Abstract
Infections with Pseudomonas aeruginosa have become a real concern in hospital-acquired infections, especially in critically ill and immunocompromised patients. The major problem leading to high mortality lies in the appearance of drug-resistant strains. Therefore, a vast number of approaches to develop novel anti-infectives is currently pursued. Diverse strategies range from killing (new antibiotics) to disarming (antivirulence) the pathogen. In this review, selected aspects of P. aeruginosa antimicrobial resistance and infection management will be addressed. Many studies have been performed to evaluate the risk factors for resistance and the potential consequences on mortality and attributable mortality. The review also looks at the mechanisms associated with resistance – P. aeruginosa is a pathogen presenting a large genome, and it can develop a large number of factors associated with antibiotic resistance involving almost all classes of antibiotics. Clinical approaches to patients with bacteremia, ventilator-associated pneumonia, urinary tract infections and skin soft tissue infections are discussed. Antibiotic combinations are reviewed as well as an analysis of pharmacokinetic and pharmacodynamic parameters to optimize P. aeruginosa treatment. Limitations of current therapies, the potential for alternative drugs and new therapeutic options are also discussed.
Keywords: bloodstream infection, ceftazidime-avibactam, ceftolozane-tazobactam, multidrug resistance, new antibiotics, Pseudomonas aeruginosa, ventilator associated pneumonia
Introduction
One of the most important challenges for physicians is the adequate treatment of infections due to Gram-negative pathogens because of the increasing antimicrobial resistance in the healthcare setting [1].
Among infections caused by Gram-negative rods, Pseudomonas aeruginosa has a leading role [2], especially in critically ill and immunocompromised patients. Antimicrobial resistance has led to a serious restriction in treatment options for P. aeruginosa infections, which has become a critical and deadly issue causing a total of 51,000 healthcare infections in the USA per year [3–6]. Despite this problem, physicians mainly rely on retrospective non-randomized controlled studies to derive conclusions about the optimal therapeutic management of these infections.
In this review, we aim to address selected aspects of P. aeruginosa antimicrobial resistance and infection management. In the first part of this review, we will focus on resistance risks factors. Many studies have been performed to evaluate the risk factors for resistance and the potential consequences on mortality and attributable mortality. We will then explore the mechanisms associated to resistance. P. aeruginosa is a pathogen presenting a large genome that can develop a large number of factors associated with antibiotic resistance involving almost all classes of antibiotics. We will then specifically focus on clinical approaches to patients with bacteremia, ventilator-associated pneumonia, urinary tract infections, and skin and soft-tissue infections. Specific syndromes such as ecthyma gangrenosum will be discussed. In the second part of our work, we will look at pharmacokinetic and pharmacodynamic parameters that may be exploited to optimize P. aeruginosa treatment. Limitations of current therapy, potential alternative drugs and new therapeutic options will also be discussed.
Risk factors for antimicrobial resistance in P. aeruginosa
Multi-drug resistance (MDR) has increased dramatically in recent years and is now recognized as a major threat worldwide [7]. Risk factors for the development of MDR strains have been evaluated in several studies. A case–control study performed in Brazil compared 142 patients infected with metallo-β-lactamases (MBLs) strains to 26 patients infected with non MBLs strains [8]. The multivariate analysis showed that ICU stay and urinary tract infection were the important factors for MBLs infections. MBLs strains were also associated with a faster onset of infection and a faster progression to death.
A retrospective study conducted over two years, starting in 2010, in Brazil evaluated 54 ICU patients with P. aeruginosa infections [9]. MDR P. aeruginosa was observed in 37% of the cases (20 of 54 patients), 20% of the isolates were positive for the blaSPM-1-like gene. Interestingly, MDR occurred in patients hospitalized for an average of 87.1 days. A case–control surveillance study performed in China showed that the prevalence of MDR P. aeruginosa was 54% among patients with P. aeruginosa infection. Independent risk factors were tracheal intubation (odds ratio [OR] 2.21) and use of carbapenems (OR 3.36). MDR strains were associated with a longer hospitalization and a higher mortality (49 versus 20%) [10]. A retrospective study on 63 episodes of carbapenems-resistant P. aeruginosa (CRPA) showed that the Acute Physiology and Chronic Health Evaluation II (APACHE II) score at the time of CRPA bacteremia and the capacity of CRPA to form biofilm were independent predictive factors for mortality in patients with CRPA bacteremia [11]. Another study also found the APACHE II score as an independent factor for colonization [12].
In a separate population of immunocompromised patients, in a matched case–control study, 31 cases colonized with extensively drug-resistant P. aeruginosa were compared with 93 controls. Four factors were associated with colonization: presence of a central venous catheter, presence of a urinary catheter, CRP>10 mg/L, and ciprofloxacin administration [13]. Another study, this time in a retrospective international cohort of P. aeruginosa nosocomial pneumonia, tried to determine the risk factors for MDR and attributable mortality [14]. From 740 patients, 226 were infected with MDR strains. Independent factors predictors of MDR were decreasing age, diabetes mellitus, and ICU admission. MDR was independently associated with in-hospital mortality (44.7 versus 31.7% for non-MDR, p=0.001). A prospective observational study compared imipenem-resistant (IR) P. aeruginosa (PA) with or without MBL-mediated resistance [15]. The researchers found that the most important predictor of prognosis was imipenem resistance itself and not MBL production – the higher mortality observed in the IR-MBL-PA group was mediated by the underlying diseases, Charlson’s index, and other factors (e.g. virulence). Another retrospective study evaluated the impact of resistance on morbidity, mortality and length of stay with 324 cases and 676 controls [16]. The authors found that mortality from all causes and 30–day mortality after infection were higher in patients with a resistant pathogen. Pseudomonas was observed in 15.1% of the cases and 19.7% of the controls (second place Gram negative after E. coli). A systematic review and meta-analysis of the association between resistance and mortality was performed in neutropenic patients [17]. A total of 30 studies were included; infections related to carbapenems-resistant Pseudomonas spp. were reported in 18 (60%) studies. Globally, mortality ranged from 33 to 71% in patients with carbapenems-resistant Pseudomonas infections. The results showed an increased mortality in carbapenems-resistant compared to carbapenems-susceptible infections (OR 4.89). This increase in mortality has been described in a previous meta-analysis [18]. Besides mortality, resistance is also associated with increased cost, using the data from 571 admissions with bacteremia, MDR P. aeruginosa bacteremia had the highest mean incremental cost (€ 44,709) [19].
Globally, these results show that development of resistance has several risk factors linked to the severity of the infection (Apache II, underlying diseases, intubation, catheter) and that resistance itself is associated with increased mortality.
Mechanisms of antibiotic resistance
Bacteria exhibit multiple resistance mechanisms to antibiotics including decreased permeability, expression of efflux systems, production of antibiotic inactivating enzymes and target modifications. P. aeruginosa exhibits most of these known resistance mechanisms through both intrinsic chromosomally encoded or genetically imported resistance determinants affecting the major classes of antibiotics such as β-lactams, aminoglycosides, quinolones and polymyxins (Table 1). Eight categories of antibiotics are mainly used to treat P. aeruginosa infections including aminoglycosides (gentamicin, tobramycin, amikacin, netilmicin), carbapenems (imipenem, meropenem), cephalosporins (ceftazidime, cefepime), fluoroquinolones (ciprofloxacin, levofloxacin), penicillin with β-lactamase inhibitors (BLI) (ticarcillin and piperacillin in combination with clavulanic acid or tazobactam), monobactams (aztreonam), fosfomycin and polymyxins (colistin, polymyxin B). The strains of P. aeruginosa are categorized as follows: (1) MDR when resistance is observed in ≥1 agent in ≥3 categories; (2) extensively drug-resistant (XDR) when a resistance is observed in ≥ agent in all but ≤ categories; and (3) pandrug-resistant (PDR) when the strain is non-susceptible to all antimicrobial agents [2]. The emergence of MDR, XDR and PDR strains occurs in a timely fashion by the modification of regulatory mechanisms controlling the expression of resistance determinants, by mutations, alteration of membrane permeability, and horizontal acquisition of antibiotic-inactivating enzymes or enzymes inducing target modifications. Noteworthy, is the multi-resistance of many strains conferred by simultaneous production of these mechanisms [3].
Table 1.
Chromosomally encoded or imported resistance mechanisms of P. aeruginosa.
| Location | Resistance mechanisms | Targeted antibiotics | Type of resistance |
|---|---|---|---|
| Intrinsic (chromosomal) | AmpC–type cephalosporinase | β-lactams | Antibiotic inactivation |
| Class D oxacillinase OXA-50 | β-lactams | Antibiotic inactivation | |
| Aminoglycosides inactivating enzymes | Aminoglycosides | Antibiotic inactivation | |
| Efflux systems (overexpression) | Multiple antibiotic classes | Efflux systems | |
| Decreased membrane permeability | Multiple antibiotic classes | Membrane impermeability and purines | |
| DNA gyrase and topoisomerase IV | Fluoroquinolones | Target modification | |
| LPS modification | Colistin | Target modification | |
| Imported (Mobile genetic elements) | Class A serine β-lactamases (PSE, CARB, TEM) | β-lactams | Antibiotic inactivation |
| Class A serine ESBL (TEM, SHV, CTX-M, PER, VEB, GES, IBC) | β-lactams | Antibiotic inactivation | |
| Class D ESBL (OXA-types) | β-lactams | Antibiotic inactivation | |
| Class B Metallo-β-lactamase (IMP, VIM, SPM, GIM) | Carbapenems | Antibiotic inactivation | |
| Class A serine carbapenemase (KPC) | Carbapenems | Antibiotic inactivation | |
| Class D carbapenemase (OXA-types: OXA-40) | Carbapenems | Antibiotic inactivation | |
| Aminoglycosides inactivating enzymes | Aminoglycosides | Antibiotic inactivation | |
| Ribosomal methyltransferase enzymes | Aminoglycosides | Target modification |
In Europe, the recent report of the eCDC published in 2016 showed that 33.9% of P. aeruginosa were resistant to at least one of the antimicrobial groups under surveillance (piperacillin ± tazobactam, fluoroquinolones, ceftazidime, aminoglycosides and carbapenems [20]). This report showed major inter-country variations for all antimicrobial groups with generally a higher percentage of resistance in southern and eastern parts of Europe compared with northern parts. For example, focusing on carbapenems resistance, 25 to 50% of invasive isolates are resistant in Latvia, Poland, Slovakia, Hungary, Croatia, Serbia, Bulgaria or Greece, and more than 50% of the strains are resistant in Romania. Looking at combined resistance to three or more of the antimicrobials previously cited, 25 to 50% of the invasive strains isolated are resistant in Slovakia, Romania, Croatia, Bulgaria and Greece.
The β-lactams
P. aeruginosa wild-type strain encodes an inducible molecular class C AmpC cephalosporinase not inhibited by BLI such as clavulanic acid, tazobactam and sulbactam [4]. The AmpC cephalosporinase usually exhibits a low level expression which, together with low membrane permeability and multiple efflux systems, confers resistance to aminopenicillins alone or in combination with BLI, first and second generation cephalosporins (C1G, C2G), cephamycins, the two third generation cephalosporins (C3G), cefotaxime and ceftriaxone, as well as the carbapenem, ertapenem [5,6]. The P. aeruginosa wild-type strain remains thus susceptible to carboxypenicillin, ureidopenicillin, the C3G ceftazidime, the C4G cefepime, aztreonam and to the carbapenems, imipenem, meropenem and doripenem (Table 2). However, induced or constitutive AmpC overexpression and/or point mutation can provide reduced susceptibility to all classes of β-lactamins except carbapenems [5,6]. Unlike the AmpC of Enterobacteriaceae, the AmpC of P. aeruginosa can also affect cefepime [5,6] (Table 2). P. aeruginosa can produce Amber Class A serine β-lactamases of types TEM (Bush functional group 2b), PSE or CARB (carbecillinase, Bush functional group 2c) [21,22] (Table 2). The substrates of these enzymes include mainly carboxypenicillin and ureidopenicillin and they can sometimes resist BLI. These enzymes show variable susceptibility to cefepime, cefpirome and aztreonam, whereas ceftazidime and carbapenem remain always active towards P. aeruginosa strains carrying these β-lactamases types [23].
Table 2.
β-lactamases activity.
| WT | PENI | ESBL | CEPH | CARBA | |||
|---|---|---|---|---|---|---|---|
| WT | TEM PSE CARB | OXA | PER VEB TEM SHV CTX-M | OXA | AmpC | IMP VIM NDM KPC | |
| Carboxypenicillins | S | R | R | R | R | R | R |
| Carboxypenicillins +BLI | S | S/I | I/R | S/I | I/R | R | R |
| Ureidopenicillins | S | I/R | R | I/R | R | I/R | R |
| Ureidopenicillins +BLI | S | S/I | I/R | S/I | I/R | I/R | R |
| Ceftazidime | S | S | S | R | I/R | I/R | R |
| Cefepime | S | S | I/R | R | I/R | I/R | R |
| Aztreonam | S | S | S | R | I/R | I/R | S |
| Imipenem | S | S | S | S | S | S | R |
BLI, β-lactamase inhibitor; CARBA, carbapenemase; CEPH, cephalosporinase AmpC; ESBL, extended-spectrum β-lactamase; I, intermediate resistance; PENI, penicillinase; R, resistance; S, susceptible; WT, wild type.
Various Class A serine extended spectrum β-lactamases (ESBL) have been described in P. aeruginosa including PER, VEB, GES and BEL types [23]. In addition, ESBL Enterobacteriacae types of enzymes such as TEM, SHV and CTX-M have been identified in P. aeruginosa, likely following horizontal gene transfer [24,25]. These Class A types of ESBL enzymes have a low genetic identity but share a similar β-lactam hydrolysis pattern with the development of resistance to carboxypenicillins, ureidopenicillins, C3G and C4G (ceftazidime, cefepime and cefpirome), and aztreonam but not to carbapenems. Moreover, these enzymes are inhibited at various degrees by the BLI clavulanic acid and tazobactam [25].
Class D β-lactamases, called oxacillinases or OXA-type enzymes belonging to the Bush functional group 2d, have been identified in P. aeruginosa [21]. OXA-type enzymes are imported in P. aeruginosa following horizontal transfer of mobile genetic elements except OXA-50, a naturally occurring oxacillinase of P. aeruginosa [26]. Classical oxacillinases (OXA-1, OXA-2, OXA-10), confer resistance to carboxypenicillins and ureidopenicillins, are usually not susceptible to BLI and may be active on cefepime [23,27]. Extended-spectrum oxacillinases, derived by point mutations from OXA-2 and OXA-10, exhibit an increased spectrum of hydrolysis to ceftazidime, cefepime and aztreonam [23,28]. Moreover, their activity is usually not suppressed by BLI, rendering their identification by conventional laboratory methods difficult. Extended-spectrum oxacillinases are usually encoded on mobile genetic elements such as plasmids and integrons, which favor their dissemination throughout bacterial species [29].
Similar to Enterobacteriacae, carbapenemase enzymes have been identified in P. aeruginosa strains. P. aeruginosa produces carbapenemases belonging to the Class A KPC or GES-2 types and MBL of Class B [21,22,29]. GES-2 is a carbapenemase deriving from the ESBL GES-1 by point mutation whereas the KPC carbapenemase has been acquired by P. aeruginosa following horizontal acquisition from Enterobacteriacae [30,31]. However, the major class of carbapenemases found in P. aeruginosa belongs to the MBL, which comprises five types: IMP, VIM, NDM, SPM, and GIM [32–34]. The IMP and VIM types comprise several variants whereas only one variant has been identified for the other types (NDM-1, SPM-1, GIM-1) [23]. These enzymes are mostly encoded on mobile genetic elements such as plasmids, integrons and cassettes that rapidly favor their dissemination. MBLs exhibit a wide spectrum of activity covering all β-lactams including the carbapenems (imipenem and meropenem) and are resistant to BLI (Table 2).
Only the monobactam, aztreonam, is not hydrolyzed by MBLs, but the susceptibility of these enzymes to aztreonam is often not observed by routine diagnostics, since MBL activities are frequently associated with other β-lactam resistance mechanisms. In addition, several MBL genes are often located on mobile genetic elements such as integrons that also carry other resistant genes such as aminoglycoside-modifying enzymes [23,35,36]
Aminoglycosides
Aminoglycosides modifying enzymes (AME) inactivate aminoglycosides by attachment of acetyl, phosphate or adenyl groups to amino and hydroxyl substituents on the antibiotic molecule. These modifications significantly reduce the affinity of aminoglycosides for the 30S ribosomal subunit target and block their activity [37]. AME are usually plasmid encoded and are classified into three families: acetyltransferases (AAC), phosphotransferases (APH) and nucleotidyltransferases (ANT) [38]. The AME mostly identified in P. aeruginosa belongs to the AAC and ANT families [39,40]. These enzymes, acting on different targets of the antibiotic molecule, do not confer cross-resistance to all aminoglycosides. However, some P. aeruginosa strains can carry several AME acting on different aminoglycosides substituents, thus providing a possible resistance towards all components of this class of antibiotic. Compared to other aminoglycosides, amikacin is usually a poor substrate for these enzymes and is known to confer a better antibiotic activity towards P. aeruginosa.
Active efflux pumps
The natural resistance of P. aeruginosa to several antibiotics classes is partly due to the combination of low membrane permeability and active efflux pumps [41]. The efflux systems involved in antibiotic resistance in P. aeruginosa belongs to the resistance-nodulation-division (RND) family [42,43]. Four main efflux systems have been described to confer resistance to several antibiotics: MexAB-OprM, MexCD-OprJ, MexEF-OprN and MexXY-OprM (Table 3). These systems are composed of three proteins: (1) an efflux pump protein located in the cytoplasmic membrane (MexB, MexD, MexF and MexY), (2) an outer membrane protein acting as a pore (OprM, OprJ, and OprN), and (3) a protein located in the periplasmic space that bridges the proteins located in the cytoplasmic and the outer membrane (MexA, MexC, MexE and MexX). The MexAB-OprM and MexXY-OprM are involved in both natural and acquired resistance whereas the other two systems are only observed in acquired resistance. Acquired resistance is observed upon overexpression of these efflux systems following mutations in the regulatory systems that can be induced by antibiotic pressure and which can confer resistance to all class of antibiotics except polymyxins (Table 3) [23]. Exposure to a single antibiotic may trigger resistance to several classes of antibiotics that are substrates of these efflux systems. Noteworthy, quinolones are substrates of all efflux systems and represent an important triggering factor that may generate cross-resistance towards several important classes of antibiotics for pseudomonal treatment, including β-lactams and aminoglycosides [41,44]. Efflux systems are known to confer a moderate level of resistance, but they usually operate simultaneously with other resistance mechanisms, participating thus to the high-level resistance that can be observed in P. aeruginosa.
Table 3.
Active efflux pumps operating in P. aeruginosa with known antibiotic substrates.
| RND system | Substrates |
|---|---|
| MexAB-OprM | β-lactams except imipenem |
| Quinolones | |
| Macrolides | |
| Tetracyclines | |
| Chloramphenicol | |
| MexCD-OprJ | Penicillin, cefepime, cefpirome, meropenem |
| Quinolones | |
| Macrolides | |
| Tetracyclines | |
| Chloramphenicol | |
| MexEF-OprN | Carbapenems |
| Quinolones | |
| MexXY-OprM | Penicillin, cefepime, cefpirome, meropenem |
| Aminoglycosides | |
| Quinolones | |
| Macrolides | |
| Tetracyclines | |
| Chloramphenicol |
Membrane impermeability and porin alteration
Membrane impermeability or reduced permeability is a mechanism known to provide resistance towards several antibiotic classes including aminoglycosides, β-lactams and quinolones [45]. For instance, this resistance mechanism is frequently encountered in cystic fibrosis isolates that are continuously under antibiotic pressure. Several mechanisms can induce membrane impermeability, such as LPS modifications, alteration of membranous proteins involved in substrate uptakes, and inactivation of enzymatic complexes involved in membrane energetic required for the activity of transporting systems [23,45].
Regarding β-lactams, the porine OprD is known to promote the internalization of imipenem and, to some extent, meropenem but not of other β-lactams. Thus, the modification of OprD structure and/or the reduction of its expression confer a reduced susceptibility to imipenem [46,47]. In addition, the alteration of OprD is often associated with overexpression of efflux systems, thus conferring a high level of resistance to imipenem, but also to other classes of antibiotics such as quinolones and aminoglycosides [48].
Target modification
Target modification is a resistance mechanism that has been described in P. aeruginosa for aminoglycosides with the methylation of 16S rRNA, β-lactams with the alteration of penicillin binding proteins (PBP), fluoroquinolones with mutations of the DNA gyrase and topoisomaerases IV, and polymyxins with alteration of the LPS. The methylation of 16S rRNA conferring a high level of resistance towards aminoglycosides is catalyzed by the RmtA or RmtB methylases [49], which are encoded by genes located on mobile genetic elements such as plasmids and transposons [23,50]. In contrast, resistance to β-lactams and quinolones is arising following modification of the target sites encoded by genes located on the bacterial chromosome. Altered target resistance of β-lactam in P. aeruginosa is rare and has been described after modification of the penicillin binding protein 4 (PBP4) or overexpression of PBP3 [47]. Similar to most bacterial species, resistance of fluoroquinolones following target modification occurs by point mutations within the quinolone resistance determining region (QRDR) of the gyrA and gyrB genes encoding subunits of the DNA gyrase and the parC and parE genes encoding subunits of the topoisomerase IV [51]. DNA gyrase (GyrA) and topoisomerase IV (ParC) are enzymes involved in DNA replication and their alteration usually affects the activity of the whole quinolones family, resulting in MICs usually above susceptibility breakpoints [51].
Polymyxins resistance is observed following modification of the bacterial lipopolysaccharides (LPS) which are the primary target of polymyxins. The alteration is characterized by a modification of the lipid A of the LPS by the addition of phosphoethanolamine (PEtN) and 4-amino-4-deoxy-L-arabinose (L-Ara4N) [52]. The addition of these components significantly reduces the negative charge of LPS and thus the binding of polymyxin. The synthesis and addition of PEtN and L-Ara4N to the lipid A is mediated by two operons, pmrCAB and arnBCADTEF, which are mainly regulated by the two component systems PhoP/PhoQ, PmrA/PmrB [53,54], but also by ParR/ParS [55], ColR/ColS and CprR/CprS [56]. Mutations in these two component systems conferring constitutive activation leads to overexpression of the LPS-modifying operons and thus to polymyxins resistance. In P. aeruginosa, polymyxins resistance is mainly conferred by the arnBCADTEF operon involved in L-Ara4N modification of the LPS. In addition, the binding of polymyxin to LPS is impaired upon overexpression of the outer membrane protein OprH [52,57,58]. OprH is a basic protein that binds to divalent cation sites of LPS, which protects the LPS from binding by polymyxins. Recent studies focusing on the characterization of the polymyxins resistome have identified additional putative genes that may be involved in resistance, such as genes encoding proteins involved in LPS biosynthesis, metabolism, transport and regulation [59].
Laboratory role
Diagnostic laboratories need to implement several methodologies and procedures to identify P. aeruginosa strains and rapidly provide antibiotic susceptibility testing (AST) for the management of antibiotic regimens. Moreover, analytical techniques allowing bacterial typing and detection of resistance mechanisms encoded on mobile genetic elements are required for the monitoring of epidemiological outbreaks in hospital environments and characterization of long-term colonization in clinical settings such as cystic fibrosis. The identification of P. aeruginosa in routine diagnostics is performed with simple procedures based on morphology and phenotypic recognition of microbial colonies growing on conventional media. The identification of suspected colonies can be rapidly performed using modern technologies such a matrix-assisted-laser-desorption-ionization time-of-flight mass spectrometry (MALDI-TOF) [60]. If needed, selective media, such as cetrimide agar, can be used to isolate P. aeruginosa from polymicrobial environmental or clinical samples [61]. P. aeruginosa strain typing, required to monitor epidemiological hospital outbreaks to prevent or to document transmission, requires additional laboratory technics with a higher discriminatory performance than conventional methods used for the identification at the species level. Several molecular techniques have been used during the past decades including pulse field gel electrophoresis, chromosomal restriction fragment length polymorphism analysis and multilocus sequence typing [62–66]. However, these techniques may be gradually replaced by high-throughput sequencing technologies allowing rapid genome sequencing with a significant higher discriminatory power than conventional molecular typing techniques [67]. This novel technology can be used in hospital epidemiology settings to prevent transmission but also to analyze strains resistomes (genes and operons involved in antibiotic resistance) and/or to follow the evolution of the population of P. aeruginosa strains in chronic diseases such as cystic fibrosis [59].
One of the major challenges of routine diagnostic laboratories is to measure antibiotic susceptibility and to identify resistance mechanisms. Conventional laboratory procedures of AST include automated antibiotic systems such as the Vitek (bioMérieux, Marcy-l’Etoile, France) and Phoenix (Becton-Dickinson, Baltimore, USA) solutions, disk diffusion (Kirby-Bauer) and Etest. The Etest is a strip with a predefined gradient of antibiotic allowing the measurement of minimum inhibitory concentrations (MIC) on agar plates. The Etest approach can be used to measure the MIC of most of the antibiotics. However, some classes of antibiotics such as polymyxins (colistin) require an alternative approach due to poor agar diffusion and limited accuracy with automated systems. Colistin MIC is measured using an antibiotic microdilution measurement, a technique less adapted for routine diagnostic than disc diffusion or Etests but considered as the gold standard for AST measurements. Noteworthy, AST are influenced by multiple experimental factors such as preparation of bacterial inoculums, culture medium (media types, ions concentrations, pH, etc), agar concentration which may affect drug diffusion and human-based measurements characterized by moderate reproducibility and accuracy. Most laboratories are using automated antibiotic systems that monitor bacterial growth exposed to different antibiotics at different concentrations for several hours to determine extrapolated or real MIC by optical density measurements. These automated systems exhibit different performances between antibiotics with unreliable AST measurement for some components such as piperacillin-tazobactam, cefepime, and carbapenems that may require confirmation with other AST approaches including MIC determination [48]. Resistance mechanisms, such as carbapenamase activity, can be detected with rapid phenotypic methods such as the CARBA NP test, which detects the hydrolysis of carbapenems by production of carboxylic acid with the pH indicator, phenol red [68]. Carbapenemase activity can also be rapidly documented by MALDI-TOF MS analysis of the antibiotic spectrum and its degradation products that are observed following antibiotic hydrolysis by these β-lactamases [69,70]. Several rapid molecular-based tests have been recently introduced to detect genes encoding carbapenemases and ESBLs [71]. The targets detected in the proposed panels cover the most common types of carbapenamases identified in Enterobacteriacae and P. aeruginsosa (KPC, VIM, NDM, OXA) and ESBLs (CTX-M); however, a negative result cannot exclude the presence of other less prevalent β-lactams resistant genes that can be found in P. aeruginosa (ESBLs PER, VEB, TEM, SHV). Thus, molecular results still require to be confronted to phenotypic measurements to exclude molecular undetected resistant mechanisms.
Diagnostic laboratories have to properly understand the advantages and limitations of the different techniques used in order to avoid the reporting of false raw measurements (disk diffusion, MIC, automated systems) that could lead to wrong interpretation criteria (Susceptible, Intermediate, Resistance) with major errors (false resistant) and very major errors (false susceptible). The AST of P. aeruginosa is challenging since (1) several resistance mechanisms can operate simultaneously at different levels, (2) some systems expressed at low basal level are difficult to detect, (3) induced systems may be not detected before antibiotic treatment initiation and (4) the inaccuracy of some laboratory AST measurement methods can generate false susceptible or resistance interpretation results. Moreover, a lack of standardization of recommendations on standard operating procedures and interpretation criteria between antimicrobial committees such as EUCAST (European Committee on Antimicrobial Susceptibility Testing; www.eucast.org) and CLSI (Clinical and Laboratory Standards Institute; https://CLSI.org) increases the lack of reproducibility between diagnostic laboratories measurements and interpretation reproducibility and accuracy.
Clinical approaches to P. aeruginosa bacteremia
P. aeruginosa bloodstream infection (BSI) is a serious disease that requires prompt attention and pertinent clinical decisions in order to achieve a satisfactory outcome. Currently, Pseudomonas spp. represent a leading cause of hospital-acquired bacteremia, accounting for 4% of all cases and being the third leading cause of gram-negative BSI [72]. Several studies indicate an increased risk of death among patients with P. aeruginosa BSI, as compared with the risk for similar patients with other gram-negative or [73] S. aureus BSI [73,74]. Therefore, the adequate management of P. aeruginosa should be considered as a significant challenge for clinicians.
Once P. aeruginosa is isolated from blood, efforts should be made to establish the source of the infection and to choose an appropriate empirical antibiotic therapy as soon as possible. As for the source of the infection, most patients have an identifiable focus of infection at the time of initial evaluation, but the source remains unknown in up to 40% of the cases [73,75]. The most common sources of P. aeuruginosa BSI are in respiratory (25%) and urinary tract (19%) followed by central venous catheter and skin and soft tissues [73]. Identification of the source is essential because its adequate control represents a critical issue in the correct management of P. aeruginosa infection (i.e. early CVC removal or surgical drainage of abscess). Accordingly, a careful patient history and a physical examination, as well as radiological and microbiological work-up are important.
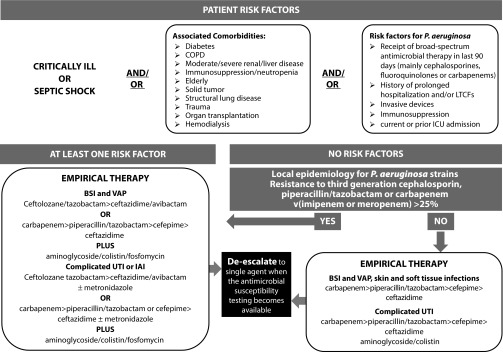
Empirical antibiotic therapy should include two agents from different classes with in vitro activity against P. aeruginosa for all serious infections known or suspected to be caused by P. aeruginosa. The rationale of the so-called ‘double coverage effect’ is to increase the likelihood that antibiotic treatment will be active against P. aeruginosa, especially in the setting of a high-risk of antimicrobial resistance. Once results of susceptibility are available, definitive therapy should be tailored accordingly, using a single in vitro active agent with the highest antimicrobial activity and the lowest propensity to select resistance. Indeed, at the time of the present review, no convincing data exist demonstrating a mortality benefit to combination therapy (Figure 1) [76].
Figure 1.
Clinical approach to patients with suspected P. aeruginosa infection.
BSI: Bloodstream infection; COPD: Chronic obstructive pulmonary disease; IAI: Intra-abdominal infections; LTCFs: Long term care facilities; UTI: Urinary tract infection; VAP: Ventilator associated pneumonia.
Management of P. aeruginosa VAP
P. aeruginosa is one of the leading causes of ventilator-associated pneumonia (VAP) in the US and Europe [77–79]. VAP due to P. aeruginosa is increasing in incidence and poses unique challenges for its clinical management. Risk factors for the development of P. aeruginosa-related VAP include prolonged mechanical ventilation [80], older age [80], prior P. aeruginosa colonization [79], prior antibiotic therapy [79,80], admission to a ward with high incidence of P. aeruginosa infections [80], solid cancer, and shock [79].
Recent data suggest that a diagnosis of P. aeruginosa-related VAP is frequently associated with the isolation of MDR pathogens [79]. MDR P. aeruginosa-related pneumonia appears to be an important determinant of excess length of stay in ICU, and prolonged mechanical ventilation, as well as a cause of increased in-hospital mortality compared to non-MDR infection [14].
We recommend prescribing two anti-pseudomonal antibiotics from different classes as the initial therapy of P. aeruginosa-related VAP, especially when patients are hospitalized in units where >20% of Gram-negative isolates are resistant to a ‘backbone’ agent considered for monotherapy. Once the antibiotic susceptibility testing is known, monotherapy using an antibiotic to which the isolate is susceptible can be considered, except for patients who have septic shock or a high risk of death. An anti-pseudomonal cephalosporin, or a carbapenem, or an anti-pseudomonal β-lactam/BLI represents potential options for definitive therapy. Aminoglycosides should not be used as monotherapy because success rates for aminoglycosides are low [81]. This may be due to the poor penetration of aminoglycosides into the lung, which require high peak serum concentrations to obtain adequate lung concentrations, thus increasing the risk of nephrotoxicity or ototoxicity [82,83]. However, because in Europe fluoroquinolone resistance rate in P. aeuruginosa exceeds 30% [84], we suggest the use of combination therapy including aminoglycosides for empirical therapy of serious VAP, if it is appropriately tailored on the basis of susceptibility data (Figure 1). As for aerosol therapy, we do not routinely recommend the use of inhaled antibiotics for the treatment of P. aeruginosa VAP. However, they may be considered as an adjunctive to intravenous therapy in cases of infections due to MDR strains.
Management of P. aeruginosa CAP
P. aeruginosa has been reported as a rare cause of community-acquired pneumonia (CAP), affecting 1.1–8.3% of the patients requiring ICU admission [85–89]. Despite this, P. aeruginosa is actually considered the pathogen with the highest attributable mortality rate, ranging from 50 to 100% [85–89]. The survival of these patients is related to early diagnosis and prompt initiation of adequate antibiotic therapy [90]. Since antibiotic therapy for P. aeruginosa is completely different from the standard therapy to treat common pathogens in CAP, current guidelines stratify therapy recommendations on the basis of pseudomonal risk factors [91]. CAP due to P. aeruginosa should be considered in immunocompromised subjects (i.e. HIV patients, solid organ transplant) who received prior antibiotic use and with structural abnormalities such as cystic fibrosis, bronchiectasis and COPD (especially those requiring frequent corticosteroid therapy and/or antibiotic use) [91–93]. Additional risk factors include male sex, low C-reactive protein and PSI risk class IV and V [90]. Risk factors associated with the isolation of MDR P. aeruginosa in CAP have been recently studied in a recent article including more than 2000 patients with CAP, where the only risk factor was previous antibiotic therapy [90].
Therefore, from a clinical point of view, we suggest use of antibiotics covering MDR P. aeruginosa in CAP only when P. aeruginosa is highly suspected.
Management of P. aeruginosa urinary tract infection
Patients with P. aeruginosa urinary tract infection (UTI) are more likely to have chronic underlying disease (e.g. hypertension, cognitive impairment, diabetes mellitus) [94], present with healthcare-associated infection and recent urinary tract instrumentation and/or chronic indwelling urinary catheters. Mortality rates are higher and reported with rates up to 20% [94]. Advanced chronic disease and inadequate definitive antimicrobial treatment are associated with worse prognosis [94]. Standard of care consists of elimination of the predisposing condition in combination with a single antibiotic therapy that is generally considered adequate for treatment [94] in absence of septic shock (Figure 1).
Management of P. aeruginosa skin and soft tissue infections
P. aeruginosa causes a variety of skin and soft tissue infections ranging from the benign (e.g. cellulitis, post-surgical infections) to the immediately life threatening. P. aeruginosa is one of the most common pathogen isolated from cellulitis in neutropenic patients, surgical site infections (SSI), infections following trauma or infections of chronic decubitus ulcers. Although combined antimicrobial and surgical debridement should be considered as standard of care, medical therapy alone maybe sufficient for some patients. For example, in acute cellulitis surgery is generally not necessary, whereas a surgical site infection or an infection of chronic decubitus ulcers requires surgical debridement to remove necrotic tissue. In all cases the importance of antimicrobial therapy is unquestioned. The optimal antibiotic regimen depends on in vitro susceptibility testing and includes an anti-pseudomonal beta-lactam, a carbapenem, or a fluoroquinolone. Although the usual duration of antibiotic therapy is 10 to 14 days, shorter courses could be considered in patients who received an adequate source control of infection and/or with prompt resolution of clinical signs and symptoms.
Among skin and soft tissue infection due to P. aeruginosa, two clinical syndromes need special considerations: (1) ecthyma gangrenosum and (2) burn wound infections.
Ecthyma gangrenosum, classically reported in the setting of P. aeruginosa BSI in neutropenic patients, is a cutaneous vasculitis caused by bacterial invasion of the media and adventitia of the vessel wall with secondary ischemic necrosis [95]. The lesion frequently begins as painless erythematous areas with papules and/or bullae that often rapidly progress becoming painful gangrenous ulcers [96]. These lesions may be single or multiple and, although they can occur at any anatomic district, they are preferentially found in the gluteal and perineal areas. Once ecthyma gangrenosum is clinically suspected, prompt collection of blood cultures, culture of exudates from an aspirate or swab of lesion or skin biopsy should be collected to isolate P. aeruginosa or other uncommon cause of viral, bacterial, mycobacterial or fungal pathogens potentially responsible [96,97].
As for therapy, we suggest administration of empirical combination therapy with beta lactams and aminoglycosides for high risk severe patients (i.e. neutropenic patients, severe immunocompromised patients or patients with septic shock) until the causative organism and its susceptibility are identified. Surgery is not generally warranted but extensive debridement could be required in patients with extensive necrosis (Figure 1) [98].
Regarding burn wound infection, P. aeruginosa represents one of the most common [99,100] pathogen-causing infection in burn injuries. Due to the severity of the patient’s condition and frequent antibiotic resistance, P. aeruginosa is a dreaded cause of infection in such populations [101–103]. Based on previous studies [104], the rate of positive swab or tissue culture results due to P. aeruginosa in burn infection was as high as 57% whereas the proportion of BSI caused by P. aeruginosa in burned populations was reported to be approximately 15% [102,103].
Superficial wound infections due to P. aeruginosa have a characteristic yellow or green color with a malodorous smell. This infection may evolve to an invasive disease causing blue-purplish lesion of the skin [105].
Similar to ecthyma gangrenosum, we also suggest, for burns wound infections, administration of empirical combination therapy until the causative organism and its susceptibility are identified. Surgical debridement can be indicated in some cases [105].
Role of combination therapy versus monotherapy
Early administration of an adequate antibiotic regimen has been associated with favorable clinical outcome, especially among critically ill patients presenting with severe P. aeruginosa infections [1]; conversely, a delay in the prescription of an adequate antibiotic therapy has been related to a significant increase in mortality.
In recent years, the progressive increase in antibiotic resistance among P. aeruginosa has been identified as the main reason for antibiotic inadequacy, with a negative impact on patient survival [106]. The available evidence suggests that the greatest benefit of a combination therapy stems from the increased likelihood of choosing an effective agent during empirical therapy rather than to prevent the resistance during definitive therapy or to benefit of in vitro synergistic activity. Therefore, to balance between early antibiotic administration and risk of resistance selection, we suggest early administration of a combination regimen when P. aeruginosa is suspected, followed by a prompt de-escalation when the antimicrobial susceptibility testing becomes available. We encourage an approach consisting of the prescription of an anti-pseudomonal β-lactam (piperacillin/tazobactam, ceftolozane/tazobactam, ceftazidime, cefepime, or a carbapenem) plus a second anti-pseudomonal agent (aminoglycoside or a fluoroquinolones).
How to optimize anti-P. aeruginosa therapy
Clinicians should be aware that in addition to adequate antimicrobial coverage, other factors including optimal dosing, interval of drug administration, and duration of therapy are key factors influencing clinical outcomes.
For example, in a recent multinational study performed in ICU patients, 16% of the patients did not achieve free antibiotic concentrations sufficiently greater than the MIC required to ensure a positive clinical outcome [107]. Another recent study performed in patients with VAP due to Gram-negative bacteria [108] showed that a serum exposure of anti-pseudomonal cephalosporins greater than 53% fT>MIC was significantly associated with a favorable outcome or presumed eradication. Therefore, these and other studies [109] support the importance of considering adequate exposure-response profiles when optimizing drug therapy in these patient groups.
In our opinion, the best way to optimize beta-lactam antibiotic dosing may be the use of prolonged or continuous infusion with the use of a loading dose to ensure early attainment of target concentration exceeding the MIC [110]. Moreover, although it is not available in most clinical laboratory, we also suggest the use of therapeutic drug monitoring (TDM).
New systemic drugs
Standard antibiotic therapy may be inferior to some new comparator agents in the treatment of serious P. aeruginosa infections, especially in the setting of increased antimicrobial resistance. Novel antibiotics with activity against P. aeruginosa have become available in Europe in recent years and others are in advanced stages of clinical development (Table 4). In some cases, indirect evidence suggests their possible superiority over standard anti-pseudomonal regimens (Table 5) [111].
Table 4.
New drugs and usual clinical dosage for new anti-Pseudomonas agents.
| Drug | Current clinical indications | Usual clinical dosage for serious infections | Other comment |
|---|---|---|---|
| Cephalosporins | |||
| Cefiderocol | Complicated UTI | 2 g intravenous every 8 hours | - |
| Cephalosporin + β-lactamase inhibitor | |||
| Ceftolozane-tazobactam | Complicated UTI and IAI | Loading dose 1.5 g or 3 g intravenous in 1 hour, followed by 1.5 g or 3 g intravenous every 8 hours | Extended infusion (over 3 h) 1.5 g or 3 g every 8 hours is recommended |
| Ceftazidime-avibactam | Complicated UTI and IAI, HAP and VAP and Gram-negative infections when other treatments might not work | Loading dose 2.5 g intravenous in 1 hour, followed by 2.5 g intravenous every 8 hours | Extended infusion (over 3 h) 2.5 g every 8 hours is recommended |
| Carbapenem + β-lactamase inhibitor | |||
| Meropenem-vaborbactam | Complicated UTI | 2 g/2 g intravenous every 8 hours | Not active against MDR strains |
| Imipenem-relebactam | Not yet approved by any regulatory authority | 500 mg/250 mg intravenous every 6 hours | Not active against MDR strains |
| Aminoglycoside | |||
| Plazomicin | Not yet approved by any regulatory authority | 15 mg/kg every 24 hours | - |
Table 5.
Advantages and disadvantages of new drugs for P. aeruginosa infections.
| Advantages |
| High activity against P. aeruginosa including MDR strains |
|
|
| Predictable PK |
|
|
| Good safety profile and tolerability |
|
|
| Carbapenem sparing |
|
|
| Rapid tissue distribution |
|
|
| Disadvantages |
|
|
| Increased costs |
|
|
| No oral formulations to allow step-down therapy |
|
|
| Superinfection with even more resistant bacteria or fungi |
Ceftolozane-tazobactam
Ceftolozane-tazobactam is being developed to overcome P. aeruginosa antimicrobial mechanisms of resistance, such as changes in porin permeability and upregulation of efflux pumps. This drug has an intrinsic potent anti-pseudomonal activity, because of a greater affinity for all essential PBPs, including PBP1b, PBP1c and PBP3 [112]. Ceftolozane/Tazobactam has proven to have a potent in vitro activity against the majority of MDR P. aeruginosa strains (including ESBL but not carbapenemase producing strains). The US FDA has proposed clinical use of ceftolozane-tazobactam in complicated intra-abdominal and urinary tract infections [113,114]. In addition, a study for treatment of HAP including VAP is currently ongoing. Data belonging to real-world studies using ceftolozane-tazobactam for the treatment of MDR P. aeruginosa infections showed positive outcomes in 71% of patients with MDR P. aeruginosa infections [115].
Ceftazidime-avibactam
Ceftazidime-avibactam is a novel β-lactam/BLI combination approved by FDA and EMA for the treatment of complicated UTIs and complicated intra-abdominal infection. In vitro studies showed that the combination of ceftazidime-avibactam is highly effective against Enterobacteriaceae producing KPCs, ESBLs OXA and AmpC enzymes. However, the drug has no activity against metallo-beta lactamases (MBL, VIM and NDM) and avibactam offers no enhanced activity against P. aeruginosa [116–119]. The effectiveness of ceftazidime-avibactam against VAP has been analyzed in a phase III studies comparing this new drug with meropenem (NTC01808092) [120]. The predominant Gram-negative pathogens isolated at baseline were K. pneumoniae and P. aeruginosa, with 28% of patients having ≥1 ceftazidime non-susceptible isolate. In the clinically evaluable population, 356 patients received ceftazidime-avibactam and 370 received meropenem. The study met the criteria for non-inferiority of ceftazidime-avibactam since there was no difference between the groups regarding the outcome. Moreover, the efficacy of ceftazidime-avibactam against ceftazidime-non-susceptible strains was similar to that against ceftazidime-susceptible pathogens and was also comparable to meropenem.
Imipenem-cilastatin-relebactam
Relebactam is a diazabicyclooctanes BLI that inhibits the activity of class A and C β-lactamase, but does not have any activity against metallo- β-lactamase [121]. The combination of imipenem-cilastatin with relebactam has shown to have synergistic activity against a wide spectrum of MDR Gram-negative pathogens including P. aeruginosa, KPC-producing K. pneumoniae and Enterobacter spp. [122,123]. This drug has been mainly studied in patients with IAI [124], complicated UTI and pyelonephritis [125] whereas a study on patients with HAP/VAP is currently ongoing.
Other new drugs such as plazomycin, meropenem-vaborbactam and aztreonam-avibactam have a limited effect on P. aeruginosa [111].
Future strategies to improve patient outcome
There is an urgent need to improve early diagnosis and empirical treatment of severe P. aeruginosa infections. First, MALDI-TOF and new molecular techniques should be systematically implemented to rapidly report the identification and susceptibility results for Pseudomonas in blood cultures and other clinical relevant cultures. However, controlled trials will be necessary to determine whether such diagnostic techniques have a real impact on length of hospitalization and patient mortality. Second, further studies aimed to identify patients at risk of MDR P. aeruginosa infections (bloodstream infections, urinary tract infections) based on clinical risk factors are urgently needed. Finally, clinical response depends on factors such as underlying diseases, severity of infection, type of infections, adequate source control and response of previous antibiotics. There is an urgent need to evaluate the real impact on patient outcomes of the new anti-Pseudomonas drugs recently approved for the treatment of these infections.
Footnotes
Disclosure and potential conflicts of interest: The authors declare no conflicts of interest. The International Committee of Medical Journal Editors (ICMJE) Potential Conflicts of Interests form for the authors are available for download at http://www.drugsincontext.com/wp-content/uploads/2018/04/dic.212527-COI.pdf
Correct attribution: Copyright © 2018 Bassetti M, Vena A, Croxatto A, Righi E, Guery B. https://doi.org/10.7573/dic.212527. Published by Drugs in Context under Creative Commons License Deed CC BY NC ND 4.0.
Provenance: invited; externally peer reviewed.
Drugs in Context is published by BioExcel Publishing Ltd. Registered office: Plaza Building, Lee High Road, London, England, SE13 5PT.
BioExcel Publishing Limited is registered in England Number 10038393. VAT GB 252772009.
For all manuscript and submissions enquiries, contact the Editorial office dic.editorial@bioexcelpublishing.com
For all permissions, rights and reprints, contact David Hughes david.hughes@bioexcelpublishing.com
Peer review comments to author: 8 March 2018
References
- 1.Pena C, Suarez C, Tubau F, Dominguez A, Sora M, Pujol M, Gudiol F, Ariza J. Carbapenem-resistant Pseudomonas aeruginosa: factors influencing multidrug-resistant acquisition in non-critically ill patients. Eur J Clin Microbiol Infect Dis. 2009;28(5):519–22. doi: 10.1007/s10096-008-0645-9. http://dx.doi.org/10.1007/s10096-008-0645-9. [DOI] [PubMed] [Google Scholar]
- 2.El Zowalaty ME, Al Thani AA, Webster TJ, El Zowalaty AE, Schweizer HP, Nasrallah GK, Marei HE, Ashour HM. Pseudomonas aeruginosa: arsenal of resistance mechanisms, decades of changing resistance profiles, and future antimicrobial therapies. Future Microbiol. 2015;10(10):1683–706. doi: 10.2217/fmb.15.48. http://dx.doi.org/10.2217/fmb.15.48. [DOI] [PubMed] [Google Scholar]
- 3.Fujii A, Seki M, Higashiguchi M, Tachibana I, Kumanogoh A, Tomono K. Community-acquired, hospital-acquired, and healthcare-associated pneumonia caused by Pseudomonas aeruginosa. Respir Med Case Rep. 2014;12:30–3. doi: 10.1016/j.rmcr.2014.03.002. http://dx.doi.org/10.1016/j.rmcr.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sligl WI, Dragan T, Smith SW. Nosocomial Gram-negative bacteremia in intensive care: epidemiology, antimicrobial susceptibilities, and outcomes. Int J Infect Dis. 2015;37:129–34. doi: 10.1016/j.ijid.2015.06.024. http://dx.doi.org/10.1016/j.ijid.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 5.Oncul O, Oksuz S, Acar A, Ulkur E, Turhan V, Uygur F, Ulcay A, Erdem H, Ozyurt M, Gorenek L. Nosocomial infection characteristics in a burn intensive care unit: analysis of an eleven-year active surveillance. Burns. 2014;40(5):835–41. doi: 10.1016/j.burns.2013.11.003. http://dx.doi.org/10.1016/j.burns.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Lund-Palau H, Turnbull AR, Bush A, Bardin E, Cameron L, Soren O, Wierre-Gore N, Alton EW, Bundy JG, Connett G, Faust SN, Filloux A, Freemont P, Jones A, Khoo V, Morales S, Murphy R, Pabary R, Simbo A, Schelenz S, Takats Z, Webb J, Williams HD, Davies JC. Pseudomonas aeruginosa infection in cystic fibrosis: pathophysiological mechanisms and therapeutic approaches. Expert Rev Respir Med. 2016;10(6):685–97. doi: 10.1080/17476348.2016.1177460. http://dx.doi.org/10.1080/17476348.2016.1177460. [DOI] [PubMed] [Google Scholar]
- 7.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii. Int J Antimicrob Agents. 2015;45(6):568–85. doi: 10.1016/j.ijantimicag.2015.03.001. http://dx.doi.org/10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Lucena A, Dalla Costa LM, Nogueira KS, Matos AP, Gales AC, Paganini MC, Castro ME, Raboni SM. Nosocomial infections with metallo-beta-lactamase-producing Pseudomonas aeruginosa: molecular epidemiology, risk factors, clinical features and outcomes. J Hosp Infect. 2015;87(4):234–40. doi: 10.1016/j.jhin.2014.05.007. http://dx.doi.org/10.1016/j.jhin.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Matos EC, Matos HJ, Conceicao ML, Rodrigues YC, Carneiro IC, Lima KV. Clinical and microbiological features of infections caused by Pseudomonas aeruginosa in patients hospitalized in intensive care units. Rev Soc Bras Med Trop. 2016;49(3):305–11. doi: 10.1590/0037-8682-0446-2015. http://dx.doi.org/10.1590/0037-8682-0446-2015. [DOI] [PubMed] [Google Scholar]
- 10.Peng Y, Bi J, Shi J, Li Y, Ye X, Chen X, Yao Z. Multidrug-resistant Pseudomonas aeruginosa infections pose growing threat to health care-associated infection control in the hospitals of Southern China: a case-control surveillance study. Am J Infect Control. 2014;42(12):1308–11. doi: 10.1016/j.ajic.2014.08.006. http://dx.doi.org/10.1016/j.ajic.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Jeong SJ, Yoon SS, Bae IK, Jeong SH, Kim JM, Lee K. Risk factors for mortality in patients with bloodstream infections caused by carbapenem-resistant Pseudomonas aeruginosa: clinical impact of bacterial virulence and strains on outcome. Diagn Microbiol Infect Dis. 2014;80(2):130–5. doi: 10.1016/j.diagmicrobio.2014.07.003. http://dx.doi.org/10.1016/j.diagmicrobio.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 12.DalBen MF, Basso M, Garcia CP, Costa SF, Toscano CM, Jarvis WR, Lobo RD, Oliveira MS, Levin AS. Colonization pressure as a risk factor for colonization by multiresistant Acinetobacter spp and carbapenem-resistant Pseudomonas aeruginosa in an intensive care unit. Levin, Clinics (Sao Paulo) 2013;68(8):1128–33. doi: 10.6061/clinics/2013(08)11. http://dx.doi.org/10.6061/clinics/2013(08)11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willmann M, Klimek AM, Vogel W, Liese J, Marschal M, Autenrieth IB, Peter S, Buhl M. Clinical and treatment-related risk factors for nosocomial colonisation with extensively drug-resistant Pseudomonas aeruginosa in a haematological patient population: a matched case control study. BMC Infect Dis. 2014;14:650. doi: 10.1186/s12879-014-0650-9. http://dx.doi.org/10.1186/s12879-014-0650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Micek ST, Wunderink RG, Kollef MH, Chen C, Rello J, Chastre J, Antonelli M, Welte T, Clair B, Ostermann H, Calbo E, Torres A, Menichetti F, Schramm GE, Menon V. An international multicenter retrospective study of Pseudomonas aeruginosa nosocomial pneumonia: impact of multidrug resistance. Crit Care. 2015;19:219. doi: 10.1186/s13054-015-0926-5. http://dx.doi.org/10.1186/s13054-015-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babu KV, Visweswaraiah DS, Kumar A. The influence of Imipenem resistant metallo-beta-lactamase positive and negative Pseudomonas aeruginosa nosocomial infections on mortality and morbidity. J Nat Sci Biol Med. 2014;5(2):345–51. doi: 10.4103/0976-9668.136181. http://dx.doi.org/10.4103/0976-9668.136181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrasa-Villar JI, Aibar-Remon C, Prieto-Andres P, Mareca-Donate R, Moliner-Lahoz J. Impact on morbidity, mortality, and length of stay of hospital-acquired infections by resistant microorganisms. Clin Infect Dis. 2017;65(4):644–52. doi: 10.1093/cid/cix411. http://dx.doi.org/10.1093/cid/cix411. [DOI] [PubMed] [Google Scholar]
- 17.Righi E, Peri AM, Harris PN, Wailan AM, Liborio M, Lane SW, Paterson DL. Global prevalence of carbapenem resistance in neutropenic patients and association with mortality and carbapenem use: systematic review and meta-analysis. J Antimicrob Chemother. 2017;72(3):668–77. doi: 10.1093/jac/dkw459. http://dx.doi.org/10.1093/jac/dkw459. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Li X, Li W, Du X, He JQ, Tao C, Feng Y. Influence of carbapenem resistance on mortality of patients with Pseudomonas aeruginosa infection: a meta-analysis. Sci Rep. 2015;5:11715. doi: 10.1038/srep11715. http://dx.doi.org/10.1038/srep11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riu M, Chiarello P, Terradas R, Sala M, Garcia-Alzorriz E, Castells X, Grau S, Cots F. Cost attributable to nosocomial bacteremia. analysis according to microorganism and antimicrobial sensitivity in a university hospital in Barcelona. PLoS One. 2016;11(4):e0153076. doi: 10.1371/journal.pone.0153076. http://dx.doi.org/10.1371/journal.pone.0153076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.European Centre for Disease Prevention and Control. Surveillance Report: Antimicrobial resistance surveillance in Europe 2015. [Last accessed: 22 May 2018]. Available at: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/antimicrobial-resistance-europe-2015.pdf.
- 21.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39(6):1211–33. doi: 10.1128/aac.39.6.1211. http://dx.doi.org/10.1128/AAC.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22(4):582–610. doi: 10.1128/CMR.00040-09. http://dx.doi.org/10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strateva T, Yordanov D. Pseudomonas aeruginosa – a phenomenon of bacterial resistance. J Med Microbiol. 2009;58(Pt 9):1133–48. doi: 10.1099/jmm.0.009142-0. http://dx.doi.org/10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 24.Chanawong A, M’Zali FH, Heritage J, Lulitanond A, Hawkey PM. SHV-12, SHV-5, SHV-2a and VEB-1 extended-spectrum beta-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J Antimicrob Chemother. 2001;48(6):839–52. doi: 10.1093/jac/48.6.839. https://doi.org/10.1093/jac/48.6.839. [DOI] [PubMed] [Google Scholar]
- 25.Picao RC, Poirel L, Gales AC, Nordmann P. Further identification of CTX-M-2 extended-spectrum beta-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(5):2225–6. doi: 10.1128/AAC.01602-08. http://dx.doi.org/10.1128/AAC.01602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girlich D, Naas T, Nordmann P. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2004;48(6):2043–8. doi: 10.1128/AAC.48.6.2043-2048.2004. http://dx.doi.org/10.1128/AAC.48.6.2043-2048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bert F, Branger C, Lambert-Zechovsky N. Identification of PSE and OXA beta-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J Antimicrob Chemother. 2002;50(1):11–8. doi: 10.1093/jac/dkf069. http://dx.doi.org/10.1093/jac/dkf069. [DOI] [PubMed] [Google Scholar]
- 28.Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–51. doi: 10.1128/CMR.14.4.933-951.2001. http://dx.doi.org/10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordmann P, Guibert M. Extended-spectrum beta-lactamases in Pseudomonas aeruginosa. J Antimicrob Chemother. 1998;42(2):128–31. doi: 10.1093/jac/42.2.128. http://dx.doi.org/10.1093/jac/42.2.128. [DOI] [PubMed] [Google Scholar]
- 30.Weldhagen GF, Prinsloo A. Molecular detection of GES-2 extended spectrum Beta-lactamase producing Pseudomonas aeruginosa in Pretoria, South Africa. Int J Antimicrob Agents. 2004;24(1):35–8. doi: 10.1016/j.ijantimicag.2003.12.012. http://dx.doi.org/10.1016/j.ijantimicag.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP Colombian Nosocomial Resistance Study Group. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother. 2007;51(4):1553–5. doi: 10.1128/AAC.01405-06. http://dx.doi.org/10.1128/AAC.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordmann P, Poirel L. Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infect. 2002;8(6):321–31. doi: 10.1046/j.1469-0691.2002.00401.x. http://dx.doi.org/10.1046/j.1469-0691.2002.00401.x. [DOI] [PubMed] [Google Scholar]
- 33.Walsh TR. The emergence and implications of metallo-beta-lactamases in Gram-negative bacteria. Clin Microbiol Infect. 2005;11(Suppl 6):2–9. doi: 10.1111/j.1469-0691.2005.01264.x. http://dx.doi.org/10.1111/j.1469-0691.2005.01264.x. [DOI] [PubMed] [Google Scholar]
- 34.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18(2):306–25. doi: 10.1128/CMR.18.2.306-325.2005. http://dx.doi.org/10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendes RE, Toleman MA, Ribeiro J, Sader HS, Jones RN, Walsh TR. Integron carrying a novel metallo-beta-lactamase gene, blaIMP-16, and a fused form of aminoglycoside-resistant gene aac(6′)-30/aac(6′)-Ib′: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. 2004;48(12):4693–702. doi: 10.1128/AAC.48.12.4693-4702.2004. http://dx.doi.org/10.1128/AAC.48.12.4693-4702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirel L, Lambert T, Turkoglu S, Ronco E, Gaillard J, Nordmann P. Characterization of Class 1 integrons from Pseudomonas aeruginosa that contain the bla(VIM-2) carbapenem-hydrolyzing beta-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob Agents Chemother. 2001;45(2):546–52. doi: 10.1128/AAC.45.2.546-552.2001. http://dx.doi.org/10.1128/AAC.45.2.546-552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Llano-Sotelo B, Azucena EF, Jr, Kotra LP, Mobashery S, Chow CS. Aminoglycosides modified by resistance enzymes display diminished binding to the bacterial ribosomal aminoacyl-tRNA site. Chem Biol. 2002;9:455–63. doi: 10.1016/s1074-5521(02)00125-4. http://dx.doi.org/10.1016/S1074-5521(02)00125-4. [DOI] [PubMed] [Google Scholar]
- 38.Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003;16(3):430–50. doi: 10.1128/CMR.16.3.430-450.2003. http://dx.doi.org/10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller GH, Sabatelli FJ, Hare RS, Glupczynski Y, Mackey P, Shlaes D, Shimizu K, Shaw KJ. The most frequent aminoglycoside resistance mechanisms – changes with time and geographic area: a reflection of aminoglycoside usage patterns? Clin Infect Dis. 1997;24(Suppl 1):S46–62. doi: 10.1093/clinids/24.supplement_1.s46. http://dx.doi.org/10.1093/clinids/24.Supplement_1.S46. [DOI] [PubMed] [Google Scholar]
- 40.Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005;49(2):479–87. doi: 10.1128/AAC.49.2.479-487.2005. http://dx.doi.org/10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56(1):20–51. doi: 10.1093/jac/dki171. http://dx.doi.org/10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 42.Livermore DM. Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother. 2001;47(3):247–50. doi: 10.1093/jac/47.3.247. http://dx.doi.org/10.1093/jac/47.3.247. [DOI] [PubMed] [Google Scholar]
- 43.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 2002;34(5):634–40. doi: 10.1086/338782. http://dx.doi.org/10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 44.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36(9):1847–51. doi: 10.1128/aac.36.9.1847. http://dx.doi.org/10.1128/AAC.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert PA. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;95( Suppl 41):22–6. [PMC free article] [PubMed] [Google Scholar]
- 46.Kohler T, Michea-Hamzehpour M, Epp SF, Pechere JC. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob Agents Chemother. 1999;43(2):424–7. doi: 10.1128/aac.43.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pechere JC, Kohler T. Patterns and modes of beta-lactam resistance in Pseudomonas aeruginosa. Clin Microbiol Infect. 1999;5(Suppl 1):S15–S18. doi: 10.1111/j.1469-0691.1999.tb00719.x. https://doi.org/10.1111/j.1469-0691.1999.tb00719.x. [DOI] [PubMed] [Google Scholar]
- 48.Mesaros N, Nordmann P, Plesiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A, Tulkens PM, Van Bambeke F. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect. 2007;13(6):560–78. doi: 10.1111/j.1469-0691.2007.01681.x. http://dx.doi.org/10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 49.Doi Y, Arakawa Y. 6S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45(1):88–94. doi: 10.1086/518605. http://dx.doi.org/10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 50.Yamane K, Doi Y, Yokoyama K, Yagi T, Kurokawa H, Shibata N, Shibayama K, Kato H, Arakawa Y. Genetic environments of the rmtA gene in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2004;48(6):2069–74. doi: 10.1128/AAC.48.6.2069-2074.2004. http://dx.doi.org/10.1128/AAC.48.6.2069-2074.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis. 2000;31(Suppl 2):S24–8. doi: 10.1086/314056. http://dx.doi.org/10.1086/314056. [DOI] [PubMed] [Google Scholar]
- 52.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. http://dx.doi.org/10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(1):205–17. doi: 10.1046/j.1365-2958.2003.03673.x. http://dx.doi.org/10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 54.Macfarlane EL, Kwasnicka A, Hancock RE. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology. 2000;146(Pt 10):2543–54. doi: 10.1099/00221287-146-10-2543. http://dx.doi.org/10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- 55.Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. Adaptive resistance to the ‘last hope’ antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother. 2010;54(8):3372–82. doi: 10.1128/AAC.00242-10. http://dx.doi.org/10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, Kaul RK, Johansen HK, Hoiby N, Moskowitz SM. Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob Agents Chemother. 2013;57(5):2204–15. doi: 10.1128/AAC.02353-12. http://dx.doi.org/10.1128/AAC.02353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol. 1999;34(2):305–16. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- 58.Young ML, Bains M, Bell A, Hancock RE. Role of Pseudomonas aeruginosa outer membrane protein OprH in polymyxin and gentamicin resistance: isolation of an OprH-deficient mutant by gene replacement techniques. Antimicrob Agents Chemother. 1992;36(11):2566–8. doi: 10.1128/aac.36.11.2566. http://dx.doi.org/10.1128/AAC.36.11.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez L, Alvarez-Ortega C, Wiegand I, Olivares J, Kocincova D, Lam JS, Martinez JL, Hancock RE. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57(1):110–9. doi: 10.1128/AAC.01583-12. http://dx.doi.org/10.1128/AAC.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Croxatto A, Prod’hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012;36:380–407. doi: 10.1111/j.1574-6976.2011.00298.x. http://dx.doi.org/10.1111/j.1574-6976.2011.00298.x. [DOI] [PubMed] [Google Scholar]
- 61.Kodaka H, Iwata M, Yumoto S, Kashitani F. Evaluation of a new agar medium containing cetrimide, kanamycin and nalidixic acid for isolation and enhancement of pigment production of Pseudomonas aeruginosa in clinical samples. J Basic Microbiol. 2003;43(5):407–13. doi: 10.1002/jobm.200310264. http://dx.doi.org/10.1002/jobm.200310264. [DOI] [PubMed] [Google Scholar]
- 62.Grothues D, Koopmann U, von der Hardt H, Tummler B. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol. 1988;26(10):1973–7. doi: 10.1128/jcm.26.10.1973-1977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolz C, Kiosz G, Ogle JW, Vasil ML, Schaad U, Botzenhart K, Doring G. Pseudomonas aeruginosa cross-colonization and persistence in patients with cystic fibrosis. Epidemiol Infect. 1989;102(2):205–14. doi: 10.1017/s0950268800029873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vernez I, Hauser P, Bernasconi MV, Blanc DS. Population genetic analysis of Pseudomonas aeruginosa using multilocus sequence typing. FEMS Immunol Med Microbiol. 2005;43(1):29–35. doi: 10.1016/j.femsim.2004.06.024. http://dx.doi.org/10.1016/j.femsim.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 65.Basset P, Blanc DS. Fast and simple epidemiological typing of Pseudomonas aeruginosa using the double-locus sequence typing (DLST) method. Eur J Clin Microbiol Infect Dis. 2014;33(6):927–32. doi: 10.1007/s10096-013-2028-0. http://dx.doi.org/10.1007/s10096-013-2028-0. [DOI] [PubMed] [Google Scholar]
- 66.Cholley P, Stojanov M, Hocquet D, Thouverez M, Bertrand X, Blanc DS. Comparison of double-locus sequence typing (DLST) and multilocus sequence typing (MLST) for the investigation of Pseudomonas aeruginosa populations. Diagn Microbiol Infect Dis. 2015;82(4):274–7. doi: 10.1016/j.diagmicrobio.2015.03.027. http://dx.doi.org/10.1016/j.diagmicrobio.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 67.Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev. 2014;27(4):783–822. doi: 10.1128/CMR.00003-14. http://dx.doi.org/10.1128/CMR.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poirel L, Nordmann P. Rapidec carba NP test for rapid detection of carbapenemase producers. J Clin Microbiol. 2015;53(9):3003–8. doi: 10.1128/JCM.00977-15. http://dx.doi.org/10.1128/JCM.00977-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mirande C, Canard I, Buffet Croix Blanche S, Charrier JP, van Belkum A, Welker M, Chatellier S. Rapid detection of carbapenemase activity: benefits and weaknesses of MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2015;34(11):2225–34. doi: 10.1007/s10096-015-2473-z. http://dx.doi.org/10.1007/s10096-015-2473-z. [DOI] [PubMed] [Google Scholar]
- 70.Chong PM, McCorrister SJ, Unger MS, Boyd DA, Mulvey MR, Westmacott GR. MALDI-TOF MS detection of carbapenemase activity in clinical isolates of Enterobacteriaceae spp., Pseudomonas aeruginosa, and Acinetobacter baumannii compared against the Carba-NP assay. J Microbiol Methods. 2015;111:21–3. doi: 10.1016/j.mimet.2015.01.024. http://dx.doi.org/10.1016/j.mimet.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 71.McMullen AR, Yarbrough ML, Wallace MA, Shupe A, Burnham CD. Evaluation of genotypic and phenotypic methods to detect carbapenemase production in gram-negative Bacilli. Clin Chem. 2017;63(3):723–30. doi: 10.1373/clinchem.2016.264804. http://dx.doi.org/10.1373/clinchem.2016.264804. [DOI] [PubMed] [Google Scholar]
- 72.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK. Emerging infections program healthcare-associated, T. antimicrobial use prevalence survey. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–208. doi: 10.1056/NEJMoa1306801. http://dx.doi.org/10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thaden JT, Park LP, Maskarinec SA, Ruffin F, Fowler VG, Jr, van Duin D. Results from a 13-year prospective cohort study show increased mortality associated with bloodstream infections caused by Pseudomonas aeruginosa compared to other bacteria. Antimicrob Agents Chemother. 2017;61(6) doi: 10.1128/AAC.02671-16. pii:e02671-16. http://dx.doi.org/10.1128/AAC.02671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang CI, Kim SH, Kim HB, Park SW, Choe YJ, Oh MD, Kim EC, Choe KW. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis. 2003;37(6):745–51. doi: 10.1086/377200. http://dx.doi.org/10.1086/377200. [DOI] [PubMed] [Google Scholar]
- 75.Tofas P, Samarkos M, Piperaki ET, Kosmidis C, Triantafyllopoulou ID, Kotsopoulou M, Pantazatou A, Perlorentzou S, Poulli A, Vagia M, Daikos GL. Pseudomonas aeruginosa bacteraemia in patients with hematologic malignancies: risk factors, treatment and outcome. Diagn Microbiol Infect Dis. 2017;88(4):335–41. doi: 10.1016/j.diagmicrobio.2017.05.003. http://dx.doi.org/10.1016/j.diagmicrobio.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 76.Pena C, Suarez C, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodriguez-Bano J, Rodriguez F, Tubau F, Oliver A, Martinez-Martinez L D. Spanish Network for Research in Infectious. Effect of adequate single-drug vs combination antimicrobial therapy on mortality in Pseudomonas aeruginosa bloodstream infections: a post Hoc analysis of a prospective cohort. Clin Infect Dis. 2013;57(2):208–16. doi: 10.1093/cid/cit223. http://dx.doi.org/10.1093/cid/cit223. [DOI] [PubMed] [Google Scholar]
- 77.Weber DJ, Rutala WA, Sickbert-Bennett EE, Samsa GP, Brown V, Niederman MS. Microbiology of ventilator-associated pneumonia compared with that of hospital-acquired pneumonia. Infect Control Hosp Epidemiol. 2007;28(7):825–31. doi: 10.1086/518460. http://dx.doi.org/10.1086/518460. [DOI] [PubMed] [Google Scholar]
- 78.Koulenti D, Tsigou E, Rello J. Nosocomial pneumonia in 27 ICUs in Europe: perspectives from the EU-VAP/CAP study. Eur J Clin Microbiol Infect Dis. 2017;36(11):1999–2006. doi: 10.1007/s10096-016-2703-z. http://dx.doi.org/10.1007/s10096-016-2703-z. [DOI] [PubMed] [Google Scholar]
- 79.Fernandez-Barat L, Ferrer M, De Rosa F, Gabarrus A, Esperatti M, Terraneo S, Rinaudo M, Li Bassi G, Torres A. Intensive care unit-acquired pneumonia due to Pseudomonas aeruginosa with and without multidrug resistance. J Infect. 2017;74(2):142–52. doi: 10.1016/j.jinf.2016.11.008. http://dx.doi.org/10.1016/j.jinf.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 80.Venier AG, Gruson D, Lavigne T, Jarno P, L’Heriteau F, Coignard B, Savey A, Rogues AM R.-R. group. Identifying new risk factors for Pseudomonas aeruginosa pneumonia in intensive care units: experience of the French national surveillance, REA-RAISIN. J Hosp Infect. 2011;79(1):44–8. doi: 10.1016/j.jhin.2011.05.007. http://dx.doi.org/10.1016/j.jhin.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 81.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O’Grady NP, Bartlett JG, Carratala J, El Solh AA, Ewig S, Fey PD, File TM, Jr, Restrepo MI, Roberts JA, Waterer GW, Cruse P, Knight SL, Brozek JL. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5):e61–e111. doi: 10.1093/cid/ciw353. http://dx.doi.org/10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carcas AJ, Garcia-Satue JL, Zapater P, Frias-Iniesta J. Tobramycin penetration into epithelial lining fluid of patients with pneumonia. Clin Pharmacol Ther. 1999;65(3):245–50. doi: 10.1016/S0009-9236(99)70103-7. http://dx.doi.org/10.1016/S0009-9236(99)70103-7. [DOI] [PubMed] [Google Scholar]
- 83.Levy J, Baran D, Klastersky J. Comparative study of the antibacterial activity of amikacin and tobramycin during Pseudomonas pulmonary infection in patients with cystic fibrosis. J Antimicrob Chemother. 1982;10(3):227–34. doi: 10.1093/jac/10.3.227. http://dx.doi.org/10.1093/jac/10.3.227. [DOI] [PubMed] [Google Scholar]
- 84.Slekovec C, Robert J, Trystram D, Delarbre JM, Merens A, van der Mee-Marquet N, de Gialluly C, Costa Y, Caillon J, Hocquet D, Bertrand X ONERBA. Pseudomonas aeruginosa in French hospitals between 2001 and 2011: back to susceptibility. Eur J Clin Microbiol Infect Dis. 2014;33(10):1713–7. doi: 10.1007/s10096-014-2125-8. http://dx.doi.org/10.1007/s10096-014-2125-8. [DOI] [PubMed] [Google Scholar]
- 85.Torres A, Serra-Batlles J, Ferrer A, Jimenez P, Celis R, Cobo E, Rodriguez-Roisin R. Severe community-acquired pneumonia. Epidemiology and prognostic factors. Am Rev Respir Dis. 1991;144(2):312–8. doi: 10.1164/ajrccm/144.2.312. http://dx.doi.org/10.1164/ajrccm/144.2.312. [DOI] [PubMed] [Google Scholar]
- 86.Cilloniz C, Polverino E, Ewig S, Aliberti S, Gabarrus A, Menendez R, Mensa J, Blasi F, Torres A. Impact of age and comorbidity on cause and outcome in community-acquired pneumonia. Chest. 2013;144(3):999–1007. doi: 10.1378/chest.13-0062. http://dx.doi.org/10.1378/chest.13-0062. [DOI] [PubMed] [Google Scholar]
- 87.Rello J, Bodi M, Mariscal D, Navarro M, Diaz E, Gallego M, Valles J. Microbiological testing and outcome of patients with severe community-acquired pneumonia. Chest. 2003;123(1):174–80. doi: 10.1378/chest.123.1.174. http://dx.doi.org/10.1378/chest.123.1.174. [DOI] [PubMed] [Google Scholar]
- 88.Paganin F, Lilienthal F, Bourdin A, Lugagne N, Tixier F, Genin R, Yvin JL. Severe community-acquired pneumonia: assessment of microbial aetiology as mortality factor. Eur Respir J. 2004;24(5):779–85. doi: 10.1183/09031936.04.00119503. http://dx.doi.org/10.1183/09031936.04.00119503. [DOI] [PubMed] [Google Scholar]
- 89.Yoshimoto A, Nakamura H, Fujimura M, Nakao S. Severe community-acquired pneumonia in an intensive care unit: risk factors for mortality. Intern Med. 2005;44(7):710–6. doi: 10.2169/internalmedicine.44.710. http://dx.doi.org/10.2169/internalmedicine.44.710. [DOI] [PubMed] [Google Scholar]
- 90.Cilloniz C, Gabarrus A, Ferrer M, Puig de la Bellacasa J, Rinaudo M, Mensa J, Niederman MS, Torres A. Community-acquired pneumonia due to multidrug- and non-multidrug-resistant Pseudomonas aeruginosa. Chest. 2016;150(2):415–25. doi: 10.1016/j.chest.2016.03.042. http://dx.doi.org/10.1016/j.chest.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 91.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM, Jr, Musher DM, Niederman MS, Torres A, Whitney CG A. Infectious Diseases Society of, S. American Thoracic. American Thoracic, S. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–72. doi: 10.1086/511159. http://dx.doi.org/10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arancibia F, Bauer TT, Ewig S, Mensa J, Gonzalez J, Niederman MS, Torres A. Community-acquired pneumonia due to gram-negative bacteria and Pseudomonas aeruginosa: incidence, risk, and prognosis. Arch Intern Med. 2002;162(16):1849–58. doi: 10.1001/archinte.162.16.1849. http://dx.doi.org/10.1001/archinte.162.16.1849. [DOI] [PubMed] [Google Scholar]
- 93.Lees CW, Heys D, Ho GT, Noble CL, Shand AG, Mowat C, Boulton-Jones R, Williams A, Church N, Satsangi J, Arnott ID Scottish Society of Gastroenterology Infliximab Group. A retrospective analysis of the efficacy and safety of infliximab as rescue therapy in acute severe ulcerative colitis. Aliment Pharmacol Ther. 2007;26(3):411–9. doi: 10.1111/j.1365-2036.2007.03383.x. http://dx.doi.org/10.1111/j.1365-2036.2007.03383.x. [DOI] [PubMed] [Google Scholar]
- 94.Lamas Ferreiro JL, Alvarez Otero J, Gonzalez Gonzalez L, Novoa Lamazares L, Arca Blanco A, Bermudez Sanjurjo JR, Rodriguez Conde I, Fernandez Soneira M, de la Fuente Aguado J. Pseudomonas aeruginosa urinary tract infections in hospitalized patients: mortality and prognostic factors. PLoS One. 2017;12(5):e0178178. doi: 10.1371/journal.pone.0178178. http://dx.doi.org/10.1371/journal.pone.0178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sarkar S, Patra AK, Mondal M. Ecthyma gangrenosum in the periorbital region in a previously healthy immunocompetent woman without bacteremia. Indian Dermatol Online J. 2016;7(1):36–9. doi: 10.4103/2229-5178.174326. http://dx.doi.org/10.4103/2229-5178.174326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stevens DL, Bisno AL, Chambers HF, Dellinger EP, Goldstein EJ, Gorbach SL, Hirschmann JV, Kaplan SL, Montoya JG, Wade JC Infectious Diseases Society of America. Practice guidelines for the diagnosis and management of skin and soft tissue infections: 2014 update by the Infectious Diseases Society of America. Clin Infect Dis. 2014;59(2):e10–52. doi: 10.1093/cid/ciu444. http://dx.doi.org/10.1093/cid/ciu444. [DOI] [PubMed] [Google Scholar]
- 97.Beasley K, Panach K, Dominguez AR. Disseminated Candida tropicalis presenting with Ecthyma-Gangrenosum-like Lesions. Dermatol Online J. 2016;22(1) pii:13030/qt7vg4n68j. [PubMed] [Google Scholar]
- 98.Khalil BA, Baillie CT, Kenny SE, Lamont GL, Turnock RR, Pizer BL, van Saene HF, Losty PD. Surgical strategies in the management of ecthyma gangrenosum in paediatric oncology patients. Pediatr Surg Int. 2008;24(7):793–7. doi: 10.1007/s00383-008-2159-z. http://dx.doi.org/10.1007/s00383-008-2159-z. [DOI] [PubMed] [Google Scholar]
- 99.Azzopardi EA, Azzopardi E, Camilleri L, Villapalos J, Boyce DE, Dziewulski P, Dickson WA, Whitaker IS. Gram negative wound infection in hospitalised adult burn patients – systematic review and metanalysis. PLoS One. 2014;9(4):e95042. doi: 10.1371/journal.pone.0095042. http://dx.doi.org/10.1371/journal.pone.0095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mayhall CG. The epidemiology of burn wound infections: then and now. Clin Infect Dis. 2003;37(4):543–50. doi: 10.1086/376993. http://dx.doi.org/10.1086/376993. [DOI] [PubMed] [Google Scholar]
- 101.Williams FN, Herndon DN, Hawkins HK, Lee JO, Cox RA, Kulp GA, Finnerty CC, Chinkes DL, Jeschke MG. The leading causes of death after burn injury in a single pediatric burn center. Crit Care. 2009;13(6):R183. doi: 10.1186/cc8170. http://dx.doi.org/10.1186/cc8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McManus AT, Mason AD, Jr, McManus WF, Pruitt BA., Jr Twenty-five year review of Pseudomonas aeruginosa bacteremia in a burn center. Eur J Clin Microbiol. 1985;4(2):219–23. doi: 10.1007/BF02013601. [DOI] [PubMed] [Google Scholar]
- 103.Gang RK, Bang RL, Sanyal SC, Mokaddas E, Lari AR. Pseudomonas aeruginosa septicaemia in burns. Burns. 1999;25(7):611–6. doi: 10.1016/s0305-4179(99)00042-x. http://dx.doi.org/10.1016/S0305-4179(99)00042-X. [DOI] [PubMed] [Google Scholar]
- 104.Estahbanati HK, Kashani PP, Ghanaatpisheh F. Frequency of Pseudomonas aeruginosa serotypes in burn wound infections and their resistance to antibiotics. Burns. 2002;28(4):340–8. doi: 10.1016/s0305-4179(02)00024-4. http://dx.doi.org/10.1016/S0305-4179(02)00024-4. [DOI] [PubMed] [Google Scholar]
- 105.Norbury W, Herndon DN, Tanksley J, Jeschke MG, Finnerty CC. Infection in burns. Surg Infect (Larchmt) 2016;17(2):250–5. doi: 10.1089/sur.2013.134. http://dx.doi.org/10.1089/sur.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garnacho-Montero J, Sa-Borges M, Sole-Violan J, Barcenilla F, Escoresca-Ortega A, Ochoa M, Cayuela A, Rello J. Optimal management therapy for Pseudomonas aeruginosa ventilator-associated pneumonia: an observational, multicenter study comparing monotherapy with combination antibiotic therapy. Crit Care Med. 2007;35(8):1888–95. doi: 10.1097/01.CCM.0000275389.31974.22. http://dx.doi.org/10.1097/01.CCM.0000275389.31974.22. [DOI] [PubMed] [Google Scholar]
- 107.Roberts JA, Paul SK, Akova M, Bassetti M, De Waele JJ, Dimopoulos G, Kaukonen KM, Koulenti D, Martin C, Montravers P, Rello J, Rhodes A, Starr T, Wallis SC, Lipman J DALI Study. DALI: defining antibiotic levels in intensive care unit patients: are current beta-lactam antibiotic doses sufficient for critically ill patients? Clin Infect Dis. 2014;58(8):1072–83. doi: 10.1093/cid/ciu027. http://dx.doi.org/10.1093/cid/ciu027. [DOI] [PubMed] [Google Scholar]
- 108.MacVane SH, Kuti JL, Nicolau DP. Clinical pharmacodynamics of antipseudomonal cephalosporins in patients with ventilator-associated pneumonia. Antimicrob Agents Chemother. 2014;58(3):1359–64. doi: 10.1128/AAC.01463-13. http://dx.doi.org/10.1128/AAC.01463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Crandon JL, Bulik CC, Kuti JL, Nicolau DP. Clinical pharmacodynamics of cefepime in patients infected with Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54(3):1111–6. doi: 10.1128/AAC.01183-09. http://dx.doi.org/10.1128/AAC.01183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vardakas KZ, Voulgaris GL, Maliaros A, Samonis G, Falagas ME. Prolonged versus short-term intravenous infusion of antipseudomonal beta-lactams for patients with sepsis: a systematic review and meta-analysis of randomised trials. Lancet Infect Dis. 2018;18(1):108–20. doi: 10.1016/S1473-3099(17)30615-1. http://dx.doi.org/10.1016/S1473-3099(17)30615-1. [DOI] [PubMed] [Google Scholar]
- 111.Wright H, Bonomo RA, Paterson DL. New agents for the treatment of infections with Gram-negative bacteria: restoring the miracle or false dawn? Clin Microbiol Infect. 2017;23(10):704–12. doi: 10.1016/j.cmi.2017.09.001. http://dx.doi.org/10.1016/j.cmi.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 112.Zhanel GG, Chung P, Adam H, Zelenitsky S, Denisuik A, Schweizer F, Lagace-Wiens PR, Rubinstein E, Gin AS, Walkty A, Hoban DJ, Lynch JP, III, Karlowsky JA. Ceftolozane/tazobactam: a novel cephalosporin/beta-lactamase inhibitor combination with activity against multidrug-resistant gram-negative bacilli. Drugs. 2014;74(1):31–51. doi: 10.1007/s40265-013-0168-2. http://dx.doi.org/10.1007/s40265-013-0168-2. [DOI] [PubMed] [Google Scholar]
- 113.Solomkin J, Hershberger E, Miller B, Popejoy M, Friedland I, Steenbergen J, Yoon M, Collins S, Yuan G, Barie PS, Eckmann C. Ceftolozane/tazobactam plus metronidazole for complicated intra-abdominal infections in an era of multidrug resistance: results from a randomized, double-blind, phase 3 trial (ASPECT-cIAI) Clin Infect Dis. 2015;60(10):1462–71. doi: 10.1093/cid/civ097. http://dx.doi.org/10.1093/cid/civ097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wagenlehner FM, Umeh O, Darouiche RO. Ceftolozane-tazobactam versus levofloxacin in urinary tract infection. Lancet. 2015;386(10000):1242. doi: 10.1016/S0140-6736(15)00263-9. http://dx.doi.org/10.1016/S0140-6736(15)00263-9. [DOI] [PubMed] [Google Scholar]
- 115.Haidar G, Philips NJ, Shields RK, Snyder D, Cheng S, Potoski BA, Doi Y, Hao B, Press EG, Cooper VS, Clancy CJ, Nguyen MH. Ceftolozane-Tazobactam for the treatment of multidrug-resistant Pseudomonas aeruginosa infections: clinical effectiveness and evolution of resistance. Clin Infect Dis. 2017;65(1):110–20. doi: 10.1093/cid/cix182. http://dx.doi.org/10.1093/cid/cix182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aktas Z, Kayacan C, Oncul O. In vitro activity of avibactam (NXL104) in combination with beta-lactams against Gram-negative bacteria, including OXA-48 beta-lactamase-producing Klebsiella pneumoniae. Int J Antimicrob Agents. 2012;39(1):86–9. doi: 10.1016/j.ijantimicag.2011.09.012. http://dx.doi.org/10.1016/j.ijantimicag.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 117.Keepers TR, Gomez M, Celeri C, Nichols WW, Krause KM. Bactericidal activity, absence of serum effect, and time-kill kinetics of ceftazidime-avibactam against beta-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58(9):5297–305. doi: 10.1128/AAC.02894-14. http://dx.doi.org/10.1128/AAC.02894-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Castanheira M, Mills JC, Costello SE, Jones RN, Sader HS. Ceftazidime-avibactam activity tested against Enterobacteriaceae isolates from U.S. hospitals (2011 to 2013) and characterization of beta-lactamase-producing strains. Antimicrob Agents Chemother. 2015;59(6):3509–17. doi: 10.1128/AAC.00163-15. http://dx.doi.org/10.1128/AAC.00163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sader HS, Castanheira M, Farrell DJ, Flamm RK, Jones RN. Ceftazidime-avibactam activity when tested against ceftazidime-nonsusceptible Citrobacter spp., Enterobacter spp., Serratia marcescens, and Pseudomonas aeruginosa from Unites States medical centers (2011–2014) Diagn Microbiol Infect Dis. 2015;83(49):389–94. doi: 10.1016/j.diagmicrobio.2015.06.008. http://dx.doi.org/10.1016/j.diagmicrobio.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 120.Torres A, Zhong N, Pachl J, Timsit JF, Kollef M, Chen Z, Song J, Taylor D, Laud PJ, Stone GG, Chow JW. Ceftazidime-avibactam versus meropenem in nosocomial pneumonia, including ventilator-associated pneumonia (REPROVE): a randomised, double-blind, phase 3 non-inferiority trial. Lancet Infect Dis. 2018;18(3):285–95. doi: 10.1016/S1473-3099(17)30747-8. http://dx.doi.org/10.1016/S1473-3099(17)30747-8. [DOI] [PubMed] [Google Scholar]
- 121.Blizzard TA, Chen H, Kim S, Wu J, Bodner R, Gude C, Imbriglio J, Young K, Park YW, Ogawa A, Raghoobar S, Hairston N, Painter RE, Wisniewski D, Scapin G, Fitzgerald P, Sharma N, Lu J, Ha S, Hermes J, Hammond ML. Discovery of MK-7655, a beta-lactamase inhibitor for combination with Primaxin(R) Bioorg Med Chem Lett. 2014;24(3):780–5. doi: 10.1016/j.bmcl.2013.12.101. http://dx.doi.org/10.1016/j.bmcl.2013.12.101. [DOI] [PubMed] [Google Scholar]
- 122.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Landman D, Quale J. Activity of imipenem with relebactam against gram-negative pathogens from New York City. Antimicrob Agents Chemother. 2015;59(8):5029–31. doi: 10.1128/AAC.00830-15. http://dx.doi.org/10.1128/AAC.00830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thaden JT, Pogue JM, Kaye KS. Role of newer and re-emerging older agents in the treatment of infections caused by carbapenem-resistant Enterobacteriaceae. Virulence. 2017;8(4):403–16. doi: 10.1080/21505594.2016.1207834. http://dx.doi.org/10.1080/21505594.2016.1207834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lucasti C, Vasile L, Sandesc D, Venskutonis D, McLeroth P, Lala M, Rizk ML, Brown ML, Losada MC, Pedley A, Kartsonis NA, Paschke A. Phase 2, Dose-Ranging study of relebactam with imipenem-cilastatin in subjects with complicated intra-abdominal infection. Antimicrob Agents Chemother. 2016;60(10):6234–43. doi: 10.1128/AAC.00633-16. http://dx.doi.org/10.1128/AAC.00633-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sims M, Mariyanovski V, McLeroth P, Akers W, Lee YC, Brown ML, Du J, Pedley A, Kartsonis NA, Paschke A. Prospective, randomized, double-blind, Phase 2 dose-ranging study comparing efficacy and safety of imipenem/cilastatin plus relebactam with imipenem/cilastatin alone in patients with complicated urinary tract infections. J Antimicrob Chemother. 2017;72(9):2616–26. doi: 10.1093/jac/dkx139. http://dx.doi.org/10.1093/jac/dkx139. [DOI] [PubMed] [Google Scholar]