium, the device was used to successfully measure an equilibrium dissociation constant for Zn2+ and human serum albumin (Kd = (5.62 ± 0.93) × 10−7 M) under physiological conditions that is statistically equal to the constants reported in the literature.
Graphical abstract
Three-dimensional (3D) printing has revolutionized measurement science by empowering scientists to rapidly create customized tools to fit individual research needs.1–3 3D printing has been used in laboratories to create platforms for pharmacokinetic/pharmacodynamic (PK/PD) analysis,4,5 microfluidic cell culture,6–8 measurements of cell secretions,6,9,10 and general labware.11–13 Polyjet printing, which is the 3D-printing method used to create the device described here, is an additive manufacturing process where an object is built by successive layer-by-layer additions of a liquid polymer that is immediately cured by ultraviolet (UV) radiation after each layer. The printer builds the device according to a design drawn by the user in a CAD software package that is submitted to the printer as a stereolithography (.stl) file. Materials with different properties (rigidity, compressibility, smoothness, etc.), can be selected prior to printing to enhance device functionality.14 The device described here, printed using multiple materials, was used to measure the binding affinity between a biologically important ligand and protein following equilibrium dialysis.
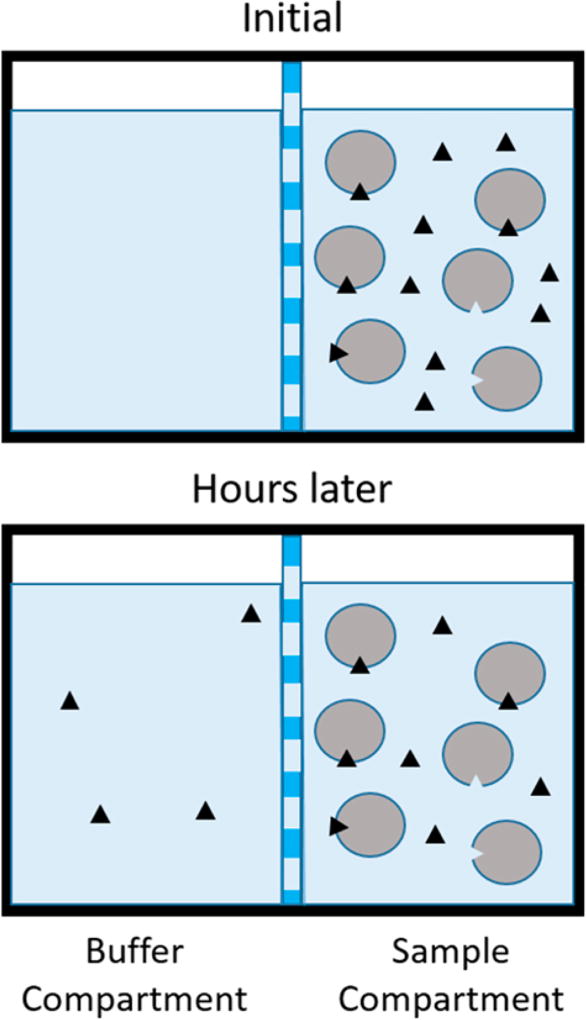
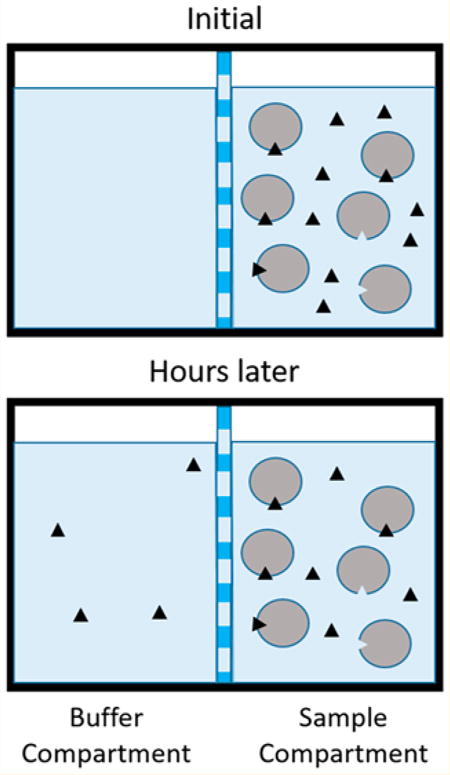
Measuring the binding affinity of drugs, ions, and hormones to serum proteins provides important information about the bioavailability of these species to cells and tissues. Various techniques have been developed to perform these measurements, including ultrafiltration,15 high-performance affinity chromatography,16,17 ultracentrifugation, surface plasmon resonance, 18,19 fluorescence spectroscopy,20 isothermal titration calorimetry, and equilibrium dialysis, the most widely used technique for measuring the fraction of a drug or ligand bound to serum proteins.19 The principle of equilibrium dialysis, shown in Figure 1, involves mixing a ligand and receptor in a buffered solution that is placed on one side of a size-exclusion membrane. The membrane contains pores small enough to exclude the receptor and ligand/receptor complex from passing through it, but large enough to allow passage of free ligand. Buffer is placed on the opposite side of the membrane and the system is allowed several hours to permit the equilibrium of free ligand concentration on both sides of the membrane to be established, typically under constant agitation by a platform shaker. The concentration of bound ligand can be calculated using eq 1, where the total number of moles of ligand are known and number of moles of free ligand can be determined by measurement of the concentration of free ligand in the buffer compartment multiplied by the total volume of the compartments. This information can then be used to calculate an equilibrium dissociation constant using a saturation-binding curve or Scatchard analysis.
| (1) |
Figure 1.
Illustration of the principle of equilibrium dialysis. The circles represent protein (albumin), and the black triangles represent ligand (Zn2+).
While enabling high accuracy in the determination of binding constants, an undesirable feature of equilibrium dialysis is the long incubation time of several hours often required for free-ligand concentration to reach equilibrium on both sides of the membrane. Therefore, to increase sampling rate and sample throughput, determinations based on equilibrium dialysis are often performed using platforms based on 96-well plate technologies.21 The ability to simultaneously determine binding constants or binding numbers using well-plate formats also enables such high throughput tools as multichannel pipettors and automated liquid handling systems. Indeed, equilibrium dialysis platforms in well-plate formats are commercially available.19 However, there are shortcomings to these commercially available systems. For example, the end user has a limited choice of membrane materials (typically cellulose) and molecular-weight cut off options. In addition, many characteristics of the dialysis membrane (such as percent porosity) are unknown, making mathematical predictive modeling of equilibrium times difficult to determine, and requiring the user to “guess and check” the equilibration time prior to performing the desired measurements. These shortcomings can be overcome by allowing the user to select a membrane of their choice.
Membranes are commonly incorporated into analytical fluidic devices for many applications.22–24 However, incorporating membranes into devices without leakage remains difficult and cumbersome, often requiring adhesives or O-rings. In a recent example of overcoming such obstacles, innovative work by Breadmore et al. described successful use of a multiple-material FDM 3D printer to integrate a polymer Lay–Felt membrane directly into a microfluidic device without leakage.25 Here, we describe a novel and simple Print–Pause–Print technique for seamlessly integrating any membrane directly into 3D-printed devices via a polyjet printer without the use of adhesives or O-rings. The device, which was developed for measuring the equilibrium-binding constants of important biological ligands to a carrier protein, enables the user to choose any membrane for their experiments. The device also has the same size and shape of a standard 96-well plate, allowing easy sample handling and pipetting. Specifically, we report the measurement of the binding affinity of the zinc metal ion (Zn2+) for human serum albumin (HSA), which is a bloodstream protein with a plasma concentration of ~600 µM, making it the most abundant plasma protein. HSA is considered an important carrier protein, because of its ability to bind different ions, hormones, and drugs and distribute them throughout the body.26 It is believed that only the unbound fraction of these species are available to cells and tissues; therefore, measuring the fraction of these species bound to HSA in vitro is essential to understanding their bioavailability and PK/PD profiles in vivo.19,26
EXPERIMENTAL SECTION
Fabrication of the Dialysis Base Plate
The device base is the same size and shape of a 96-well plate; the wells are 2.5 mm in width, allowing convenient pipetting of sample out of the device with the use of a multichannel pipet for rapid transport to an instrument for quantitation of the analyte ligand. The device was created by an additive manufacturing technique using an Objet Connex 350 3D printer housed in the department of Electrical and Computer Engineering at Michigan State University. The printer is capable of 16 µm resolution in the Z-axis and 600 dpi in the XY plane. An .stl file containing the blueprints of the device, as created in CAD software (Autodesk Inventor Professional), was submitted to the printer. The printer interprets the .stl file and successively deposits ~30 µm layers of a chosen liquid polymer, followed by curing of the polymer via UV light to create a rigid object according to the specifications of the .stl file. This 3D printer is capable of printing multiple materials with different properties into the same device for added functionality. In this way, a dialysis base plate (Figure 2) was printed in a rigid material (VeroClear, Stratasys, Ltd., Eden Prairie, MN) containing 12 wells that were lined with 0.6 mm of a rubberlike material (Tangoblack, Stratasys, Ltd.). The rubberlike properties of the Tangoblack material provide a watertight seal in the well, which is important to prevent leaking when hosting the membranes and membrane holders, which are described below.
Figure 2.
(Left) Autodesk CAD drawing of the equilibrium dialysis base plate. The base plate has the exact same dimensions as a standard 96-well plate and is printed with two different materials. The TangoBlack material printed within the wells provides compression against the inserted membrane holders, which is a necessary feature to prevent unwanted leakage of liquid between compartments. (Right) Photograph of the finished device, as printed by a Connex 350 3D printer, depicting a membrane holder about to be slid into position.
Print–Pause–Print Technique for Integrating Membranes Directly into 3D-Printed Devices
The membrane holders (3.3 mm wide, 32.8 mm long) were designed to slide tightly into the wells of the dialysis base plate to create two separate compartments. The technique for integrating the membranes directly into the membrane holders is illustrated in Figure 3. The window-shaped membrane-holders, shown in Figures 4A–D, were fabricated with 0.55 mm outer layers of the rubberlike Tangoblack in order to create a water-tight seal when slid into the dialysis base plate. The interior of the membrane holders was a 2.0 mm layer of Verowhite material for rigidity, with a thin (0.2 mm) additional layer of Tangoblack printed directly in the middle of the Verowhite, and is shown in Figure 4A. The print process is monitored by the operator as the printer creates layers of material to build the membrane holder from the bottom up. When exactly half of the membrane holder was printed (1.65 mm from the bottom), the print process was paused, the cover of the Objet Connex 350-printer (Figure 4B) was opened, and precut commercially purchased 3.5 kDa MWCO regenerated-cellulose dialysis membranes (Spectrum Laboratories, Inc., Rancho Dominquex, CA) were carefully placed onto the freshly printed middle layer of Tangoblack on each membrane holder, as shown in Figure 4C (the thin layer of Tangoblack is barely visible; therefore, the object appears light blue). The Tangoblack is somewhat adhesive, thus facilitating membrane placement throughout the process. The membranes were hand-cut by the operator and placed onto the membrane holder to cover the void area in the holder. The printing resumed, laying polymer on the edges of the membrane and sealing it directly into the holder. The printer was then allowed to finish the print process to create the finished product, as shown in Figure 4D. The membrane containing holders are then placed by hand into the base plate (Figure 2) with a small amount of silicone vacuum grease (Dow Corning, Midland, MI) on the edges of the Tangoblack material on the membrane holder.
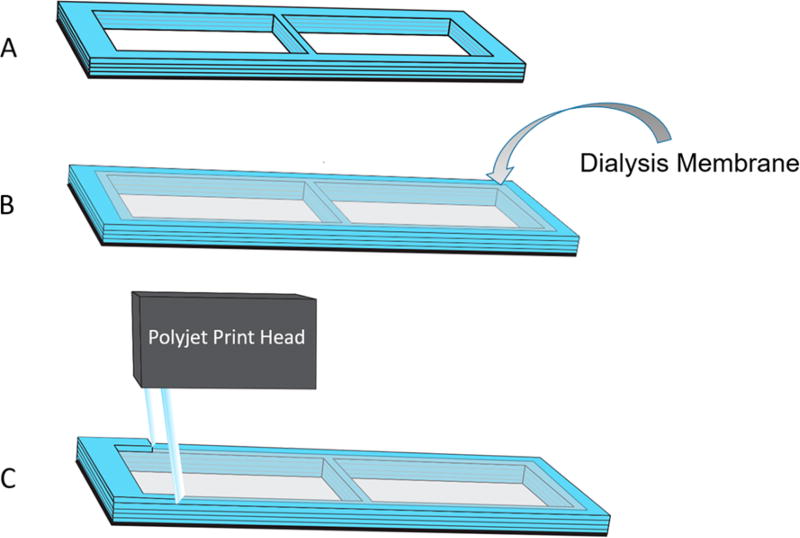
Figure 3.
A Connex 3D printer was used to integrate membranes directly into a device. First, in panel (A), a device containing a window was printed halfway. The printer was paused, and in panel (B), the operator laid down a porous membrane of choice over the window. In panel (C), the printer resumes and the polyjet print head lays polymer over the edges of the membrane to seamlessly seal it into the device.
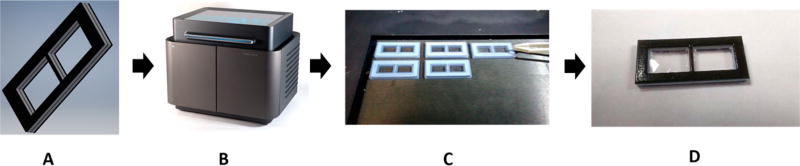
Figure 4.
In panel (A), a model is drawn using Autodesk Inventor Professional 2017 CAD software. The model is saved as an .stl file and sent to the 3D printer (panel (B)). The membrane holder is printed with multiple materials. The interior is a rigid Verowhite material, and the exterior is a compressible TangoBlack material, to prevent leaking. In panel (C), the operator places the membranes into the device halfway through the print process. Panel D shows the final product: a membrane holder with a membrane seamlessly sealed into the device.
Reagents
Albumin from human serum (HSA; lyophilized powder, free of fatty acid and globulin, ≥99% pure, via agarose gel electrophoresis) was purchased from Sigma (St. Louis, MO, USA). The Pierce bicinchoninic acid (BCA) protein assay kit was purchased from ThermoScientific. Radio-labeled zinc was purchased as 65ZnCl2 from PerkinElmer with a radionuclide purity of 99.0% (half-life = 244 days) and diluted in distilled and deionized H2O (DDW, 18 MΩ) to prepare a stock solution whose concentration of 65Zn2+ was calculated daily.
Sample Preparation
A stock solution of HSA was prepared daily by diluting the lyophilized HSA powder with dialysis buffer (150 mM NaCl, 21 mM Tris, pH 7.40 in DDW, sterile filtered through 0.22 µm filters from Sigma) to make a 5 mg/mL HSA solution, as confirmed by the BCA assay. From this stock solution, samples (each 750 µL total) were prepared in dialysis buffer to contain 15.0 µM HSA and varying concentrations of 65ZnCl2 ranging from 1 µM to 25 µM.
Characterization of Incubation Time
The membrane holders were placed into the dialysis base plate and the compartments and membranes rinsed three times with DDW to remove any contamination or debris. Excess liquid was removed with a pipet. A 700 µL aliquot containing 15 µM HSA and 5 µM 65ZnCl2 was then pipetted into a sample compartment on the device. Subsequently, 700 µL of dialysis buffer were pipetted into the other compartment on the device. This exact procedure was repeated in 4 other wells on the device, followed by placement of a 96-well plate adhesive plate sealer to prevent evaporation during incubation. The entire device was agitated at 220 rpm at 37 °C on an incubating orbital shaker (Talboys) to provide constant mixing and maintain physiological temperature. Lastly, 500 µL of sample were removed from the buffer compartment of an individual well following 2, 4, 5.5, 8, and 10 h of incubation for quantitation of free 65Zn2+.
Quantitation of Free 65Zn2+ via Liquid Scintillation Counting
Samples containing 15.0 µM HSA and different concentrations of 65ZnCl2 (1, 2.5, 5, 10, 16, 19, and 25 µM 65Zn2+) were pipetted into separate sample compartments of the device, as described in the previous section, with equal volumes of diluted dialysis buffer in the adjacent buffer compartments. The device was agitated/incubated at 37 °C for 6 h. Then, immediately following the 6 h of incubation, 500 µL of sample from the buffer compartments were removed for quantitation of free 65Zn2+ using a PerkinElmer MicroBeta TriLux 1450 liquid scintillation counter. A 90 µL aliquot of sample was pipetted into a well of a 96-well plate, followed by the addition of 90 µL of scintillation cocktail (OptiPhase Supermix, PerkinElmer) to the sample-containing well. Standards of known 65Zn2+ concentrations in dialysis buffer were treated in the same manner to generate an external standards calibration curve. The plate was placed into the instrument for counting, following a 30 min delay.
Calculations and Data Analysis
After the concentration of free 65Zn2+ was quantitatively determined, the concentration of bound 65Zn2+ was calculated by subtracting the moles of free 65Zn2+ from the total moles of 65Zn2+ originally added and accounting for the volume in the sample-compartment (see eq 1). The concentrations of free versus bound 65Zn2+ were used to create a saturation binding curve. This curve was then analyzed by nonlinear regression software (SigmaPlot 13.0) to calculate an equilibrium dissociation constant (Kd) and the binding stoichiometry (n) for the system.
RESULTS
The initial challenge of fabricating the device was creating a system that did not leak. That is, a device containing two separate compartments separated by a membrane in which the only molecular transport between the two compartments is by diffusion through the membrane pores. Initial studies using this device with glued-in membranes failed to create a leak-free system and the process of gluing membranes into the device was cumbersome and time-consuming with low precision between devices. Also, initial studies featured a device using a single material (Verowhite) that resulted in leakage of liquid samples from one compartment into the other around the outside of the membrane holder. The incorporation of multiple materials of different properties, rigid Verowhite and rubbery Tangoblack, allowed the membrane insert to form a watertight seal when inserted into the dialysis base plate. The Tangoblack material is able to compress under pressure, much like an O-ring, thus providing the leak-free seal. The Print–Pause–Print approach was developed to seamlessly integrate membranes in a time-efficient manner into 3D-printed devices without leakage. The membrane seal was evaluated by simply placing water on one side of the membrane and nothing else on the other and shaking the device for 6 h, while regularly monitoring the adjacent membrane compartment visually for leaked water.
Once a device had been constructed that had no leaking around the membrane holders, the binding of ligand to protein was implemented. Size-exclusion dialysis membranes with a 3.5 kDa molecular weight cutoff (MWCO) were chosen in order to allow the passage of free Zn2+ through the membrane pores, while efficiently blocking the passage of HSA (molar mass ≈ 66.5 kDa) and the Zn2+-HSA complex. This allows the quantitative determination of free and bound Zn2+ concentration at equilibrium, which allows the calculation of an equilibrium binding constant.
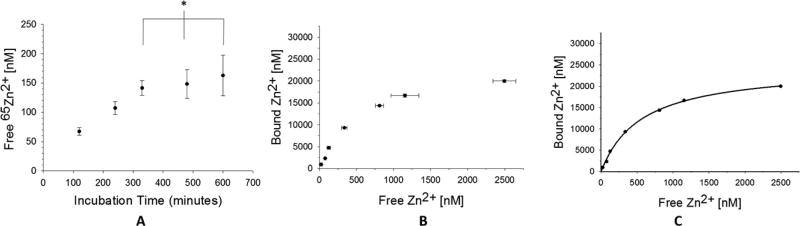
Preliminary experiments were required to measure the time needed for the concentration of free Zn2+ to reach equilibrium on both sides of the membrane. The incubation time needed to reach equilibrium was indicated by the time which the free concentration of Zn2+ stopped, increasing in the buffer compartment. As seen in Figure 5A, the difference in the concentration of free Zn2+ after 5.5 h of incubation was not statistically different than the concentration at 8 or 10 h, indicating a minimum incubation time of 5.5 h to be sufficient for the system to reach equilibrium. Thus, future studies used a conservative incubation time of 6 h.
Figure 5.
In panel (A), the time needed to reach equilibrium in the device is estimated as the time when the free [65Zn2+] ceases to increase. These data suggest that equilibrium is reached between 5 and 6 h (n = 3, error = standard deviation × p = 0.606). In panel (A), seven samples were made containing 15 µM HSA and varying concentrations of [65Zn2+] (1–25 µM). The samples were placed in separate wells of the device and incubated for 6 h (n = 3, error = standard deviation). In panel (C), the mean free and bound [65Zn2+] concentrations were analyzed using nonlinear regression software (SigmaPlot 13.0), assuming a one-site binding model and accounting for nonspecific binding (goodness-of-fit (R2) = 0.9985).
Experiments were then performed in order to produce a saturation binding curve between HSA and Zn2+ by holding the concentration of HSA constant and varying the concentration of Zn2+, so that the free concentration of Zn2+ was varied across several orders of magnitude. The experiment was replicated three times and the results can be seen in Figure 5B, showing a reproducible binding curve. The three data points collected for each different concentration of Zn2+ were averaged, plotted, and analyzed by nonlinear regression software (SigmaPlot 13.0), to produce the graph in Figure 5C. The data fit well to a one-site binding model accounting for nonspecific binding with a goodness-of-fit (R2) of 0.9985. An equilibrium dissociation constant (Kd) and binding stoichiometry (n) was calculated as (5.62 ± 0.93) × 10−7 molar and 1.6 ± 0.2, respectively. The Kd can be converted to a log K value of 6.2 ± 0.1, and can be compared to a previously reported log K value of 6.4 ± 0.8.27 The data agrees well with previously reported literature values, therefore confirming the capability of the device to be used to measure the binding affinity between a small ligand and a large protein.
CONCLUSIONS
Here, a 3D-printed device was fabricated and employed to perform equilibrium dialysis experiments to measure the binding affinity of Zn2+ to HSA. In contrast to commercially available technologies, this device is fully customizable and allows the user to select any membrane of their choice to perform the dialysis experiment. This is important for several reasons, one being that certain membrane materials can affect binding measurements by causing nonspecific binding of the analyte ligand. Also, some dialysis membranes have more stringent MWCO properties than others, which may be of interest to the user who requires high specificity separations based on size exclusion. This also enables the user to optimize the incubation time of their equilibrium dialysis experiments by selecting membranes with known properties, such as percent porosity. The ability to incorporate any membrane into the device is made possible by the Print–Pause–Print polyjet 3D-printing approach reported here. The Print–Pause–Print method empowers the user to integrate membranes seamlessly into 3D-printed fluidic devices without leakage. The potential of the application of this technique in analytical chemistry and engineering is high, as integrating membranes into microfluidic devices is a necessary but previously cumbersome, time-consuming, and often low precision task. The 3D-printed device described here can be used for measuring the binding affinity of small ligands to larger proteins or even cells. By allowing the use of any membrane, the binding experiments that the user can perform via equilibrium dialysis are no longer limited by commercial availability.
Acknowledgments
The authors would like to thank Scot Stanulis and Jack Buhl for technical assistance, and Brian Wright (Department of Electrical and Computer Engineering, Michigan State University) for assistance with the design and fabrication of the 3D-printed devices. Financial support for this project is from the National Institutes of Health (Nos. DK110665 and GM110406).
Footnotes
The authors declare no competing financial interest.
References
- 1.Gross B, Lockwood SY, Spence DM. Anal. Chem. 2017;89:57–70. doi: 10.1021/acs.analchem.6b04344. [DOI] [PubMed] [Google Scholar]
- 2.Gross BC, Erkal JL, Lockwood SY, Chen C, Spence DM. Anal. Chem. 2014;86:3240–3253. doi: 10.1021/ac403397r. [DOI] [PubMed] [Google Scholar]
- 3.Waheed S, Cabot JM, Macdonald NP, Lewis T, Guijt RM, Paull B, Breadmore MC. Lab Chip. 2016;16:1993–2013. doi: 10.1039/c6lc00284f. [DOI] [PubMed] [Google Scholar]
- 4.Lockwood SY, Meisel JE, Monsma FJ, Jr, Spence DM. Anal. Chem. 2016;88:1864–1870. doi: 10.1021/acs.analchem.5b04270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaBonia GJ, Lockwood SY, Heller AA, Spence DM, Hummon AB. Proteomics. 2016;16:1814–1821. doi: 10.1002/pmic.201500524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Chen C, Summers S, Medawala W, Spence DM. Integr. Biol. 2015;7:534–543. doi: 10.1039/c4ib00243a. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Mehl BT, Sell SA, Martin RS. Analyst. 2016;141:5311–5320. doi: 10.1039/c6an01282e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharjee N, Urrios A, Kang S, Folch A. Lab Chip. 2016;16:1720–1742. doi: 10.1039/c6lc00163g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Wang Y, Lockwood SY, Spence DM. Analyst. 2014;139:3219–3226. doi: 10.1039/c3an02357e. [DOI] [PubMed] [Google Scholar]
- 10.Erkal JL, Selimovic A, Gross BC, Lockwood SY, Walton EL, McNamara S, Martin RS, Spence DM. Lab Chip. 2014;14:2023–2032. doi: 10.1039/c4lc00171k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Au AK, Bhattacharjee N, Horowitz LF, Chang TC, Folch A. Lab Chip. 2015;15:1934–1941. doi: 10.1039/c5lc00126a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macdonald NP, Bunton GL, Park AY, Breadmore MC, Kilah NL. Anal. Chem. 2017;89:4405–4408. doi: 10.1021/acs.analchem.7b00443. [DOI] [PubMed] [Google Scholar]
- 13.Belka M, Ulenberg S, Baczek T. Anal. Chem. 2017;89:4373–4376. doi: 10.1021/acs.analchem.6b04390. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, Mehl BT, Munshi AS, Townsend AD, Spence DM, Martin RS. Anal. Methods. 2016;8:6005–6012. doi: 10.1039/C6AY01671E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melten JW, Wittebrood AJ, Willems HJ, Faber GH, Wemer J, Faber DB. J. Pharm. Sci. 1985;74:692–694. doi: 10.1002/jps.2600740625. [DOI] [PubMed] [Google Scholar]
- 16.Mallik R, Yoo MJ, Briscoe CJ, Hage DS. J. Chromatogr. A. 2010;1217:2796–2803. doi: 10.1016/j.chroma.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hage DS. Clin. Chem. 2017;63:1083. doi: 10.1373/clinchem.2016.262253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshannessy DJ, Brighamburke M, Soneson K, Hensley P, Brooks I. Anal. Biochem. 1993;212:457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- 19.Vuignier K, Schappler J, Veuthey J-L, Carrupt P-A, Martel S. Anal. Bioanal. Chem. 2010;398:53–66. doi: 10.1007/s00216-010-3737-1. [DOI] [PubMed] [Google Scholar]
- 20.Epps DE, Raub TJ, Caiolfa V, Chiari A, Zamai M. J. Pharm. Pharmacol. 1999;51:41–48. doi: 10.1211/0022357991772079. [DOI] [PubMed] [Google Scholar]
- 21.Waters NJ, Jones R, Williams G, Sohal B. J. Pharm. Sci. 2008;97:4586–4595. doi: 10.1002/jps.21317. [DOI] [PubMed] [Google Scholar]
- 22.Cannon DM, Kuo T-C, Bohn PW, Sweedler JV. Anal. Chem. 2003;75:2224–2230. doi: 10.1021/ac020629f. [DOI] [PubMed] [Google Scholar]
- 23.Genes LI, Tolan NV, Hulvey MK, Martin RS, Spence DM. Lab Chip. 2007;7:1256–1259. doi: 10.1039/b712619k. [DOI] [PubMed] [Google Scholar]
- 24.Li F, Guijt RM, Breadmore MC. Anal. Chem. 2016;88:8257–8263. doi: 10.1021/acs.analchem.6b02096. [DOI] [PubMed] [Google Scholar]
- 25.Li F, Smejkal P, Macdonald NP, Guijt RM, Breadmore MC. Anal. Chem. 2017;89:4701–4707. doi: 10.1021/acs.analchem.7b00409. [DOI] [PubMed] [Google Scholar]
- 26.Peters T., Jr . All about Albumin: Biochemistry, Genetics, and Medical Applications. Academic Press; San Diego, CA: 1995. [Google Scholar]
- 27.Goumakos W, Laussac J-P, Sarkar B. Biochem. Cell Biol. 1991;69:809–820. doi: 10.1139/o91-121. [DOI] [PubMed] [Google Scholar]