Abstract
O-antigens are cell surface polysaccharides of many Gram-negative pathogens that aid in escaping innate immune responses.1 A widespread O-antigen biosynthesis mechanism involves the synthesis of the lipid-anchored polymer on the inner membrane’s (IM’s) cytosolic face, followed by transport to the periplasmic side where it is ligated to the lipid A core to complete a lipopolysaccharide (LPS) molecule2. The O antigen’s transport to the periplasm is mediated by an ATP-binding cassette (ABC) transporter, called Wzm/Wzt, Extended Data Fig. 1. We present the crystal structure of the Wzm/Wzt homolog from Aquifex aeolicus in an open conformation. The transporter forms a transmembrane (TM) channel sufficiently wide to accommodate a linear polysaccharide. It’s nucleotide binding domain (NBD) and a periplasmic loop form ‘gate helices’ at the cytosolic and periplasmic membrane interfaces, likely serving as substrate entry and exit points. Site-directed mutagenesis of the gates impairs in vivo O antigen secretion in the E. coli prototype. Combined with a closed structure of the isolated NBDs, our structural and functional analyses suggest a processive O antigen translocation mechanism, which stands in contrast to the classical alternating access mechanism of ABC transporters.
Main
Microorganisms commonly use cell surface polysaccharides to establish extended barriers that protect against the defense machineries of their hosts.1 O antigens help bacteria to evade innate immune responses including phagocytosis and complement-mediated lysis.3–5 The polymers are hypervariable polysaccharides up to ~100 sugar units long and most reach the periplasm by one of two convergent pathways.2 In the widespread ABC transporter dependent pathway, the O antigen is fully synthesized as an undecaprenyl diphosphate (Und-PP)-linked intermediate, before being transported to the IM’s periplasmic leaflet by Wzm/Wzt and ligated to lipid A-core, Extended Data Fig. 1. PglK, an oligosaccharide ABC transporter from a bacterial protein N-glycosylaton system, provided the first example of an exporter translocating Und-PP-linked substrates.6
Some systems signal completion of O antigen biosynthesis by modifying the growing (non-reducing) end of the polysaccharide chain with, for example, phosphate, methyl or sugar moieties.7 The corresponding ABC transporter recognizes the modified terminus via a carbohydrate-binding domain (CBD) fused to the C terminus of its NBD to accomplish transport, Extended Data Fig. 1. 8,9–10 In other systems, export of uncapped glycans, such as O antigens and teichoic acids, occurs without the involvement of CBDs 7.
ABC transporters usually cycle between inward and outward facing conformations to facilitate substrate transport. However, this ‘alternating access’ model may not apply to transporters translocating high molecular weight polymers, such as polypeptides, O antigens, and capsular polysaccharides.
To elucidate the O antigen translocation mechanism, we determined the crystal structure of Aquifex aeolicus (Aa) Wzm/Wzt, which is homologous to the prototypical E. coli Wzm/Wzt and S. aureus wall teichoic acid transporters,11 Extended Data Fig. 2. In a nucleotide-free state, Wzm/Wzt forms a continuous channel across the membrane. The transporter’s similarity to the E. coli O9a transporter allowed testing of functional predictions in vivo with an established prototype. Combined with structures of the transporter’s isolated NBDs in a closed conformation, our structural and functional analyses suggest substrate entry and exit pathways and a model for O antigen membrane translocation.
To gain structural insights into O antigen translocation, we expressed and purified AaWzm/Wzt, with Wzm and Wzt forming the TM domain (TMD) and NBD, respectively. For crystallization, the C terminal Wzt-CBD was removed, generating a construct including residues 1 through 235 (WztN). Similar constructs of E. coli O9a and K. pneumoniae O12 Wzt proteins are fully functional in vivo if the CBD is expressed in trans.8,10 The 3.85 Å-resolution Wzm/WztN structure includes residues 2 to 255 of Wzm and 2 to 235 of WztN, Extended Data Table and Extended Data Fig. 3.
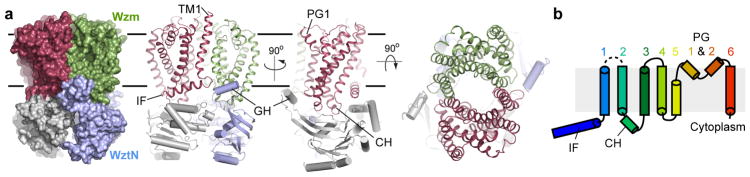
In a nucleotide-free state, Wzm/WztN adopts a compact structure containing a Wzm dimer interacting with a WztN dimer in an open conformation, Fig. 1a. The TMDs closely interact over the entire length of their TM regions and surround a central TM channel, formed by TM helices 1, 2 and 5, that is open to the intra- and extracellular sides (see below).
Figure 1. Architecture of the Wzm/Wzt O antigen transporter.
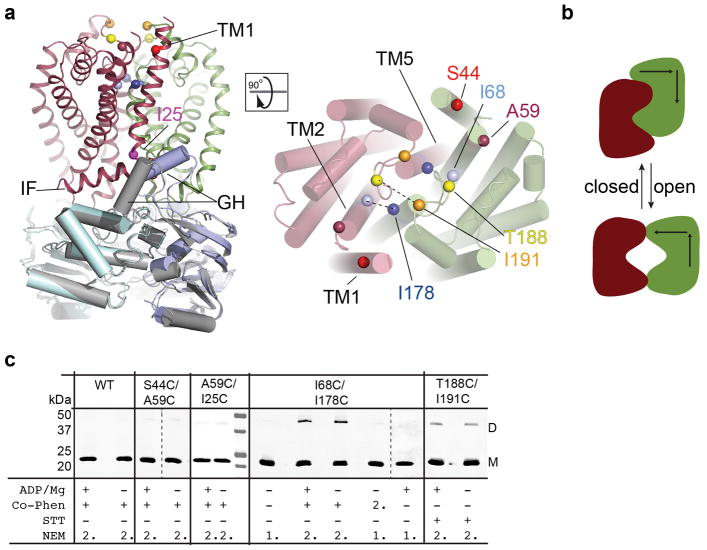
(a) The Wzm protomers are shown in green and red and the nucleotide-binding WztN domains are shown in blue and gray, respectively. WztN forms a short gate helix (GH) near the Wzm protomer interface. Wzm contains an N-terminal interface helix (IF). (b) Transmembrane topology of Wzm. Wzm forms six transmembrane helices and the cytosolic TM2/3 loop forms the coupling helix (CH) Wzm’s periplasmic TM5/6 loop generates two periplasmic gate helices (PG1 and 2). Horizontal lines indicate likely membrane boundaries.
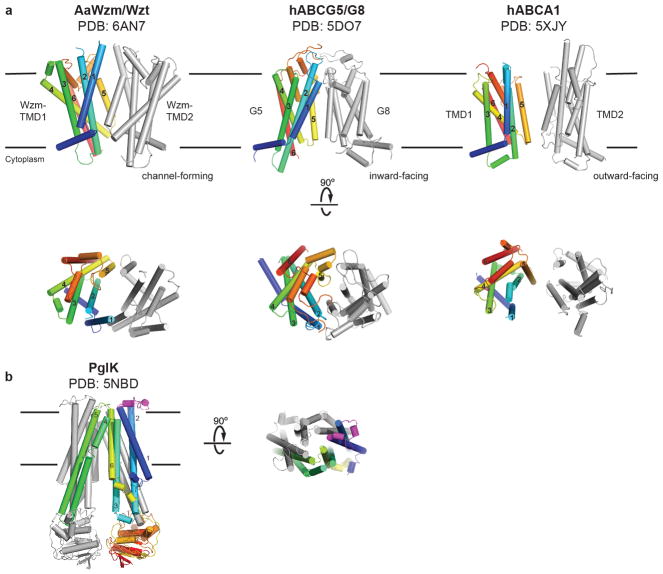
Wzm does not contain any cross-over helices that would interact with WztN of the neighboring half-transporter, unlike previously described bacterial exporters.12 At its N terminus, Wzm forms an amphipathic interface helix (IF) that runs parallel to the Wzm-WztN interface, followed by six TM helices. The loop connecting TM2 and TM3 couples Wzm with WztN (coupling helix) and the periplasmic connection between TM5 and TM6 forms two reentrant helices, PG1 and PG2. Overall, the Wzm architecture resembles the Type-II ABC exporter topology, to date only observed in human lipid exporters, Fig. 1b and Extended Data Fig. 4a.13,14 Of note, the Wzm/WztN architecture and thus likely its translocation mechanism differs significantly from PglK, which translocates Und-PP-linked oligosaccharides, Extended Data Fig. 4b.6
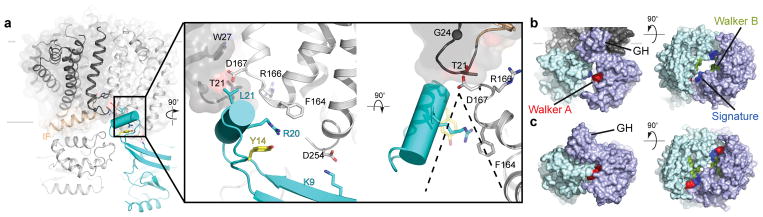
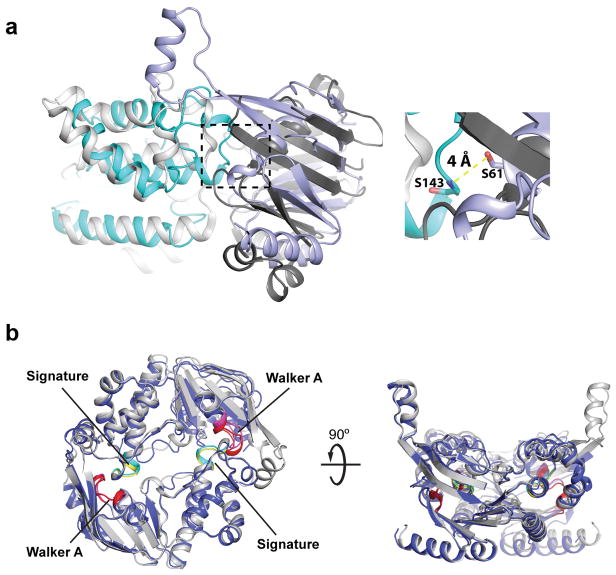
The transporter’s NBDs are separated by about 8 Å between the Walker A and Signature motifs, sufficient for nucleotide diffusion, Fig. 1a and 2b.12 A defining feature of WztN is an extension of the β-strand 1/2 loop (residues 13–32), which forms a short ‘gate helix’ (residues 18–26) that rests near the Wzm-Wzm protomer interface at the putative water-lipid boundary, Fig. 2a. The gate helix packs against a loop connecting Wzm’s TM4 and TM5 of the same half transporter. This loop contains a conserved F-X-R/K-D motif, of which Phe164 interacts with Arg20 of the gate helix and Asp167 sits directly at the Wzm-Wzm interface, Fig. 2a. Additional interactions occur with the backbone of the IF-TM1 loop in the opposing Wzm subunit.
Figure 2. Wzt forms a unique interface with Wzm.
(a) Position of the gate helix (GH) at the Wzm protomer interface. The GH packs against Wzm’s TM4/5 loop and forms a wedge-shaped opening towards the cytosolic water-lipid interface. The transporter is shown as a cartoon and one Wzm protomer is shown as a semi-transparent surface. (b) Open conformation of the WztN NBDs. Surface representation of the transporter’s NBDs colored blue and cyan, respectively. Conserved regions are labeled. (c) Surface representation of the isolated WztN structure in a closed conformation colored as in (b).
Because the C terminus of the gate helix is rotated away from the TM region, the helix creates a wedge-shaped path towards the Wzm dimer interface, likely forming a substrate-binding pocket, Fig. 2a. At its center, the gate helix contains a conserved positively charged residue (Arg20), which could be implicated in binding the Und-PP diphosphate, Fig. 2a and Extended Data Fig. 2a. Strikingly, a preceding Tyr residue (Tyr14) packs against and stabilizes the gate helix on its membrane distal side, Fig. 2a. Primary sequence alignments of homologous transporters reveal that the gate helix and the Tyr residue are characteristic features of all known O antigen and wall teichoic acid ABC transporters, Extended Data Fig. 2a and b, which accept substrates synthesized as Und-PP-linked intermediates.11,15
We also determined the structure of the isolated WztN NBD in two different crystal forms at 2.05 and 3.5 Å resolution, Extended Data Table. Despite the absence of a stabilizing nucleotide, both structures represent a WztN dimer in a closed conformation, with only about 4.0 Å between the hydroxyl group of Ser61 (Walker A) and the backbone amide nitrogen of Ser143 (Signature), Fig. 2c and Extended Data Fig. 5. This closed conformation is in agreement with the AMPPNP-stabilized closed state of the maltose transporter’s NBDs,16 and likely reflects the nucleotide-bound conformation of Wzm/Wzt, Fig. 2b and c.
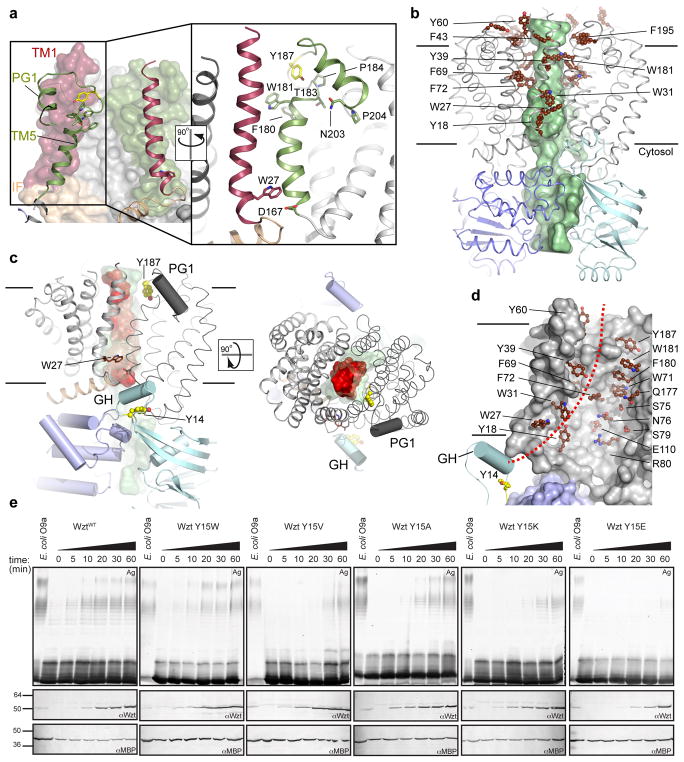
The transporter’s TMD is formed by two closely interacting Wzm protomers that contact each other through TM-helices 1 and 5, Fig. 3a. TM5 is capped at the C terminus by a cluster of conserved aromatic residues that pack against the C-terminal end of TM1 of the opposing Wzm protomer. Strikingly, the Wzm protomers enclose a large channel spanning the entire membrane, Fig. 3b. The channel is constricted near the periplasmic exit as well as the Wzm-WztN interface, yet continuously accessible to a 3.5 Å radius probe, thus capable of accommodating a polysaccharide. The structure of AaWzm/Wzt’s native substrate is currently unknown but a model of the E. coli O9a polymannose antigen can be accommodated, with 8–10 sugar units spanning the channel, Fig. 3c.
Figure 3. The polysaccharide translocation channel.
(a) The Wzm interface. One Wzm protomer is shown as a surface and the opposing subunit as a cartoon. Both subunits are shown as cartoons in the close-up view. TM1 and TM5 are colored red and green, respectively, and the IF is colored beige. Conserved residues are shown as sticks. (b) Surface representation of the Wzm/WztN channel. The channel volume accessible to a 3.5 Å radius probe is shown as a green surface and aromatic residues lining the channel are shown as brown spheres. Selected residues are labeled. (c) Cytosolic and periplasmic gate helices at the Wzm protomer interface. A model of the E. coli O9a antigen containing 10 mannose units was manually placed in the channel and is shown as a red surface. (d) Putative translocation path (red dashed line). Channel-exposed aromatic and polar residues are shown as brown sticks. (e) In vivo O antigen translocation. The indicated point mutations were introduced into E. coli O9a Wzm/Wzt. O antigen export was detected after inducing transporter expression by silver staining (Ag) of whole-cell lysates, detecting exported and LPS-linked O antigens only. Western blots detecting Wzt and MBP were performed to monitor transporter expression and as a loading control, respectively. All experiments were repeated independently at least three times with similar results. Time=Period after inducing Wzt/Wzm expression.
The channel is lined with aromatic residues organized in three layers. First, Tyr18, Trp27, and Trp31 reside at the cytosolic Wzm-Wzm interface where the channel is widest. Second, Tyr39, Phe69, Trp71, Phe72, Phe180, and Trp181 form a central layer halfway across the membrane, and Phe43, Tyr60, Tyr63, and Phe195 surround the periplasmic channel exit, Fig. 3b and d. Protein-carbohydrate interactions are frequently mediated by CH-π stacking interactions between aromatic residues and the sugar rings.17 Clustering of these residues within the channel strongly suggests a role in O antigen coordination during transport. Indeed, a continuous and mostly conserved ‘aromatic path’ runs from the putative cytosolic substrate entrance to the periplasmic channel exit, Fig. 3d and Extended Data Fig. 2. Similar paths have been described in cellulose synthase, cellobiohydrolase 1 and maltoporin.6,18–19,20 As discussed for the maltoporin channel, hydrophobic interactions with aromatics are often combined with a continuous pattern of H-bond donors and acceptors that contact the polymers’ hydroxyl groups and likely minimize translocation energy barriers.20 In Wzm/WztN, Tyr39, Ser75, Asn76, Ser79, Arg80, Glu110, and Gln177 may serve this purpose, Fig. 3d.
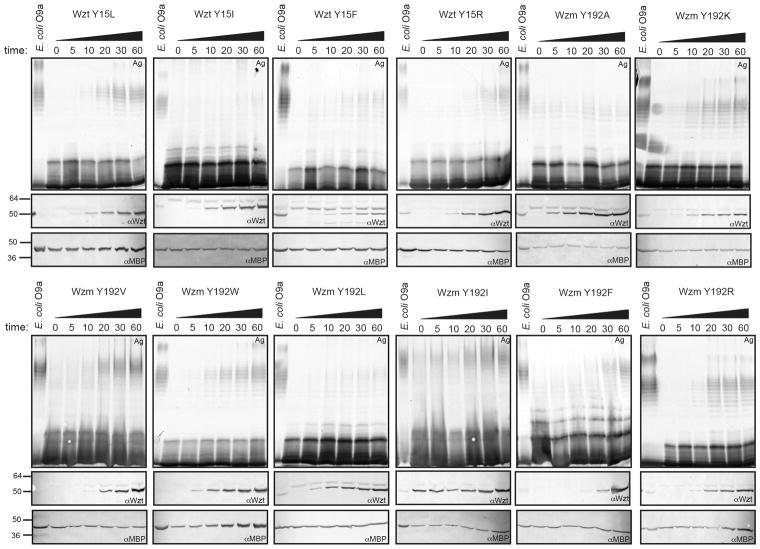
At the periplasmic channel exit, the PG1 helix is also preceded by a conserved aromatic residue, usually a tyrosine (Tyr187), similar to the gate helix on the cytosolic side, Fig. 3c. It is thus likely that PG1 forms the gate towards the periplasmic membrane leaflet. The functional importance of the gate helices was addressed by introducing point mutations into the E. coli Wzm/Wzt O9a transporter and monitoring O antigen export in vivo.8,9 Tyr14 and 187 of AaWzm/Wzt correspond to Tyr15 and 192 in E. coli Wzm/Wzt, respectively (Extended data Fig. 2). As shown in Fig. 3e and Extended Data Fig. 6, replacing Wzt-Tyr15 at the cytosolic gate with Trp or Phe supports O antigen export similar to wild type levels, while replacing it with the hydrophobic β-branched residues Val and Ile abolishes export. Among the charged residues, Lys and Arg support some export, requiring longer incubation periods (post-induction) before reaching detectable levels, while the Y15E mutant is inactive. Replacing Tyr15 with Ala only shows a kinetic effect, the exported O antigen levels reach WT levels about 30 min after initiating transport. This is possibly due to a second Tyr directly N terminal to the conserved residue, which could functionally replace Tyr15 E. coli Wzt. We were unable to express a Wzt double Tyr mutant to test this hypothesis. These results are consistent with the cytosolic gate forming the substrate-binding pocket, perhaps through CH-π stacking interactions of the tyrosine with the substrate’s first sugar moiety. On the periplasmic side, Wzm-Tyr192 is less critical for export as most of the tested substitutions supported O antigen secretion, Extended Data Fig. 6. It is likely that once initiated, translocation is completed even with a compromised periplasmic gate and/or that other aromatic residues nearby functionally replace Tyr192.
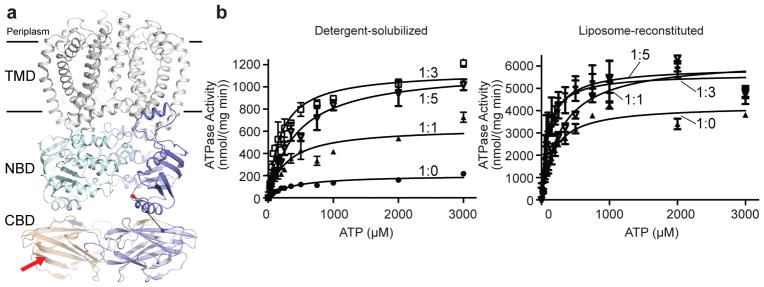
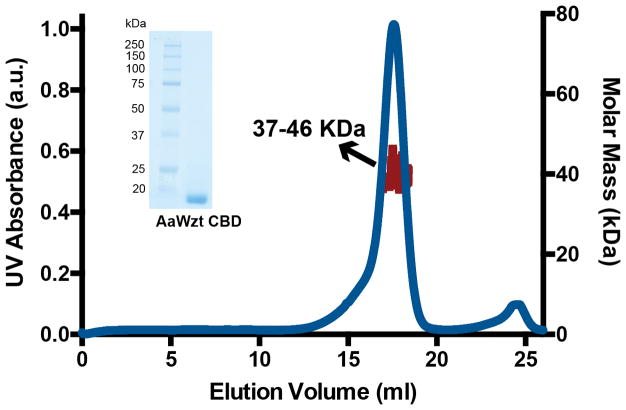
The CBD extending the transporter’s NBD (Extended Data Fig. 2c) interacts with the modified terminus of the O antigen substrate.8,9 Crystal structures of isolated CBDs from K. pneumoniae O12 and E. coli O9a reveal CBD dimers that are stabilized by intermolecular β-strand exchange9,10 and, accordingly, the AaWzt-CBD also purifies as a dimer, Extended Data Fig. 7. Binding of the O antigen cap likely occurs on the surface of the jelly-roll fold, Fig. 4a,9,10 but the CBD’s precise orientation relative to the NBD remains unknown.
Figure 4. The CBD stimulates Wzm/WztN’s hydrolytic activity.
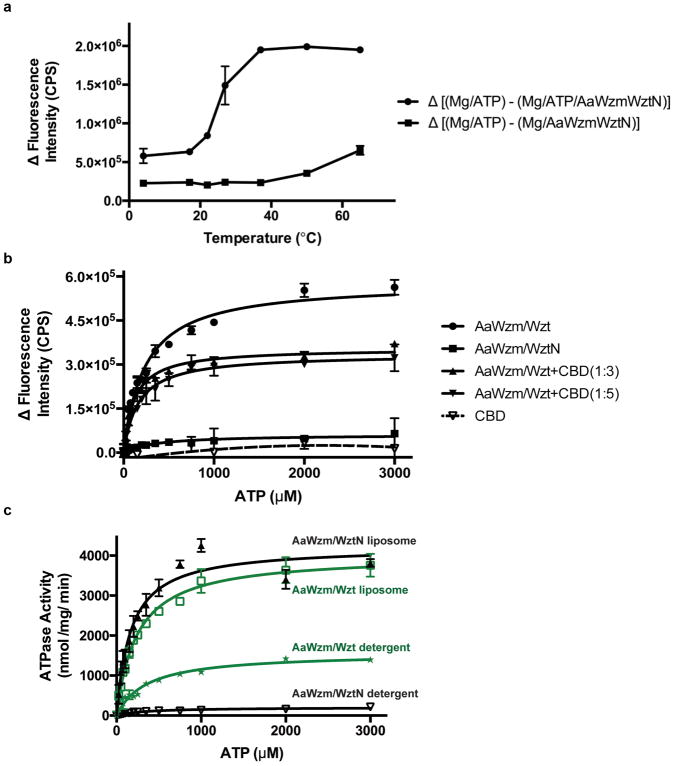
(a) Putative organization of the full-length Wzm/Wzt transporter. Alignment of the E. coli O9a Wzt CBD structure (PDB: 2R5O) with the Wzm/WztN transporter. C and N termini of WztN and CBD are shown as red and blue spheres, respectively. Red arrow: putative binding site of the modified O antigen cap. (b) Hydrolytic activity of Wzm/WztN in detergent-solubilized and lipid-reconstituted states, respectively. ATP hydrolysis was performed under increasing CBD concentrations as indicated (Wzm/WztN:CBD, molar ratio). Error bars represent the standard deviations from the means of at least three independent replicas.
In the absence of the CBD, Wzm/WztN hydrolyzes ATP in a detergent-solubilized state in a tested temperature range from 27 to 65°C, with an apparent Km for ATP of about 350 μM at 27°C, Fig. 4b and Extended Data Fig. 8. Strikingly, the full-length transporter hydrolyzes ATP about 7-times faster than the truncated version, but a similar apparent Km for ATP suggests that the CBD accelerates the rate-liming step of ATP hydrolysis, Extended Data Fig. 8c.
To investigate a direct interaction of Wzm/WztN with the CBD, we measured its hydrolytic activity in the presence of increasing CBD concentrations in detergent-solubilized and liposome-reconstituted states. Wzm/WztN’s ATPase activity increases with increasing CBD concentrations and reaches maximum rates at an approximately 3-fold molar excess of CBD over Wzm/WztN, consistent with a direct CBD-NBD interaction, Fig. 4b. Control experiments in which purified CBD was added to the full-length transporter did not increase its hydrolytic activity. Instead, we observed a slight reduction in ATPase activity, perhaps due to non-specific interactions of the isolated and NBD-attached CBDs, Extended Data Fig. 8b.
Compared to detergent-solubilized states, the transporters’ hydrolytic activities increase significantly upon reconstitution into liposomes, Extended Data Fig. 8c. Assuming similar concentrations of catalytically active transporters, the apparent catalytic rates in liposomes increase about 3 and 20-fold for full-length and truncated Wzm/Wzt, respectively, relative to detergent-solubilized transporter. These data suggest that the transporter adopts a different, perhaps closed, conformation in a lipid bilayer environment or the presence of the CBD, thereby affecting its hydrolytic activity. These properties could be modulated by the O antigen to facilitate translocation.
In the absence of a translocating substrate in vivo, the transporter’s TM channel must be closed to prevent leakage of small solutes across the membrane. Channel closure likely correlates with closing of the transporter’s NBDs, perhaps through rigid body movements of the Wzm/Wzt half transporters relative to one another. This model can be generated by superimposing the NBDs of the Wzm/WztN transporter halves with the closed structure of the isolated WztN-NBD dimer, Fig. 2c. In this model, the TM channel is closed because the Wzm subunits pack tightly against each other without any significant backbone clashes, Extended Data Fig. 9. However, significant overlaps occur at the cytosolic substrate-binding site, where the NBD’s gate helix contacts the IF-TM1 loop of the opposing Wzm subunit, Fig. 2. This region likely undergoes additional conformational changes during channel closure to facilitate substrate translocation (discussed below). The predicted rigid body movement of the transporter halves is supported by disulfide cross-linking of the Wzm subunits. Cys residues introduced into periplasmic loops predicted to be in close proximity in the closed conformation indeed form disulfide bridges under oxidizing conditions, Extended Data Fig. 9c.
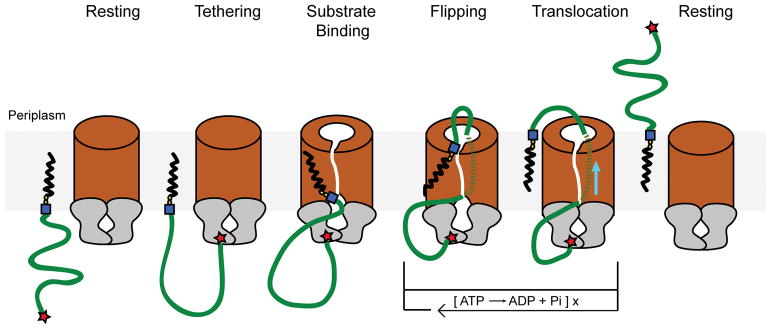
The channel-forming conformation of the ABC transporter is consistent with its biological function. However, in the absence of a polysaccharide, mechanisms must exist that prevent spontaneous transporter opening. It is possible that channel formation is tightly coupled to substrate recognition and insertion, such that the translocating polymer seals the channel, Fig. 5.
Figure 5. Model of O antigen membrane translocation.
In a resting state, the transporter’s TM channel and the NBDs are in a closed conformation. Tethering the substrate to the transporter via interactions of the CBD with the modified O antigen terminus increases its local concentration. Binding of the Und-PP lipid anchor to the cytosolic gate induces NBD and TM channel opening. The lipid head group inserts into the channel and reorients spontaneously to the periplasmic side. The now channel-inserted polysaccharide is translocated through repeated cycles of ATP binding and hydrolysis. Upon polymer release to the periplasmic side, the transporter returns to the resting conformation with a closed TM channel. Blue square: N-acetylglucosamine; yellow spheres: phosphate; red star: modified terminus.
It is unknown which end of the O antigen enters the transporter first. Our structural and functional data argue that the NBD’s gate helix functions in substrate binding, most likely by recognizing Und-PP’s pyrophosphate group, together with the first sugar unit. Accordingly, some ABC transporters for O antigens23 and teichoic acids21 that operate without the fine specificity imposed by CBDs can export glycans with different repeat unit structures. Yet, all substrates contain an acetylated amino sugar as the connector between Und-PP and the repeat-unit glycan7. Our data suggest that Wzm/Wzt specifically recognizes this motif, in contrast to PglK, which has been proposed to recognize the Und-moiety, Extended Data Fig. 4b.6
Substrate binding to the cytosolic entrance likely leads to opening of the transporter and insertion of the lipid head group into the channel through a gate between the Wzm subunits, Fig. 5. Following insertion, the lipid anchor spontaneously reorients to the periplasmic side, possibly facilitated by the proton-motive force across the IM. During this transition, the hydrophobic part of the lipid anchor likely remains in the membrane, similar to the model proposed for PglK.6 After this passive flipping, the transporter contains the polysaccharide in the channel proper. Export could be achieved in a single cycle or require several steps of ATP hydrolysis, yet these alternatives are currently impossible to distinguish.
We speculate that the cytosolic gate’s IF-TM1 loop, Fig. 1 and 2, contacts the polysaccharide during NBD closure. The gate helix, upon ATP binding, likely pushes against the IF-TM1 loop, such that it moves horizontally towards the channel, Fig. 2. The loop contains several polar residues, including Thr21, that could interact with and move the polysaccharide during this transition, similar to the translocation mechanism proposed for cellulose synthase.22 Conformational changes at the gate could mediate the translocation of ~1–2 sugar units at a time. As such, Wzm/Wzt would combine the functions of a lipid flippase and polysaccharide translocase.
ABC transporters exporting uncapped O antigens (e.g. K. pneumoniae O2a) do not contain C-terminal CBDs.23 In these systems, polymer export is dependent on simultaneous synthesis, whereas in E. coli O9a, O antigen synthesis and export can be temporally uncoupled.9,23 Both types of transporter share structural features key to our model (Extended Data Fig. 2). While uncapped O antigens may be synthesized and exported by multi-subunit complexes including the transporter, CBD-containing ABC transporters likely function independently during or after O antigen biosynthesis. In this scenario, the CBD may ensure a sufficient local substrate concentration.
Methods
Cloning and protein expression
The wzm and wzt genes were PCR amplified from genomic Aquifex aeolicus VF5 DNA and sequentially cloned into an engineered pETDuet expression vector (Novagen) with a C-terminal histidine tag on Wzt. A second construct containing only residues 1 to 235 of Wzt (WztN) was cloned in a similar manner. The transporters were expressed in E. coli C43 cells in LB medium upon induction with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at an optical density at 600 nm of 0.6. Cells were harvested by centrifugation after incubation at 37°C for 4 hrs. The cells were resuspended in RB-1 buffer containing 20 mM Tris HCl pH 7.5, 0.1 M NaCl and 5 mM β-mercaptoethanol (β-ME) and then lysed in a microfluidizer. The crude membranes were collected by centrifugation for 60 min at 200,000 g in a Beckman Ti45 rotor and solubilized for 60 min at 4 °C in RB-2 buffer containing 50 mM sodium phosphate pH 7.2, 0.1 M NaCl, 20 mM imidazole, 5 mM β-ME and 2% polyoxyethylene(8)-dodecyl ether (C12E8). The insoluble material was cleared by centrifugation for 30 min at 200,000 g in a Beckman Ti45 rotor and the membrane extract was batch incubated with Ni-NTA agarose (Qiagen) for 60 min at 4 °C. The resin was packed in a gravity flow chromatography column, washed with 50 ml WB1-buffer (RB-1 buffer containing 20 mM imidazole and 5 mM dodecyl-N,N-dimethylamine-N-oxide, LDAO), 50 ml WB2-buffer (RB-1 buffer containing 40 mM imidazole and 5 mM LDAO), 50 ml WB3-buffer (RB-1 buffer containing a total of 1.5 M NaCl, 20 mM imidazole and 5 mM LDAO) and the transporter was eluted in 50 ml EB-buffer containing 20 mM Tris HCl pH 7.5, 0.1 M NaCl, 300 mM imidazole, 5 mM β-ME and 5 mM LDAO. The eluted protein was purified over an analytical S200 gel filtration column (GE Healthcare) equilibrated in buffer containing 20 mM Tris HCl pH 7.5, 0.1 M NaCl, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) and 5 mM LDAO. The peak fraction was concentrated to 15 mg/mL final concentration in a 50-kDa cut-off centrifugal filter (Amicon) before crystallization. To guide model building, Thr128 in TM3 of Wzm was replaced with a Cys residue for derivatization with ethylmercurithiosialicylic acid. This mutant was generated by QuikChange mutagenesis and purified as described for the wild type transporter. Selenomethionine-derivatized Wzm/Wzt N was prepared as described above with the exception that the cells were grown in the M9 minimal medium supplemented with 60 μg ml−1 L-selenomethionine (Se-Met).
The Wzt CBD (residues 235–394) was expressed from a pET30a vector (Novagen) in E. coli BL21 (DE3) cells (Invitrogen). The E. coli cells were cultured in LB medium at 37 °C and protein expression was induced at an optical density at 600 nm of 0.6 with 0.5 mM IPTG. The cells were harvested by centrifugation after 4 hrs of incubation at 37 °C. Subsequently, the cells were resuspended in RB-1 buffer containing 20 mM sodium phosphate pH 7.5, 0.05 M NaCl and 5 mM β-ME and then lysed in a microfluidizer. The insoluble material was cleared by centrifugation for 30 min at 200,000 g in a Beckman Ti45 rotor. The supernatant was batch incubated with Ni-NTA agarose (Qiagen) for 60 min at 4 °C. The protein was then purified by Ni-NTA affinity chromatography at 4 °C, washed with 50 ml WB1-buffer containing 25 mM Tris HCl pH 8.5, 0.5 M NaCl, 30 mM imidazole and 5 mM β-ME, 50 ml WB2-buffer (WB-1 buffer containing a total of 50 mM imidazole and 50 mM NaCl) and eluted with EB-buffer, consisting of 25 mM Tris HCl pH 8.5, 50 mM NaCl, 300 mM imidazole and 5 mM β-ME. The protein was further purified by gel filtration chromatography (Superdex-200) in 25 mM Tris, pH 8.5, 50 mM NaCl, 5 mM β-ME.
Crystallization
The truncated Wzm/WztN transporter was crystallized by combining 1 μL of well (32% PEG400, 0.05 M sodium acetate pH 5.4, and 0.1 M magnesium acetate) with 1 μL of protein solution and sitting-drop vapor diffusion at 22 °C. Addition of 3.6 mM decyl-β-D-maltoside (DM) to the crystallization solution significantly improved diffraction. Crystallization trials with full-length Wzm/Wzt have been unsuccessful. WztN crystallized from a Wzm/WztN sample set up under different conditions, each producing a P3121 crystal form but with different unit cell dimensions. Crystals with a smaller unit cell contained a WztN monomer in the crystallographic asymmetric unit (ASU) and were obtained by sitting drop vapor diffusion in the presence of 41% PEG400, 0.05 M sodium acetate pH 5.4, 0.15 M magnesium acetate, and 47 mM octyl-glucoside at 17 °C. Crystals with a larger unit cell contained a WztN dimer per ASU and grew in the presence of 0.4 M magnesium nitrate, 17.5 % PEG8000, and 0.1 M Tris pH 8.5 by sitting-drop vapor diffusion at 17 °C.
Initial crystals were observed after approximately 3 days and reached their final size within 2 weeks for all samples. The crystals were collected and directly cryo-cooled in liquid nitrogen. WztN crystals grown in PEG8000 were cryo-protected in the presence of 25 % glycerol in the crystallization solution. Wild-type and Wzm-T128C crystals were soaked with 5–20 mM ethylmercurithiosialicylic acid for 2–20 hrs before harvesting.
Structure determination
Diffraction data were collected at the Argonne National Laboratory beam lines SER- and GM/CA-CAT as well as the AMX and FMX beam lines at the Brookhaven National Laboratories (NSLS-II). Data was integrated in XDS and reduced in Aimless, as part of the CCP4 program suite.24 The isolated WztN structure was determined after single anomalous dispersion (SAD) phasing with an ethylmercurithiosialicylic acid-derivatized crystal containing a WztN monomer per ASU in Phenix.autosol with subsequent model building.25,26 The obtained model was of sufficient quality for molecular replacement using Phaser with the native data set,27 after which the model was manually completed in Coot28 and refined in Phenix.refine25 using TLS parameters.29 In this crystal form, the protomers of a WztN dimer are related by crystallographic two-fold symmetry. The obtained model was used for molecular replacement of the crystal form containing a WztN dimer per ASU and the obtained model was completed in Coot and refined as described above. In this structure, residues 12 to 33 of chain A are disordered and not included in the final model. The final monomeric and dimeric WztN models contain 99.2/0.4/0.4% and 98.4/1.6/0% in the preferred, allowed and outlier regions of the Ramachandran plot.
The AaWzm/WztN structure was determined by SAD phasing with a ethylmercurithiosialicylic acid-derivatized wild type Wzm/WztN crystal. Phasing was performed in Phenix.autosol without automated model building and based on 5 mercury sites near Cys residues in the transporter’s NBDs. The obtained experimental phases to ~9 Å resolution were improved by non-crystallographic symmetry (NCS) averaging and solvent flattening in DM with manually built averaging and solvent masks.30,31 The improved phases allowed manual docking of the WztN NBDs as well as 3 partial TM helices. The obtained poly-alanine model was used to improve averaging and solvent masks for subsequent rounds of NCS averaging. Following placement of another 2 partial TM helices, the initial model was of sufficient quality for molecular replacement with the native and highest resolution data set. Subsequently, NCS and cross-crystal averaging in DMMulti31 was used to improve and gradually extend the phases to 3.85 Å resolution using a solvent mask that covered the entire transporter and an averaging mask covering a Wzm/WztN half-transporter. The greatly improved density maps were used to place additional regions as poly-alanine traces, refine the averaging and solvent masks, and iterative rounds of NCS and cross-crystal averaging until the backbone of the entire transporter could be traced and bulky residues were discernible. We confirmed the annotation of TM helices and the assigned registers based on seleno-methionines at positions 147 and 156 in TM helix 4 and 205 in PG2, as well as a mercury-derivatized Cys-residue introduced at position 128 at the C terminal end of TM helix 3. The initial model was built as a poly-alanine model and refined in Phenix.refine with NCS constraints. To generate a model containing all amino acid side chains, the high-resolution structure of the NBD was docked into the Wzm/WztN density and manually refined. The Wzm subunit was manually built starting with aromatic amino acids and methionines, and cycles of NCS refinement in Phenix.refine. Additionally, refinement in Refmac5 with jelly-body refinement greatly improved the model.32 Towards the end of the refinement, NCS restraints were used instead of constraints. The final Wzm/WztN model contains residues 2 to 235 of Wzt plus a KLHH sequence corresponding to an engineered HindIII restriction site and C terminal His-tag. Wzm contains residues 2 to 255 (of 256 in total) with a short gap between residues 51 and 55 in chain C and 49 to 55 in chain D. The final model contains 93.5, 6.3, and 0.2% of the residues in favored, allowed and disallowed regions of the Ramachandran plot, respectively. The coordinates and structure factors have been deposited at the Protein-Data-Bank. All figures were generated using Pymol33 and channel dimensions were analyzed using HOLLOW.34 Primary sequence alignments were generated in CLUSTALW Omega and displayed in Jalview35.
ATPase activity measurements
The ATPase activity was measured using an NADH-consuming coupled method as described.36 In a first step, the transporter was incubated in reaction buffer containing 10 mM MgCl2 and the indicated amounts of ATP at 27°C (or otherwise indicated temperatures) for 10 min, after which the samples were snap frozen in liquid nitrogen until all data points had been collected. In a second step, the thawed samples were incubated with NADH-buffer containing 50 mM HEPES pH7.5, 8 u pyruvate kinase, 8 u lactate dehydrogenase, 4 mM phosphoenolpyruvate, 1 mM MgCl2 and 0.05 mM NADH at 22°C for 5 minutes in the dark. At 22°C, Aquifex Wzm/Wzt does not exhibit any detectable ATPase activity. ADP formation was quantified by following the decrease in NADH fluorescence at 450 nm (excitation at 340 nm) using a FluoroMax 3(Horiba) fluorimeter at 22°C. For ATPase assays in proteoliposomes, purified Wzm/Wzt was reconstituted into 3 mg/mL E. coli total lipid extract at a protein concentration of 0.3 mg/mL. Vesicles were formed upon detergent removal using SM-2 Bio-beads (Bio-rad). For CBD titration experiments, the Wzt-CBD was pre-mixed with transporter for 3 hours at 4°C before adding ATP and MgCl2. Decrease in NADH fluorescence was converted to molar concentrations based on measurements of known standards. All experiments were repeated at least three times and data were fit to Michaelis–Menten kinetics to calculate Km and Vmax values using GraphPad Prism 6. Error bars represent deviations from the means.
Disulfide cross-linking of Wzm/WztN
For disulfide cross-linking experiments, Wzm and WztN were co-expressed from pETDuet and pACYC vectors, respectively, with an N-terminal FLAG tag on Wzm and C-terminal His-tag on WztN. Protein purification was as described above with the exception that the gel filtration buffer contained 1 mM DTT instead of TCEP. Disulfide cross-linking experiments confirming the modeled closed conformation of Wzm/WztN were performed with purified detergent-solubilized transporter. Oxidation was induced with copper-phenanthroline (Co-Phen) or sodium tetrathionate (STT). A Co-Phen stock solution was prepared by combining 0.36 M 1,10-phenanthroline monohydrate (VWR) (in 50% ethanol) with 0.24 M copper sulfate (Sigma) at a 2:1 volume ratio. STT was dissolved in ddH2O at 80 mM concentration and N-ethylmaleimide (NEM) to block free cysteines was prepared at 1 M concentration in DMSO. Wild type or Cys-introduced Wzm/WztN at 0.09 mM concentration was incubated with 4 mM Co-Phen or STT and incubated for 40 min at room temperature followed by addition of 25 mM NEM and incubation at 4°C for 30 min. Samples oxidized in the presence of ADP/Mg2+ were pre-incubated with 2 mM ADP and 2 mM MgCl2 for 20 min at room temperature. The oxidized protein was resolved by non-reducing SDS-PAGE and protein bands were visualized by Western blotting against an N-terminal FLAG-tag on Wzm.
Size-exclusion multi angle light scattering
Mass measurements on Wzt-CBD were performed on a Dionex UltiMate3000 HPLC system with a UV detection module (ThermoFisher, Waltham, MA), connected to a miniDAWN TREOS static light scattering detector (Wyatt Technology, Santa Barbara, CA) and Optilab T-rEX differential refractometer (Wyatt Technology). A 100 μL sample at 0.1 mM concentration was loaded onto a Superdex 200 HR 10/300 GL column (GE Healthcare) equilibrated in 25 mM Tris, pH 8.5, 50 mM NaCl, 5 mM β-ME at a flow rate of 0.4 mL/min. Data were recorded and processed using ASTRA software (Wyatt Technology).
E. coli O9 antigen modeling
An E. coli O9a antigen containing 10 mannose units was modeled using the GLYCAM carbohydrate builder and manually placed into the AaWzm/WztN transmembrane channel. (http://glycam.org/tools/molecular-dynamics/oligosaccharide-builder/build-glycan?id=1).
In vivo O antigen export assays
Growth conditions
Bacterial cultures (Supplementary Table) were grown with aeration in Luria Broth Base (Invitrogen) at 37 °C. Broth was supplemented with 100 mg/mL ampicillin, 0.4% D-mannose and/or 0.1% L-arabinose where appropriate. Unless otherwise stated, cells were grown in the presence of 0.4% D-glucose to repress mannose uptake.
DNA methods
Oligonucleotide primers were custom designed and obtained from Sigma Aldrich (Supplementary Table). PfuUltra DNA polymerase (Agilent) was used for PCR reactions, according to manufacturers instructions, and PCR product was treated with DpnI (New England Biolabs). DNA sequencing was performed by the Genomics Facility at the University of Guelph Advanced Analysis Center.
Complementation
E. coli CWG638 transformants containing plasmids containing WT or variant wzt, along with wzm in the native chromosomal organization were used to ensure equal protein expression levels. Cultures were grown overnight in the presence of 0.4% D-glucose. E. coli CWG638 cannot produce its own GDP-mannose, the substrate of glycosyltransferases responsible for O9a O-antigen assembly, due to a deletion of manA and, therefore, O-antigen production relies upon mannose uptake. Accumulation of Und-PP-O9a intermediates in the absence of export results in growth defects that are alleviated by second-site mutations which repress O-antigen synthesis. Growth in glucose represses uptake of any trace amounts of mannose in the medium and prevents harmful O9a synthesis. Overnight cultures were diluted 1/10 in fresh LB supplemented with 0.4% glucose and grown for 4 hours to an A600nm of ~1.0. Cells were subjected to centrifugation at 5,000 g for 10 min, resuspended in fresh LB containing 0.4% D-mannose to induce O-antigen biosynthesis and grown for 15 min at 37 °C with aeration. After 15 min, 0.1% L-arabinose was added to induce protein expression. Aliquots of culture were taken immediately and after 5, 10, 20, 30 and 60 min and put immediately on ice to suppress further cell growth and O-antigen export. An equivalent of 1 OD cells were harvested by centrifugation at 13,000 g and resuspended in 100 μL of SDS-PAGE loading buffer. Cells were lysed by boiling for 10 minutes. For Western immunoblots, samples were subjected to SDS-PAGE using 12% acrylamide resolving gels in Tris-glycine buffer.37,38 For LPS analysis, samples were first treated with 500 μg/mL proteinase K for 1 hour at 55 °C prior to SDS-PAGE. LPS was visualized by silver staining.39
For immunoblot analyses, material resolved by SDS-PAGE was transferred to nitrocellulose membranes (Protran; PerkinElmer Life Sciences). Wzt was detected using anti-Wzt primary antisera, generated in rabbits and cross-reactive material adsorbed against E. coli CWG708 whole cell lysate. Goat-anti-rabbit secondary antibody conjugated to alkaline phosphatase (Cedrar Lane Laboratoreis) was used to facilitate detection with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indoyl phosphate (BCIP) (Roche Applied Science). Maltose-binding protein was detected using monoclonal anti-MBP mouse primary antisera (New England Biolabs) with secondary alkaline phosphatase-conjugated goat-anti-mouse (Jackson Immuno-Research Laboratories, Inc.) for detection with BCIP and NBT.
Extended Data
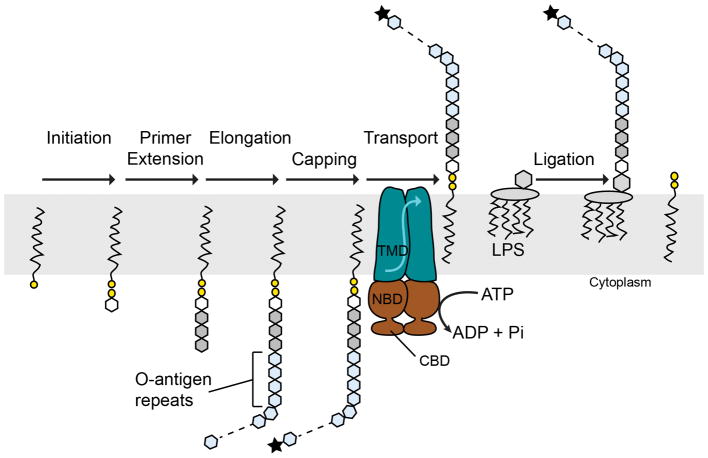
Extended Data Figure 1. ABC transporter-dependent O-antigen biosynthesis.
In this pathway, O antigens are completely synthesized on the cytosolic leaflet of the plasma membrane. Undecaprenyl-phosphate (black line and yellow circle) serves as the lipid acceptor and is modified by the addition of an acetylated amino sugar phosphate (frequently N-acetylglucosamine-1-P, white hexagon) as well as 2 or more additional sugar residues (gray hexagons) to generate a biosynthesis primer. The polymerizing enzyme(s) extend the primer with tens to hundreds of O-antigen repeat units (light blue hexagons). In some species, termination of O antigen biosynthesis is achieved by modifying the polymer’s non-reducing end (black star). An ABC transporter translocates the Und-PP-linked O antigen intermediate to the membrane’s periplasmic side, where it forms a substrate for glycosylation of the lipopolysaccharide (LPS) core. Only transporters translocating terminally-modified O-antigens contain carbohydrate-binding domains (CBD) that bind the polysaccharide’s modified terminus. TMD/NBD: Transmembrane and nucleotide binding domains.
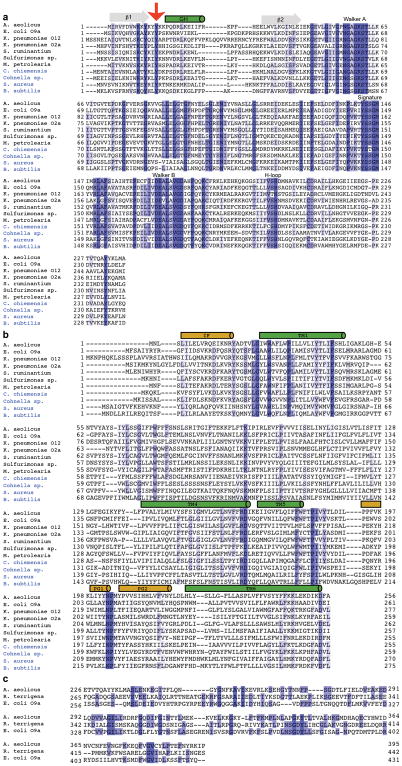
Extended Data Figure 2. Sequence alignment of O antigen and wall teichoic acid transporters.
Alignments of the nucleotide binding and transmembrane domains are shown in panels (a) and (b), respectively. The conserved tyrosine preceding the NBD’s cytosolic gate helix and in the periplasmic gate are highlighted with a red arrow and red box in panels a and b, respectively. Transmembrane helices and cytosolic and periplasmic gate helices are shown as green and beige cylinders, respectively. Blue sequence labels indicate predicted teichoic acid transporters. All O antigen transporter NBDs except for K. pneumoniae O2a contain predicted CBDs at their C termini, which are not shown. (c) Alignment of the C-terminal region of AaWzt with the corresponding domains from the E. coli O9a (PDB: 2R5O) and R. terrigena/K. pneumoniae O12 (PDB: 5HNO) transporters. Sequences were aligned in CLUSTAL Omega and displayed in Jalview colored by sequence identity.
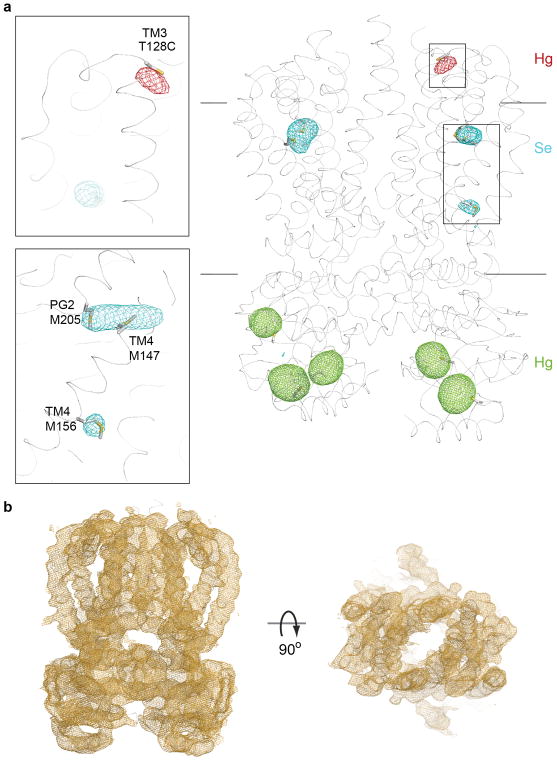
Extended Data Figure 3. Anomalous difference and experimental electron density maps.
(a) Heavy atom positions used for experimental phasing and model building. Five native cysteines in the NBDs as well as an engineered Cys at the C terminus of TM3 (T128C) were modified with ethylmercurithiosialicylic acid, shown as green and red meshes and contoured at 4.5 and 3σ, respectively. Only the Hg sites shown in green were used for Hg-SAD phasing. The TMD contains three native Met residues, which were identified upon substitution with seleno-methionine (cyan mesh, contoured at 3σ). Shown are SigmaA-weighted anomalous difference electron densities, AaWzm/WztN is shown as a gray ribbon. (b) Unbiased experimental SigmaA-weighted electron density after NCS and cross-crystal averaging and phase extension to 3.85Å, contoured at 1σ.
Extended Data Figure 4. Comparison of Type-II ABC exporter folds.
(a) The structures of the transmembrane domains of A. aeolicus Wzm, H. sapiens ABCG5 and H. sapiens ABCA1 are shown as cylindrical cartoons. One subunit of the dimers is colored in rainbow colors from blue to red, N to C terminus. (b) Structure of PglK, an ABC transporter translocating Und-PP-linked oligosaccharides across the plasma membrane. PglK likely recognizes the substrate’s polyprenyl moiety via a conserved periplasmic helix (shown in magenta), which is missing in Wzm.
Extended Data Figure 5. Closed conformation of the isolated WztN NBD.
(a) The isolated WztN dimer structure was aligned by secondary matching in Coot with the NBDs of the AMPPNP-stabilized maltose transporter, PDB entry 3RLF. The WztN dimer is shown in cyan and light blue and the maltose transporter’s NBDs are shown in light and dark gray. Right panel: The Walker A (S61) and Signature (S143) motifs in the closed WztN dimer structure are separated by approx. 4 Å. (b) Comparison of WztN dimer structures. The structure shown in dark blue was obtained from a crystal form containing a WztN dimer in the crystallographic asymmetric unit. The structure shown in gray was obtained from a crystal form with a monomeric WztN per crystallographic asymmetric unit related to the other protomer by two-fold crystallographic symmetry. The Signature motifs are colored cyan and yellow and the Walker-A motifs are colored magenta and red for the crystallographic monomeric and dimeric WztN structures, respectively.
Extended Data Figure 6. Impact of conserved tyrosine residues of the cytosolic and periplasmic gates on O antigen translocation.
The indicated point mutations were introduced into the E. coli O9a Wzt/Wzm transporter and O antigen transport was assayed by silver staining of the whole-cell lysate. Ag: Silver stained SDS-PAGE. Wzt and MBP were detected immunologically to monitor transporter expression and as a loading control, respectively. All experiments were repeated independently at least three times with similar results. Time: Period after inducing Wzt/Wzm expression in minutes.
Extended Data Figure 7. Dimerization of the isolated Wzt carbohydrate-binding domain.
Multi-angle static light scattering coupled to size-exclusion chromatography was used to determine the molecular weight of the purified Wzt carbohydrate-binding domain (one representative experiment is shown). The molecular weight of a monomeric Wzt-CBD is 20 kDa, including a C-terminal 6x-His-tag and linker region. Inset: Coomassie-stained SDS-PAGE of purified Wzt-CBD.
Extended Data Figure 8. Hydrolytic activity of the Wzm/Wzt ABC transporter.
ATP hydrolytic activity was measured by following the decrease of NADH fluorescence in an enzyme-coupled assay upon excitation at 340 nm and emission at 450 nm in a temperature range from 4 to 65°C. (a) Temperature dependence of Wzm/WztN’s ATPase activity. Shown is the difference in NADH fluorescence between control reactions in the absence of Wzm/WztN and reactions in its presence. (b) Hydrolytic activity of full-length Wzm/Wzt in the presence of isolated Wzt-CBD measured at 27°C. Shown are fluorescence intensity differences (calculated as for Fig. 4b) but not converted to apparent catalytic rates. Dashed line: ATP titration in the presence of only the Wzt-CBD. Hydrolytic activity of Wzm/WztN in the absence of Wzt-CBD is shown for comparison. (c) Comparison of ATPase activities of full-length (green) and truncated (black) Wzm/Wzt. Shown are apparent catalytic rates in detergent-solubilized and liposome-reconstitute states. Data points represent the mean of a three independent repeats with standard deviations. CPS: Counts per second.
Extended Data Figure 9. Model of the Wzm/WztN closed conformation.
(a) Rigid body alignment of the Wzm/WztN transporter halves with the corresponding NBDs of the closed WztN dimer structure. The closed WztN dimer is shown in gray and Wzm/WztN is colored in red and green for Wzm and cyan and blue for WztN. Residues replaced with Cys are shown as spheres for their C-alpha carbons and labeled. Observed disulfide cross-links are indicated with a dashed line. (b) Cartoon illustration of the transporter’s open to closed transition. (c) Disulfide cross-linking of Wzm protomers. Purified Wzm/WztN transporters harboring the indicated Cys mutations were oxidized either with copper-phenanthroline (Co-Phen) or sodium tetrathionate (STT), blocked with N-ethylmaleimide (NEM), and analyzed by Western blotting against the N-terminal Wzm FLAG-tag. Experiments were repeated three times with similar results. M and D: Wzm monomer and dimer.
Extended Data Table.
Crystallographic data collection and refinement statistics.
| WzmWztN | WzmWztN (Hg) | WzmWztN-T128C(Hg) | WzmWztN (Se-Met) | WztNBD (monomer) | WztNBD (Hg) | WztNBD (dimer) | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P4132 | P4132 | P4132 | P4132 | P3121 | P3121 | P3121 |
| Wavelength (Å) | 0.9895 | 1.0052 | 1.0052 | 0.9895 | 0.9895 | 1.0052 | 0.9895 |
| Cell dimensions | |||||||
| a, b, c (Å) | 228.1, 228.1, 228.1 | 233.5, 233.5, 233.5 | 232.0, 232.0, 232.0 | 230.8, 230.8, 230.8 | 96.2, 96.2, 60.9 | 97.0, 97.0, 60.9 | 97.8, 97.8, 104.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 24.9–3.85 (4.22–3.85)* | 30.5–8.50 (9.51–8.5) | 29.46–7.09 (7.93–7.09) | 24.89–5.21 (5.82–5.21) | 31.5–2.05 (2.11–2.05)* | 42.0–3.69 (4.04–3.69) | 39.25–3.51 (3.84–3.51) |
| Rmerge | 0.23 (2.19) | 0.12 (0.18) | 0.12 (1.16) | 0.19 (1.38) | 0.08 (1.22) | 0.09 (0.15) | 0.16 (0.72) |
| Rpim | 0.08 (0.73) | 0.01 (0.02) | 0.03 (0.26) | 0.04 (0.31) | 0.04 (0.61) | 0.02 (0.03) | 0.08 (0.35) |
| I/σI | 7.6 (1.3) | 45.0 (33.5) | 20.4 (3.0) | 12.8 (3.0) | 9.5 (1.5) | 30.3 (20.8) | 8.0 (2.8) |
| Completeness (%) | 99.6 (100.0) | 97.8 (100.0) | 97.5 (96.1) | 99.1 (100.0) | 98.3 (99.3) | 99.9 (100.0) | 99.9 (99.9) |
| Redundancy | 9.6 (9.9) | 76.7 (81.6) | 20.3 (19.5) | 19.0 (20.1) | 4.8 (4.7) | 20.9 (20.9) | 5.3 (5.4) |
| Refinement | |||||||
| Resolution (Å) | 24.9–3.85 | 27.8–2.05 | 27.1–3.5 | ||||
| No. reflections | |||||||
| Total | 36222 | 37942 | 13640 | ||||
| Rfree | 1824 | 1766 | 722 | ||||
| Rwork/Rfree (%) | 25.8/32.1 | 20.2/23.4 | 24.5/30.5 | ||||
| No. atoms | |||||||
| Protein | 7962 | 1919 | 3579 | ||||
| PEG-400 | 204 | ||||||
| B-factors (Å2) | |||||||
| Chain A | 172.9 | 67.3 | 107.8 | ||||
| Chain B | 184.5 | 128.2 | |||||
| Chain C | 130.3 | ||||||
| Chain D | 145.8 | ||||||
| PEG-400 | 68.4 | ||||||
| R.m.s deviations | |||||||
| Bond lengths (Å) | 0.005 | 0.008 | 0.005 | ||||
| Bond angles (°) | 0.856 | 0.859 | 0.813 | ||||
Values in parentheses refer to the highest-resolution shell.
^ Correlation between intensities from random half-data sets.42
Supplementary Material
Acknowledgments
We thank Jacob Morgan and Gabriel Lipkowitz for cloning the full-length Wzm/Wzt transporter, Chris Caffalette for critical comments on the manuscript, Owen Pornillos for access to SEC-MALS, and the staffs at the APS SER- and GM/CA-CAT as well as NSLS-II AMX and FMX beam lines. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Northeastern Collaborative Access Team beam lines are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). Data for this research was also in part collected at the Southeast Regional (SER) Collaborative Access Team beam line at the Advanced Photon Source, Argonne National Laboratory. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This research also used resources at the AMX and FMX beamlines of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. Y.B. and J.Z. were supported by NIH grant 5R01GM110143. E.M. is a recipient of an Alexander Graham Bell Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council. C.W. was supported by funding from the Canadian Institutes of Health Research. C.W. is a recipient of a Canada Research Chair.
Footnotes
Author contribution
Y.B. and J.Z. designed all structural and in vitro biochemical experiments and E.M and C.W. designed the in vivo functional assays. Y.B. and E.M. performed all structural and in vivo functional experiments, respectively. Y.B. and J.Z. wrote and all authors edited the manuscript.
Competing financial interests
The authors declare no competing financial interests
Data Availability
Atomic coordinates for the atomic models have been deposited in the Protein Data Bank (PDB) under accession numbers 6AN7 for Wzm/WztN, and 6AMX and 6AN5 for WztN.
References
- 1.Whitfield C, Szymanski CM, Aebi M. In: Essentials of Glycobiology. Varki A, editor. Chapter 21. 2017. pp. 265–282. [Google Scholar]
- 2.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caboni M, et al. An O antigen capsule modulates bacterial pathogenesis in Shigella sonnei. PLoS Pathog. 2015;11:e1004749. doi: 10.1371/journal.ppat.1004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goebel EM, Wolfe DN, Elder K, Stibitz S, Harvill ET. O antigen protects Bordetella parapertussis from complement. Infect Immun. 2008;76:1774–1780. doi: 10.1128/IAI.01629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skurnik M, Bengoechea JA. The biosynthesis and biological role of lipopolysaccharide O-antigens of pathogenic Yersiniae. Carbohydr Res. 2003;338:2521–2529. doi: 10.1016/s0008-6215(03)00305-7. [DOI] [PubMed] [Google Scholar]
- 6.Perez C, et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 2015;524:433–438. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]
- 7.Liston SD, Mann E, Whitfield C. Glycolipid substrates for ABC transporters required for the assembly of bacterial cell-envelope and cell-surface glycoconjugates. Biochim Biophys Acta. 2017;1862:1394–1403. doi: 10.1016/j.bbalip.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Cuthbertson L, Powers J, Whitfield C. The C-terminal domain of the nucleotide-binding domain protein Wzt determines substrate specificity in the ATP-binding cassette transporter for the lipopolysaccharide O-antigens in Escherichia coli serotypes O8 and O9a. J Biol Chem. 2005;280:30310–30319. doi: 10.1074/jbc.M504371200. [DOI] [PubMed] [Google Scholar]
- 9.Cuthbertson L, Kimber MS, Whitfield C. Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc Natl Acad Sci U S A. 2007;104:19529–19534. doi: 10.1073/pnas.0705709104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann E, Mallette E, Clarke BR, Kimber MS, Whitfield C. The Klebsiella pneumoniae O12 ATP-binding Cassette (ABC) Transporter Recognizes the Terminal Residue of Its O-antigen Polysaccharide Substrate. J Biol Chem. 2016;291:9748–9761. doi: 10.1074/jbc.M116.719344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Es D, Hogendorf WF, Overkleeft HS, van der Marel GA, Codee JD. Teichoic acids: synthesis and applications. Chem Soc Rev. 2017;46:1464–1482. doi: 10.1039/c6cs00270f. [DOI] [PubMed] [Google Scholar]
- 12.Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 13.Lee JY, et al. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature. 2016;533:561–564. doi: 10.1038/nature17666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian H, et al. Structure of the Human Lipid Exporter ABCA1. Cell. 2017;169:1228–1239. e1210. doi: 10.1016/j.cell.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Hug I, Feldman MF. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 2011;21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- 16.Oldham ML, Chen J. Snapshots of the maltose transporter during ATP hydrolysis. Proc Natl Acad Sci U S A. 2011;108:15152–15156. doi: 10.1073/pnas.1108858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spiwok V. CH/pi Interactions in Carbohydrate Recognition. Molecules. 2017;22 doi: 10.3390/molecules22071038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan J, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Divne C, Ståhlberg J, Teeri TT, Jones TA. High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei. J Mol Biol. 1998;275:309–325. doi: 10.1006/jmbi.1997.1437. [DOI] [PubMed] [Google Scholar]
- 20.Meyer JE, Schulz GE. Energy profile of maltooligosaccharide permeation through maltoporin as derived from the structure and from a statistical analysis of saccharide-protein interactions. Protein science: a publication of the Protein Society. 1997;6:1084–1091. doi: 10.1002/pro.5560060515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirner K, Stone LK, Walker S. ABC transporters required for export of wall teichoic acids do not discriminate between different main chain polymers. ACS Chem Biol. 2011;6:407–412. doi: 10.1021/cb100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan JL, et al. Observing cellulose biosynthesis and membrane translocation in crystallo. Nature. 2016;531:329–334. doi: 10.1038/nature16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kos V, Cuthbertson L, Whitfield C. The Klebsiella pneumoniae O2a antigen defines a second mechanism for O antigen ATP-binding cassette transporters. J Biol Chem. 2009;284:2947–2956. doi: 10.1074/jbc.M807213200. [DOI] [PubMed] [Google Scholar]
- 24.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Adams P, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zwart P, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 27.McCoy A, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Painter J, Merritt E. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 30.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Progr Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 31.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012;68:404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.PyMol. DeLano Scientific. Sancarlos CA: [Google Scholar]
- 34.Ho B, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin DY, Huang S, Chen J. Crystal structures of a polypeptide processing and secretion transporter. Nature. 2015;523:425–430. doi: 10.1038/nature14623. [DOI] [PubMed] [Google Scholar]
- 37.Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 40.Clarke BR, Cuthbertson L, Whitfield C. Nonreducing terminal modifications determine the chain length of polymannose O antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. J Biol Chem. 2004;279:35709–35718. doi: 10.1074/jbc.M404738200. [DOI] [PubMed] [Google Scholar]
- 41.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karplus PA, Diederichs K. Linking Crystallographic Model and Data Quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.