Summary
Characterization of the function of stress‐related genes helps to understand the mechanisms of plant responses to environmental conditions. The findings of this work defined the role of the wheat TaHDZipI‐5 gene, encoding a stress‐responsive homeodomain–leucine zipper class I (HD‐Zip I) transcription factor, during the development of plant tolerance to frost and drought. Strong induction of TaHDZipI‐5 expression by low temperatures, and the elevated TaHDZipI‐5 levels of expression in flowers and early developing grains in the absence of stress, suggests that TaHDZipI‐5 is involved in the regulation of frost tolerance at flowering. The TaHDZipI‐5 protein behaved as an activator in a yeast transactivation assay, and the TaHDZipI‐5 activation domain was localized to its C‐terminus. The TaHDZipI‐5 protein homo‐ and hetero‐dimerizes with related TaHDZipI‐3, and differences between DNA interactions in both dimers were specified at 3D molecular levels. The constitutive overexpression of TaHDZipI‐5 in bread wheat significantly enhanced frost and drought tolerance of transgenic wheat lines with the appearance of undesired phenotypic features, which included a reduced plant size and biomass, delayed flowering and a grain yield decrease. An attempt to improve the phenotype of transgenic wheat by the application of stress‐inducible promoters with contrasting properties did not lead to the elimination of undesired phenotype, apparently due to strict spatial requirements for TaHDZipI‐5 overexpression.
Keywords: 3D protein modelling, abiotic stress, activation domain, phenotypic features, protein homo‐ and hetero‐dimerization, stress‐inducible promoters
Introduction
Drought and frost are significant limitations to plant growth and development and substantially decrease crop yields globally, including in Australia. Depending on seasonal conditions, a sudden frost at flowering can be a major cause of wheat and barley grain yield losses. Overnight frost events during flowering will damage the sensitive reproductive tissues, often resulting in a near‐total loss of grain. If crops are sown late with the aim to avoid productivity losses due to frost, severe yield losses may occur in hot and dry periods at the end of the growing season. In addition, late planting often leads to reduced grain size, yield and quality. The cost to Australian wheat and barley industries caused by frost is estimated to be around AUS$ 360 million in direct and indirect losses annually (GRDC National Frost Initiative, goo.gl/hKWf34). Thus, identification, characterization and application of candidate genes for the molecular breeding of crop acclimation to both frost and drought are of the utmost importance.
Environmental stresses such as frost or drought trigger specific signal transduction pathways, which activate the expression of stress‐responsive genes (Braam et al., 1997; Bray, 1997; Hwang et al., 2002; Tena et al., 2001; Zhu, 2016). Gene expression starts from the modulation of transcription by stress‐related transcription factors (TFs), which regulate a number of physiological processes under stress, including cuticular wax biosynthesis (Aharoni et al., 2004; Bi et al., 2016, 2017; Borisjuk et al., 2014; Seo et al., 2011), stomatal closure (Ren et al., 2010; Tan et al., 2017), reactive oxygen species (ROS) detoxification (Jiang and Deyholos, 2009) and structural alterations in plasma membranes (Pearce, 1999). Manipulation using genes encoding stress‐related TFs offers the possibility to regulate large groups of genes involved in the same physiological processes, and therefore, this intervention draws the attention of plant biotechnologists (Agarwal et al., 2017; Gahlaut et al., 2016; Hrmova and Lopato, 2014).
An attractive target for this approach is the family of homeodomain–leucine zipper (HD‐Zip) TFs, which contains proteins regulating plant development after plants are exposed to environmental stimuli and stresses (Brandt et al., 2014; Harris et al., 2011; Perotti et al., 2017). All HD‐Zip TFs possess a highly conserved homeodomain (HD) and leucine zipper (Zip or LZ) motifs (Ariel et al., 2007; Harris et al., 2016; Mattsson et al., 1992; Ruberti et al., 1991; Schena and Davis, 1992, 1994). An HD is a folded helix‐turn‐helix motif, which contains 60 amino acid residues (Gehring et al., 1990; Laughon and Scott, 1984; Otting et al., 1990) and functions during the recognition of specific DNA sequences (Gehring et al., 1990; Shepherd et al., 1984). LZ, adjacent to HD, participates in dimerization of HD‐Zip TFs by forming a coiled coil structure (Harris et al., 2016; Ruberti et al., 1991; Szilák et al., 1997). Dimerization could affect the affinity of HD‐Zip proteins to specific DNA binding sites and hence potentially regulate the strength of activation of target genes (Chew et al., 2013; Harris et al., 2016; Palena and Gonzalez, 1999; Szilák et al., 1997).
The HD‐Zip family of proteins has been classified into four subfamilies, designated HD‐Zip classes I to IV, based on unique features in the domain structure and specificity of cis‐element binding (Ariel et al., 2007). The members of HD‐Zip class I differ from the other family members by the absence of common domains and/or motifs besides the HD and Zip domains (Ariel et al., 2007; Chan et al., 1998; Mukherjee and Bürglin, 2006; Ponting and Aravind, 1999; Schrick et al., 2004). The members of the HD‐Zip I family recognize a specific 9‐bp pseudo‐palindromic binding site: CAATNATTG (Meijer et al., 1997; Sessa et al., 1993). No obvious requirements for the central nucleotide of the cis‐element have been observed for wheat HD‐Zip I TFs (Harris et al., 2016; Kovalchuk et al., 2016). However, it is not clear how homo‐ or hetero‐dimerization of HD‐Zip I TFs influences DNA binding and the activation of target genes (Chew et al., 2013; Harris et al., 2016; Hrmova and Lopato, 2014).
Homeodomain–leucine zipper class I TFs were isolated from a variety of species such as Arabidopsis thaliana (Ariel et al., 2007; Schena and Davis, 1992), resurrection plant Craterostigma plantagineum (Deng et al., 2002; Frank et al., 1998), sunflower (Cabello and Chan, 2012; Cabello et al., 2012), rice (Agalou et al., 2008), maize (Zhao et al., 2011) and wheat (Harris et al., 2016; Lopato et al., 2006). Some of these TFs have been reported to respond to various abiotic stresses on transcriptional and/or post‐translational levels (Bhattacharjee et al., 2016; Harris et al., 2016; Kovalchuk et al., 2016; Olsson et al., 2004; Wu et al., 2016; Zhao et al., 2014). For instance, transcription of Athb7 and Athb12 from Arabidopsis was induced by elevated levels of abscisic acid (ABA) and by water deficiency (Olsson et al., 2004; Söderman et al., 1996). Transcription of Hahb1 from sunflower was responsive to low temperatures (Cabello et al., 2012), while Hahb4 was activated by desiccation (Dezar et al., 2005a,b). Transcription of wheat TaHDZipI‐2 was not influenced by ABA and was partially suppressed by low temperatures; however, the transactivation activity of the TaHDZipI‐2 protein was strongly increased by the addition of exogenous ABA (Kovalchuk et al., 2016). In contrast, TaHDZipI‐4 and TaHDZipI‐5 were activated by ABA on both transcriptional and post‐translational levels (Harris et al., 2016).
The effect of overexpression of HD‐Zip TFs on the ability of transgenic plants to survive severe stress conditions has also been demonstrated (Bhattacharjee et al., 2016; Cabello et al., 2012; Cabello and Chan, 2012; Kovalchuk et al., 2016; Wu et al., 2016; Zhang et al., 2012). For instance, overexpression of the Oshox22 and Oshox24 genes (HD‐Zip I γ‐clade) in transgenic rice and Arabidopsis led to an increased ABA content and increased sensitivity to drought and high salinity (Bhattacharjee et al., 2016, 2017; Zhang et al., 2012). In contrast, overexpression of the similar ZmHDZ4 gene (HD‐Zip I γ‐clade) from maize in transgenic rice enhanced plant tolerance to drought, despite an increased sensitivity to ABA. ZmHDZ4‐expressing transgenic plants had a lower relative electrolyte leakage, lower malondialdehyde levels and increased proline contents under drought compared to wild‐type (WT) plants (Wu et al., 2016). All of these changes could potentially contribute to enhanced drought tolerance.
Improvement of cold/frost tolerance was demonstrated only for the representatives of the HD‐Zip I α‐clade. Constitutive overexpression of AtHB13 from Arabidopsis and HaHB1 from sunflower had little influence on the growth and yield of transgenic Arabidopsis, but stabilized cell membrane integrity under cold, drought and high salinity conditions and increased plant stress tolerance (Cabello and Chan, 2012; Cabello et al., 2012). Frost tolerance enhancement of transgenic barley seedlings was achieved by constitutive overexpression of TaHDZipI‐2, the wheat orthologue of AtHB13. However, it was accompanied by negative changes in the phenotype of transgenic plants and a significant yield loss compared to those of control plants (Kovalchuk et al., 2016).
In our previous projects, five genes encoding the members of HD‐Zip subfamily I TFs, designated TaHDZipI‐1 to TaHDZipI‐5, were isolated from wheat and partially characterized (Harris et al., 2016; Kovalchuk et al., 2016; Lopato et al., 2006). TaHDZipI‐1 (ζ‐clade) expression was detected in seedlings and mature vegetative tissues, while TaHDZipI‐2 (α‐clade) was predominantly expressed in shoots of seedlings and during early grain development, with no expression detected in mature tissues (Lopato et al., 2006). TaHDZipI‐2 was demonstrated to function as a regulator of plant growth, flowering time and frost tolerance (Kovalchuk et al., 2016). Overexpression of TaHDZipI‐2 in transgenic barley directly or indirectly regulated a number of genes responsible for barley adaptation to cold, vernalization, flowering time and shape of spikes (Kovalchuk et al., 2016). The TaHDZipI‐3 gene (γ‐clade) was initially identified as a close homologue of AtHB7 and AtHB12 from Arabidopsis, and its induction of transcription by drought was demonstrated by Harris et al. (2016). In contrast to the homologous genes from Arabidopsis, TaHDZipI‐3 was not activated by cold and was not able to function as an activator in yeast or in wheat cells. Therefore, the full‐length coding region of this protein provided ideal bait for a yeast 2‐hybrid (Y2H) screen. The screen identified two interacting partners, which were identified to be the monocot‐specific members of the γ‐clade, designated TaHDZipI‐4 and TaHDZipI‐5. In contrast to TaHDZipI‐3, transcription of both monocot‐specific genes was ABA‐dependent and was strongly up‐regulated by both cold and drought (Harris et al., 2016).
This study is directed to identify regulatory genes that could be used for the improvement of frost and drought tolerance in economically important plants, such as wheat and barley. TaHDZipI‐5 was selected for further characterization because it was more strongly induced by cold and drought than the two other genes from the wheat HD‐Zip I γ‐clade (Harris et al., 2016). In this work, we studied the expression levels of TaHDZipI‐5 in a variety of wheat tissues, analysed the TaHDZipI‐5 transactivation properties and revealed the determinants of homo‐ and hetero‐dimerized TaHDZipI‐5 and TaHDZipI‐3 in complex with a defined cis‐element at the 3D molecular level. TaHDZipI‐5 was initially constitutively overexpressed in transgenic wheat, and comparative evaluations of transgenic and WT plants for growth characteristics and yield components, and tolerance to extreme stress conditions were performed. Overexpression of TaHDZipI‐5 significantly improved plant tolerance to both stresses; however, it negatively influenced plant growth and grain yields. Stress‐inducible expression of TaHDZipI‐5 was applied in an attempt to reduce the negative influence of the transgene on plant development, the onset of flowering and yield.
Results
A reconstruction of the phylogenetic relationship of HD‐Zip I γ‐clade proteins
A phylogenetic tree was constructed using sequences of HD‐Zip I γ‐clade proteins from the dicot model plant Arabidopsis and from several monocots including sorghum, rice, maize and wheat. Protein sequences were either derived from a previous study (Henriksson et al., 2005) or were taken from NCBI databases and compared with the translated sequence of TaHDZipI‐5 (Table S2). The phylogenetic tree (Figure 1) shows that TaHDZipI‐5 shares a closer evolutionary relationship with Oshox22 from rice (69% sequence identity) and Zmhdz4 from maize (63% sequence identity), than with other entries in the tree.
Figure 1.
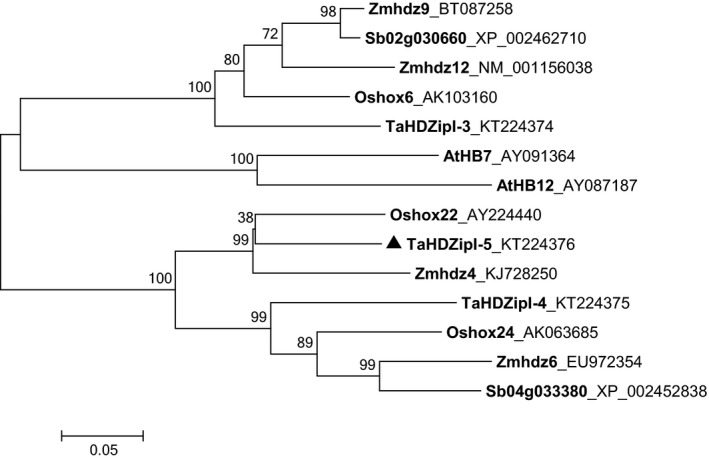
A rectangular phylogenetic tree displaying the evolutionary relationships of HD‐Zip I γ‐clade TFs from Arabidopsis and selected monocots. Abbreviations of species: At, Arabidopsis thaliana; Os, Oryza sativa; Sb, Sorghum bicolor; Ta, Triticum aestivum; Zm, Zea mays.
Endogenous TaHDZipI‐5 expression in a variety of unstressed bread wheat tissues
Expression of the endogenous TaHDZipI‐5 gene was analysed in a variety of tissues of unstressed wheat plants. TaHDZipI‐5 had the highest expression level in endosperm (Figure 2a). Additionally, high expression levels were found in roots and reproductive plant tissues sampled around fertilization. The lowest expression levels of TaHDZipI‐5 were detected in coleoptiles; these were about 30‐fold lower than those in the endosperm.
Figure 2.
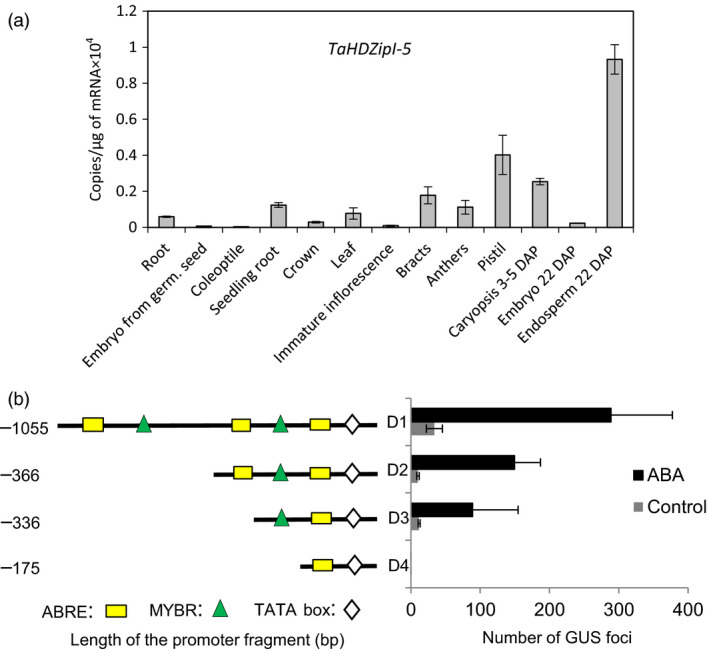
Characterization of the TaHDZipI‐5 gene. (a) Transcript numbers of the TaHDZipI‐5 gene in wheat tissues were estimated by Q‐PCR. (b) Mapping of cis‐elements responsible for the abscisic acid (ABA)‐dependent activation of the TdHDZipI‐5A promoter, using a transient expression assay in wheat cell culture. Depicted is a schematic representation of ABA‐responsive element (ABRE) and MYB responsive (MYBR) cis‐elements in four promoter deletions (D1–D4) of the TdHDZipI‐5A promoter, and the graph shows activation of GUS expression by the deletions detected in a transient expression assay, in the presence (black bars) or absence (control; grey bars) of 0.5 mm ABA in the culture medium. Error bars were calculated from three technical replicates.
Functional cis‐elements responsible for ABA‐dependent TdHDZipI‐5 promoter activation
Promoter sequences of two homeologous genes, TdHDZipI‐5A and TdHDZipI‐5B, were isolated from a durum wheat BAC library (Cenci et al., 2003), because the respective bread wheat sequences were not yet available at the time when this work commenced. The comparison of durum wheat promoter sequences (TdHDZipI‐5A and TdHDZipI‐5B) with corresponding sequences from bread wheat, identified in the Whole Genome Reference Assembly Pseudomolecules v1.0 databases of the International Wheat Genome Sequencing Consortium (IWGSC), revealed more than 99% sequence identity in a region containing functional cis‐elements (Figure S3). Sequences of the promoters (each approximately 1300 bp long) were aligned using LALIGN (Huang and Miller, 1991) to find the best local alignments (Figure S4). Several ABRE and MYB responsive elements were predicted in conserved positions in both TdHDZipI‐5 promoter regions (PLACE software; Higo et al., 1999). TdHDZipI‐5A promoter deletions were generated based on putative cis‐acting elements at −1055, −366, −336 and −175 bp positions, and these were named D1, D2, D3 and D4 (Figures 2b and S4). To define the functional cis‐elements, 0.5 mm ABA was used to induce the activation of the GUS reporter gene by four promoter deletions in a transient expression assay performed in cultured wheat cells. Transformation with D1, D2 and D3 led to step‐by‐step decreasing numbers of GUS foci, while D4 could not activate GUS gene expression (Figure 2b). Therefore, the putative cis‐element responsible for the ABA‐dependent activation of TdHDZipI‐5A is the MYB responsive (MYBR) element GGATA, which is located in the 161‐bp region between D3 (−336 np) and D4 (−175 np), upstream of the transcription initiation site (Figure 2b). Two upstream ABA‐responsive elements (ABREs) and/or one MYBR element enhanced the ABA‐inducible promoter activation.
Identification of the TaHDZipI‐5 activation domain using an in‐yeast activation assay
Results of the in‐yeast activation assay showed that the yeast strains carrying pGBKT7‐TaHDZipI‐5 grew well on SD/‐Trp medium (confirming transformation) and the SD/‐Trp/‐His medium containing 5 mm 3‐AT (confirming the yeast HIS3 reporter gene activation by a plant activation domain‐AD) (Figure 3a). To locate the position of the TaHDZipI‐5 AD, different truncated variants of the TaHDZipI‐5 protein were tested with the in‐yeast activation assay. The empty pGBKT7 vector was transformed into yeast and used as a negative control. After 4 days of cultivation, the yeast carrying the 26–255 residue fragment grew well on SD/‐Trp/‐His medium, while the yeast carrying the 1–229 aa and 1–214 residue fragments were not able to grow on the selective medium, suggesting that the putative AD localizes to the C‐terminal part of the protein (amino acid residues 229–255) (Figure 3a). The identified AD region is represented by a C‐terminal sequence that is conserved in TaHDZipI‐5 homologues from other monocot plants (Figure 3b).
Figure 3.
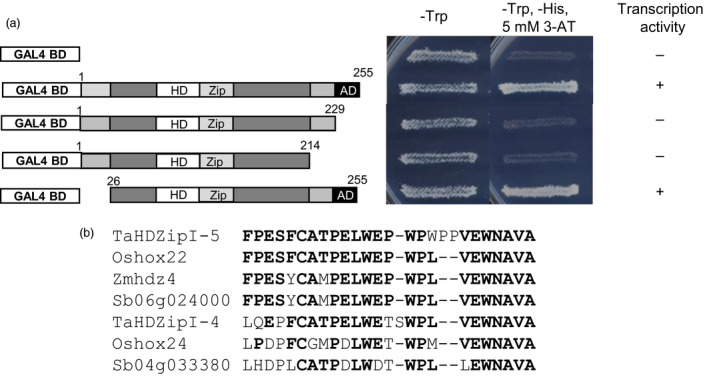
Identification of the TaHDZipI‐5 activation domain (AD) using an in‐yeast activation assay. (a) Transcription activity determined through the in‐yeast activation assay, where homeodomain (HD), Zip and AD designate putative homeodomain, leucine zipper and AD, respectively. Amino acid residues at the beginning and end of each truncated protein are indicated with numbers. (b) Conserved sequences of the identified AD in TaHDZipI‐5 and close homologues from other monocots. Ta, Triticum aestivum; Zm, Zea mays; Os, Oryza sativa; Sb, Sorghum bicolor. Amino acid residues, which are the same in more than the half of the investigated sequences, are in bold.
Based on molecular model predictions of TaHDZipI‐5, homo‐dimerization and hetero‐dimerization influence the DNA binding specificity
Harris et al. (2016) have recently shown that the expression levels of TaHDZipI‐4 and TaHDZipI‐5 increased under cyclic drought conditions, while those of TaHDZipI‐3 remained low. These authors proposed a model explaining how TaHDZipI‐3 binds to DNA cis‐elements in a homo‐dimeric form and that a hetero‐dimeric form of TaHDZipI‐4 and TaHDZipI‐5 (also members of the class I γ‐clade HDs) would initiate a stress response. In the current work, we compared 3D models of homo‐dimeric TaHDZipI‐3 and TaHDZipI‐5, and of hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5, in complex with a HDZ1 cis‐element to seek whether DNA interactions differed between homo‐ and hetero‐dimeric structural models.
Structural bioinformatic comparisons of wheat TaHDZipI‐3 and TaHDZipI‐5 proteins showed the presence of HD domains with well‐defined boundaries. An alignment of the template (PDB: 1JGG) used for structural modelling and TaHDZipI‐3 and TaHDZipI‐5 indicated that there was not a high level of sequence identity between the investigated proteins (Figure 4, top panel). HDs of TaHDZipI‐3 shared respective 26% identity and 53% similarity to the template (PDB: 1JGG), while TaHDZipI‐3 and TaHDZipI‐5 between themselves shared 31% identity and 52% similarity. The positions of 15 identical residues between the template and target sequences of HDs (Figure 4a) indicated which residues might participate in DNA binding. Secondary structure element predictions in TaHDZipI‐3 and TaHDZipI‐5 indicated the presence of three α‐helices (marked as ‘h’ in Figure 4, top panel) that carried most of these identical residues.
Figure 4.
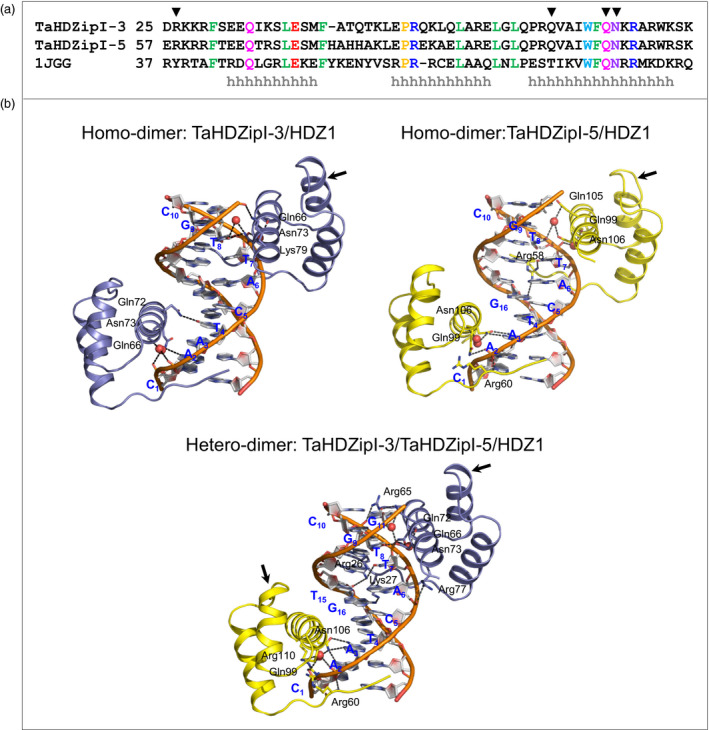
Molecular features of homeodomains (HDs) of TaHDZipI‐3 and TaHDZipI‐5 in homo‐ and hetero‐dimeric forms in complex with the HDZ1 cis‐element. (a) A sequence alignment of TaHDZipI‐3 and TaHDZipI‐5 HDs and of even‐skipped HD from Drosophila melanogaster (PDB: 1JGG). Identical amino acid residues are coloured based on their properties. α‐Helical secondary structural elements are indicated with ‘h’ below the sequences. HD residues that interact with DNA cis‐elements are indicated by inverted triangles (▼). (b) Ribbon representations of homo‐dimeric TaHDZipI‐3 and TaHDZipI‐5, and hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5 models in complex with the HDZ1 cis‐element; blue descriptions and atomic colour representations are used for HDZ1. The ribbons of TaHDZipI‐3 and TaHDZipI‐5 are coloured in blue and yellow, respectively. DNA‐interacting residues are shown in sticks, and DNA sugar‐phosphate backbones are coloured in cpk‐orange. Water molecules are shown as red spheres. Interactions (less than 3.5 Å; Table S3) between residues and HDZ1 are shown in black dashed lines. Arrows point to differences in folding of α‐helices between TaHDZipI‐3 and TaHDZipI‐5.
Homo‐dimeric (TaHDZipI‐3/TaHDZipI‐3 or TaHDZipI‐5/TaHDZipI‐5) and hetero‐dimeric (TaHDZipI‐3/TaHDZipI‐5) HD models in complex with HDZ1 (5′‐CAATCATTGC‐3′/5′‐GCAATGATTG‐3′; interacting nucleobases are underlined) (Sali and Blundell, 1993) were constructed to understand the differences in binding of DNA. The structural models of HDs of TaHDZipI‐5 showed the presence of three α‐helices, interconnected with loops similarly to TaHDZipI‐3 (Figure 4, bottom panel); however, minor structural differences were observed between TaHDZipI‐3 and TaHDZipI‐5 (cf. arrows in Figure 4, bottom panel). Data from structural modelling indicated that differences in DNA binding to cis‐elements resulted from differences in the presence of charged and polar residues at the N‐terminus and at the third α‐helix of each HD monomer that contacted DNA at a major groove (Figure 4, bottom panel; Table S3).
More specifically, our molecular analysis showed that Arg58 in TaHDZipI‐5 at the N‐terminus of the first HD monomer bound directly to two nucleobases T7 and G16, while Gln105 and Asn106 contacted the T8 nucleobase indirectly through a water molecule (Figure 4 bottom panel). In addition, Asn106 in TaHDZipI‐5, which corresponds to Asn73 in TaHDZipI‐3, bound to the nucleobase A3. Further, the analyses of HDZ1‐binding modes in TaHDZipI‐3/TaHDZipI‐5 showed that two positively charged residues of the TaHDZipI‐3 monomer at the N‐terminus bound to the nucleobases T7 and T15 via Arg26 and to the nucleobase G16 through Lys27. Additionally, Asn73 at the major groove α‐helix bound to the nucleobase T8, while Gln72 contacted T8 via a water molecule and could also form a hydrogen bond to the nucleobase G11. On the other hand, the interactions of the TaHDZipI‐5 HD monomer with HDZ1 were similar to those of the second HD monomer in the homo‐dimeric form.
A detailed analysis of hydrogen bond patterns in homo‐dimeric (TaHDZipI‐5/TaHDZipI‐5) and hetero‐dimeric (TaHDZipI‐3/TaHDZipI‐5) DNA complexes (Table S3) showed that three nucleobases (A2, T4 and T8) were bound to Asn73 (participating through both monomers) and Gln72 (Harris et al., 2016). Besides, four nucleobases (A3, T7, T8 and G16) were bound to Arg58, Gln105 and Asn106 (participating through both monomers) in homo‐dimeric TaHDZipI‐5. The analysis of interactions in hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5 showed the participation of seven nucleobases (A2, A3, T7, T8, G11, T15 and G16) that were contacted through Arg26, Lys27, Gln72, Asn73 of TaHDZipI‐3 and through Asn106 of TaHDZipI‐5; Arg26 and Gln72 residues formed bidentate interactions with DNA. Free energies of homo‐dimeric TaHDZipI‐5 (290 kcal/mol) and hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5 (244 kcal/mol), calculated through FoldX (Schymkowitz et al., 2005), indicated that hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5 was more stable than its TaHDZipI‐5 homo‐dimeric form.
Evaluation of transgenic wheat plants constitutively expressing TaHDZipI‐5
Initially, transgenic wheat plants (cv. Gladius) were generated using a construct where expression of the transgene TaHDZipI‐5 was driven by a constitutive maize polyubiquitin promoter. Three independent transgenic T1 lines, L1, L2 and L4 containing a single copy of the transgene (Figure S5), were used for characterization of plant phenotypes and yield components under well‐watered conditions. Two of three transgenic lines showed a very similar phenotype to that of WT plants (Figure S6). However, L1 plants were significantly shorter than WT plants; they had a significantly lower seed number per spike and a lower grain yield than WT plants (Figure S6). All transgenic lines showed delay in flowering time compared to that of WT plants (Figure S6).
The T3 progeny of transgenic and WT plants were grown in two large containers with different watering regimes (Figure S1; Table S4). Using pilot experiments described in the Materials and Methods, all three starting T2 lines, L1‐3‐9, L2‐7‐9 and L4‐8‐9 (pUbi‐TaHDZipI‐5), were identified to be homozygous for the transgene. The phenotypic data of transgenic wheat lines were compared to those of WT plants (Figure 5). The data obtained from the well‐watered bin correlated well with those obtained for T1 transgenic plants grown in pots. The progeny of L1‐3‐9 had reduced plant height, less dry biomass, fewer tillers, spikes and seeds, and about 90% lower grain yield (seed weight per plant) than WT plants. The other two lines showed up to a 25%–30% decrease in all parameters compared with those of WT plants, except for flowering time where differences did not exceed 2 days (Figure 5). In contrast to the data obtained under well‐watered conditions, the differences in growth characteristics and yield parameters between progenies of these two lines (L2‐7‐9 and L4‐8‐9) and WT plants were small under drought (Figure 5).
Figure 5.
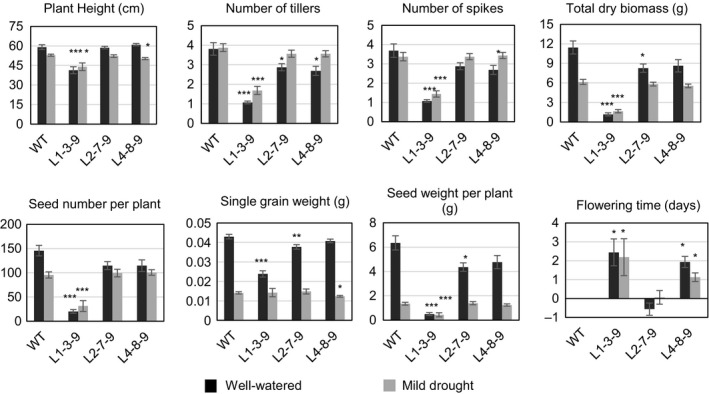
Growth characteristics and yield components of control wild‐type (WT) (Triticum aestivum cv. Gladius) and T3 transgenic wheat transformed with pUbi‐TaHDZipI‐5 under well‐watered (black boxes) and under drought conditions (grey boxes). Flowering time of transgenic plants was compared to the average flowering time of 16 control WT plants, which is represented as day 0. Differences between transgenic lines and WT plants in each of the well‐watered and drought conditions were tested using unpaired Student's t‐tests (*P < 0.05, **P < 0.01, ***P < 0.001).
Drought tolerance (survival) of transgenic wheat seedlings was significantly higher than that of WT plants
Three‐week‐old transgenic wheat seedlings (T2 progenies of sublines L2‐7 and L4‐8 transformed with pUbi‐TaHDZipI‐5, and WT plants, ten plants for each class) were used in three independent drought tolerance experiments. We only examined lines with minimal phenotypic differences compared to those of WT plants. Two control plants and two transgenic plants from each line were planted in the same pot with the aim to minimize influence of differences in seedling sizes and respective differences in water consumption on water availability in the soil. Only 10% of WT plants survived the applied drought conditions and recovered after rewatering (Figure 6a). In contrast, both tested transgenic lines showed a significantly stronger ability to recover than WT plants, with over half of all tested plants surviving (Figure 6a).
Figure 6.
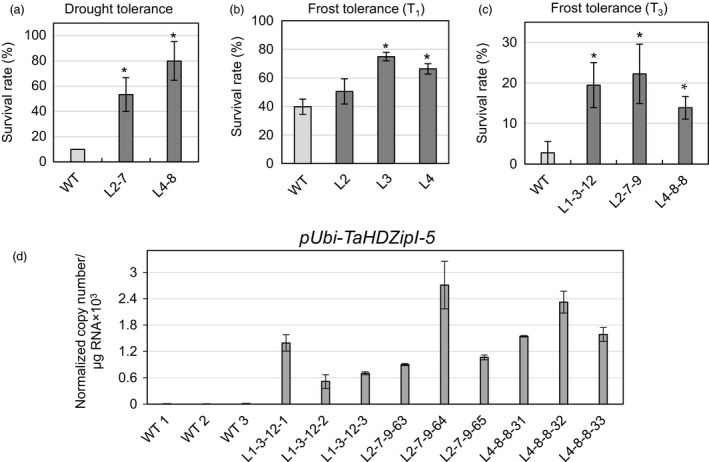
Comparison of drought and frost tolerance of wild‐type (WT) (Triticum aestivum cv. Gladius) and transgenic wheat transformed with pUbi‐TaHDZipI‐5. (a) Drought tolerance of two independent transgenic lines (T2 progeny), sizes of which were similar to those of the control WT plants’, is shown as the survival rate of plants recovered after the terminal drought stress, followed by rewatering. (b, c) Survival rates of plants recovered after seedling‐stage frost: (b) T1 progeny and (c) T3 progeny of transgenic plants. Error bars represent ± SD of three independent experiments. Values represent means ± SE (n varies for each column and is shown in each case directly on the graphs). Significant differences between transgenic lines and WT plants were tested using an unpaired Student's t‐test (*P < 0.05). (d) The expression levels of the TaHDZipI‐5 transgene in leaves of T3 plants in unstressed control WT and transgenic plants, sampled prior to frost treatment. Error bars represent ± SD of three technical replicates.
Transgenic wheat seedlings tolerate frost better than WT plants
Three‐week‐old T1 (Figure 6b) and T3 (Figure 6c) generations of transgenic seedlings were used in frost tolerance tests. Twelve plants of each transgenic line and twelve WT plants were treated in a semi‐automated cold cabinet, using an updated program (Figure S2) based on the earlier established protocol for barley (Kovalchuk et al., 2013). This included 6.5‐h exposure to −7 °C for the T1 generation and the same exposure time to −8 °C for T3 plants; both generations were tested in three independent experiments. All tested transgenic lines showed a higher survival rate than those of WT plants in both experiments (Figure 6b,c). Levels of transgene expression were determined by Q‐PCR using RNA isolated from unstressed wheat leaves collected from transgenic and control plants before frost tolerance tests were conducted (Figure 6d).
Evaluation of transgenic wheat plants with stress‐inducible expression of TaHDZipI‐5
With the aim to avoid or minimize the influence of TaHDZipI‐5 overexpression on the developmental phenotype of transgenic wheat, we replaced the ZmUbiquitin constitutive promoter with one of the stress‐inducible promoters from OsWRKY71 and TdCor39 genes. These stress‐inducible promoters were previously employed to avoid the negative influences of TaDREB3 gene expression on phenotype and yield in transgenic barley (Kovalchuk et al., 2013). Generation of transgenic plants, selection of homozygous lines, assessment of yield components and evaluation of frost tolerance were performed similarly as for transgenic wheat plants transformed with the pUbi‐TaHDZipI‐5 construct (Figures 7 and 8). Surprisingly, cold‐inducible expression of the transgene led to a reduced difference between flowering time of transgenic and WT plants only for the plants with the pWRKY71 promoter, whereas both stress‐inducible promoters, pWRKY71 and pCor39, stabilized the single grain weight in transgenics (Figures 7a and 8a). Although both promoters showed the low levels of basal promoter activity in unstressed transgenic wheat plants (Figures 7b and 8b), many of the negative phenotype and yield characteristics failed to improve, compared to transgenic plants with constitutive transgene expression. Nevertheless, both transgenics demonstrated the substantial enhancement of frost tolerance during the vegetative developmental stage (Figures 7c and 8c).
Figure 7.
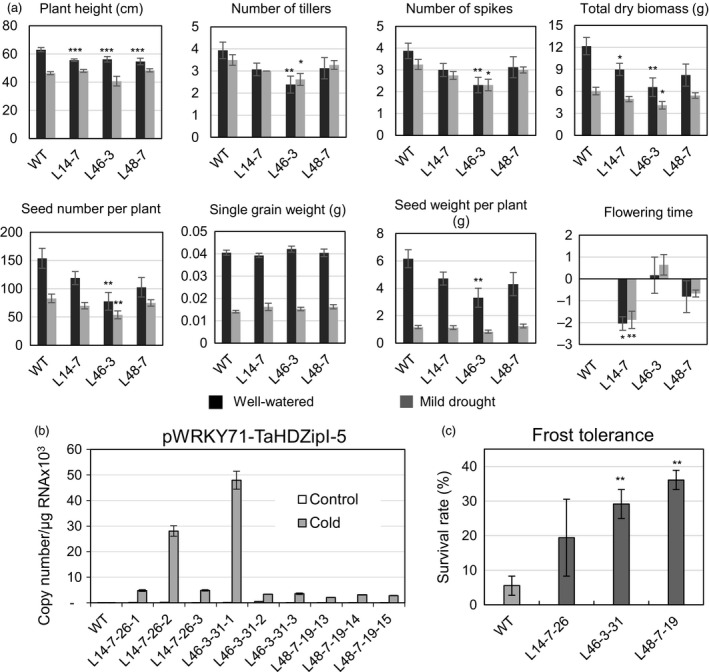
Characteristics of transgenic wheat transformed with pWRKY71‐TaHDZipI‐5. (a) Comparisons of plant growth and yield characteristics of wild‐type (WT) and transgenic T2 plants under well‐watered conditions (black boxes) and mild drought (grey boxes). (b) Transgene expression levels in control WT and transgenic T4 plants at 23 °C (control) and 4 °C (cold). (c) Frost tolerances of WT and transgenic wheat transformed with pWRKY71‐TaHDZipI‐5 are shown as the survival rate of plants recovered after the terminal frost treatment. Error bars represent ± SD for three independent experiments. Differences between transgenic lines and WT plants were tested in the unpaired Student's t‐test (*P < 0.05, **P < 0.01, *** for P < 0.001).
Figure 8.
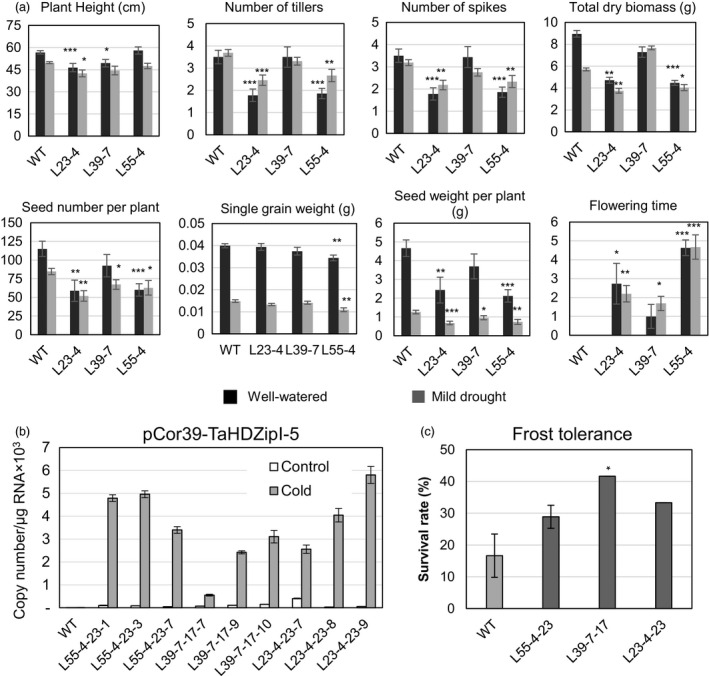
Characteristics of transgenic wheat transformed with pCor39‐TaHDZipI‐5. (a) Comparison of plant growth and yield characteristics of wild‐type (WT) and transgenic T2 plants, under well‐watered conditions (black boxes) and mild drought (grey boxes). (b) Transgene expression levels in WT and transgenic T4 plants at 23 °C (control) and 4 °C (cold). (c) Frost tolerance of WT and transgenic wheat transformed with pCor39‐TaHDZipI‐5 is shown as the survival rate of plants recovered after the terminal frost treatment. Error bars represent ± SD for three independent experiments. Differences between transgenic lines and WT plants were tested in the unpaired Student's t‐test (*P < 0.05, **P < 0.01, *** for P < 0.001).
Discussion
The TaHDZipI‐5 cDNA was isolated in a Y2H screen using TaHDZipI‐3 as bait, from a cDNA library prepared from flag leaves and spikes of Triticum aestivum L. genotype RAC875, subjected to drought and heat stresses. Subsequently, the gene and gene product were characterized at the molecular level (Harris et al., 2016). TaHDZipI‐5 expression was induced by drought, frost and ABA treatment. Based on a close evolutionary relationship with the Arabidopsis γ‐clade of the HD‐Zip I subfamily and conserved intron/exon structure, the TaHDZipI‐5 gene was identified as a monocot‐specific member of the γ‐clade (Harris et al., 2016). The closest homologues of TaHDZipI‐5 from rice and maize, Oshox22 and Zmhdz4, participate in ABA‐mediated drought response, and expression of these genes is up‐regulated by water deficiency (Wu et al., 2016; Zhang et al., 2012).
The analysis of transgenic rice and Arabidopsis plants revealed that constitutive overexpression of Oshox22 and another monocot‐specific γ‐clade member from rice, Oshox24, leads to negative regulation of response to drought/dehydration and, hence, to increased sensitivity of transgenic plants to drought (Bhattacharjee et al., 2016, 2017; Zhang et al., 2012). In contrast, overexpression of the very similar gene from maize, Zmhdz4, positively regulated plant responses to stress in transgenic Arabidopsis and conferred tolerance to drought in transgenic rice (Wu et al., 2016).
Regulation of HD‐Zip I gene expression by low temperatures was demonstrated for Arabidopsis (Cabello et al., 2012), tomato (Zhang et al., 2014), paper mulberry (Peng et al., 2015), sunflower (Cabello et al., 2012), rice (Zhang et al., 2012) and wheat (Harris et al., 2016). The overexpression of α‐clade HD‐Zip class I TFs conferred cold/frost tolerance to transgenic Arabidopsis and barley (Cabello et al., 2012; Kovalchuk et al., 2016); however, the influence on the overexpression of monocot‐specific γ‐clade members on cold or frost tolerance of transgenic plants remains to be determined.
The molecular characterization of the TaHDZipI‐5 gene and its protein product by Harris et al. (2016) and in this work demonstrated its role in wheat tolerance to drought and frost. These studies were conducted through overexpression of the TaHDZipI‐5 gene in transgenic wheat and the evaluation of transgenic plants. We compared phenotypes and yield components of transgenic and control WT wheat plants under optimal growth conditions and under a slowly increasing drought. The ultimate aims of this work were to optimize transgene performance, decrease negative influences of the transgene on plant development using stress‐inducible promoters and select the optimal homozygous lines for field trials in Australian agricultural regions that are prone to seasonal frosts and long periods of drought (annual and monthly potential frost days; http://www.bom.gov.au/jsp/ncc/climate_averages/frost/index.jsp).
The analysis of TaHDZipI‐5 expression in a variety of wheat tissues revealed that the level of this gene was elevated in flowers, developing grain and particularly in the endosperm, a plant tissue that contains increased ABA. In contrast, the number of TaHDZipI‐5 transcripts in vegetative tissues was low (Figure 2a). Expression of TaHDZipI‐5 in flowers shortly before fertilization, and during the early stages of grain development, and the strong induction of TaHDZipI‐5 expression by low temperatures possibly suggest the involvement of this gene in the protection of wheat tissues that are most vulnerable to night frosts.
To understand the function of the TaHDZipI‐5 gene, we isolated gene promoters and revealed DNA‐specific cis‐elements responsible for the ABA‐dependent promoter activation. As the gene/promoter sequences of TaHDZipI‐5 were not available in databases, we analysed the TdHDZipI‐5A and TdHDZipI‐5B promoters of homeologous genes from durum wheat. Firstly, several concentrations of ABA were tested to select the minimal endogenous concentration (0.5 mm) leading to a strong promoter activation (data not shown). Secondly, we analysed the 1055‐bp sequence of the TaHDZipI‐5A promoter (including 5′UTR), which could activate ABA‐dependent TFs. Mapping revealed that the promoter was activated through the proximal MYBR element GGATA (−310 bp from the translation start), which was identified by Baranowskij et al. (1994) as the binding site for MYBSt1, a MYB‐like protein with an endosperm‐related function (Mercy et al., 2003). Additionally, the activity of the TdHDZipI‐5A promoter was enhanced by two ABREs and/or one MYBR elements, situated upstream of the proximal MYBR element (Figures 2b and S3). All mapped cis‐elements were found in the promoters of both homeologous durum genes in conserved positions. Predicted ABRE situated in the D4 fragment of the promoter close to the TATA box was not involved in promoter activation by ABA.
Using a transient expression assay in wheat cells and a reporter construct with synthetic promoter, Harris et al. (2016) demonstrated that TaHDZipI‐5 acted as a transcriptional activator. In this work, the transactivation TaHDZipI‐5 domain was defined in an in‐yeast activation assay. A series of CDS deletions encoding truncated variants of TaHDZipI‐5 were generated, and the constructs were transformed in yeast cells to detect transactivation activity. Similar to Zmhdz4 (Wu et al., 2016) and Oshox22 (Zhang et al., 2012), TaHDZipI‐5 was found to contain an AD at the C‐terminal region of the protein (Figure 3a), which is a highly conserved sequence in homologous proteins from various grasses (Figure 3b).
According to our previous study (Harris et al., 2016), all wheat HD‐Zip I γ‐clade members homo‐dimerize and also interact with other members through hetero‐dimerization. We revealed that TaHDZipI‐5 displayed an equal propensity to form homo‐dimeric TaHDZipI‐5 and hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5 complexes. However, the DNA interaction differences between TaHDZipI‐5 homo‐ and hetero‐dimers remained unclear. Hence, we constructed the 3D models of homo‐dimers of TaHDZipI‐5 or TaHDZipI‐3 and hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5, in complex with HDZ1 cis‐elements, to explore the differences in DNA binding between homo‐ and hetero‐dimeric structures. 3D models showed that DNA interactions in the TaHDZipI‐3/TaHDZipI‐5 hetero‐dimer were more stable than those in the TaHDZipI‐5 homo‐dimer. These models suggested that the TaHDZipI‐3/TaHDZipI‐5 hetero‐dimer is more efficient in binding DNA. This may indicate that the TaHDZipI‐3/TaHDZipI‐5 hetero‐dimer could be more efficient also in vivo, during the activation of target promoters, than the TaHDZipI‐5 homo‐dimer.
Initially, we overexpressed the TaHDZipI‐5 gene in transgenic wheat plants using a constitutive polyubiquitin promoter from maize. Homozygous T1 or T2 sublines were selected in a pilot experiment using T2 and T3 generations, and seeds of selected sublines were used for the analyses of growth characteristics and yield components under sufficient and limited watering (Figure 5). This experiment was performed in large containers to better reflect interplant competition for water, light and nutrients that might occur in the field. We observed that the TaHDZipI‐5 transgene negatively influenced plant phenotypes by decreasing the numbers of tillers and spikes per plant and consequently decreasing the total plant biomass and seed number compared to those of WT plants. Under well‐watered conditions, the differences in all characteristics were significantly higher than under drought (Figure 5). Differences in flowering time resulted in 1‐ to 3‐day delays in T3 sublines compared to the average flowering times of control plants. The differences in flowering times of selected T1 plants amounted up to 2 weeks (Figure S6), but these differences decreased in two subsequent generations of transgenic lines. In contrast, the remainder of growth and yield characteristics in the T1 generation of transgenic plants diverged less, most probably because they represented a mixture of homo‐ and heterozygous plants, including those of null segregants, which were not excluded from this analysis.
Under well‐watered conditions, lines L2 and L4 had more similar phenotypes compared to those of WT plants and L1 in T1 and T3 generations. Thus, these two lines were selected for drought tolerance evaluation, which could be defined as plant's ability to survive severe drought at the vegetative developmental stage. Seedlings of transgenic and control plants were grown and assessed in the same pot, to accommodate differences in seedling sizes. These data, using T2 plants, suggested approximately fivefold to eightfold enhancement of drought tolerance in both transgenic lines compared to control plants (Figure 6a). This observation is in contradiction with that obtained for Oshox22, overexpressed in transgenic rice (Zhang et al., 2012), but correlates with the data obtained for Zmhdz4, overexpressed in transgenic Arabidopsis and rice (Wu et al., 2016). For these discrepancies, we offer the following explanation. Both Zmhdz4 and TaHDZipI‐5 originate from plants which have different physiological responses to drought compared to rice. Therefore, Zmhdz4 and TaHDZipI‐5 may have different biological roles to Oshox22, which might be connected to small, but functionally important differences in protein structure or to differences in spatial or temporal patterns of gene expression. These hypotheses require further investigation.
Frost tolerance experiments were performed once with T1 transgenic plants (Figure 6b) with null segregants identified and excluded from the analysis and twice using T3 homozygous transgenic lines (Figure 6c,d). Both experiments were performed similarly; however, in the second experiment (using T3 homozygous transgenic lines), the minimal incubation temperature was 1 °C lower than that in the first experiment (using T1 plants). This resulted in a lower survival rate of WT plants in the second experiment. An enhancement of frost tolerance was observed in all tested lines in both experiments, confirming that TaHDZipI‐5 is a promising candidate gene for improvement of wheat frost tolerance. The analysis of potential downstream stress‐inducible LEA (late embryogenesis abundant)/COR (cold‐responsive)/DHN (dehydrin) genes in control WT and transgenic lines with the constitutive overexpression of the TaHDZipI‐5 transgene revealed the up‐regulation of TaCOR14B (GenBank: AF207546) and TaRAB15 (GenBank: X59133) transcripts in several transgenic lines (Figure S7). However, this up‐regulation did not correlate with the levels of TaHDZipI‐5 transgene transcripts (Figure 6d).
Constitutive overexpression of TaHDZipI‐5 led to a negative effect of the transgene on the plant phenotype similar to overexpression of Oshox22 in transgenic rice; this also resulted in a stunted phenotype of transgenic plants, a smaller size of plants and fewer tillers (Zhang et al., 2012). To eliminate or decrease the negative effect of the transgene on the phenotype of transgenic wheat lines, the constitutive promoter was replaced with stress‐inducible pWRKY71 and pCor39 promoters, which were previously used to optimize the phenotype of transgenic barley (Kovalchuk et al., 2013). Both promoters are active in vegetative and flowering parts of the plant. The first promoter originates from the rice OsWRKY71 gene, and in barley, it was moderately activated by cold and weakly activated by drought. The activity of the promoter was ABA‐independent, and the basal level of activity in transgenic barley was low. In contrast, the second promoter, which originates from the TdCor39 gene, was strongly activated in barley by frost and drought. TdCor39 was strongly induced by ABA and had a moderate basal level of activity in barley (Kovalchuk et al., 2013).
Transgenic wheat lines with the TaHDZipI‐5 transgene driven by pWRKY71 and pCor39 promoters showed activation of both promoters under low temperature (Figures 7b and 8b), and low levels of transgene expression in the absence of stress. Inducible transgene expression improved single grain weight compared to when the constitutive polyubiquitin promoter was used. Single grain weights did not decrease in plants with either stress‐inducible promoters compared to WT. Differences in flowering times between transgenic and control plants were smaller in plants with the pWRKY71 promoter, compared to those containing pUbi promoter constructs. However, the improvement or elimination of other negative changes in phenotypes of transgenic lines growing under conditions of sufficient watering was not achieved. This disappointing result was somewhat surprising because the levels of pWRKY71 and pCor39 promoter activities in the absence of stress (basal levels) were low. One feasible explanation for these results could be that the application of heterologous promoters led to a ‘poisonous’ effect of the transgene on plant development because of the wrong spatial pattern of TaHDZipI‐5 expression. This problem could be ameliorated by (i) using the native TaHDZipI‐5 promoter boosted by the addition of enhancing elements, so that the spatial activity of the promoter remains unchanged (Chen et al., 2017), (ii) enhancing the efficiency of translation of the native TaHDZipI‐5 gene or (iii) manipulation of native TaHDZipI‐5 protein structure to increase the strength of binding to target cis‐elements or the efficacy of ABA‐dependent protein activation. These options could be explored with the recently developed technology of genome editing using engineered nucleases, without the generation of transgenic plants with an extra copy of the target gene.
Experimental procedures
Plasmid construction and plant transformation
The generation of vectors for plant transformation has been described previously (Eini et al., 2013; Kovalchuk et al., 2013; Morran et al., 2011). Briefly, the 2x35S promoter was excised from the pMDC32 vector (Curtis and Grossniklaus, 2003) and replaced with one of three promoters: the constitutive ZmUbiquitin promoter (Eini et al., 2013), or stress‐inducible OsWRKY71 or TdCor39 promoters (Kovalchuk et al., 2013), resulting in pUbi, pWRKY71 and pCor39 vectors, respectively. A 767‐bp fragment of the TaHDZipI‐5 coding sequence (CDS) (GenBank accession KT224376) (Harris et al., 2016) was isolated from T. aestivum L. cv. RAC875 and cloned into a pENTR‐D‐TOPO vector (Invitrogen, Melbourne, Victoria, Australia). The cloned insert was verified by sequencing and subcloned into the pUbi, pWRKY71 and pCor39 vectors, resulting in pUbi‐TaHDZipI‐5, pWRKY71‐TaHDZipI‐5 and pCor39‐TaHDZipI‐5 constructs.
Constructs were transformed in the Australian elite bread wheat cv. Gladius using a biolistic bombardment method (Ismagul et al., 2014; Kovalchuk et al., 2009). Genomic DNA was isolated from leaf tissue using a freeze‐drying method described by Shavrukov et al. (2010). Transgene integration was confirmed by PCR using a forward primer from the 3′ end of the TaHDZipI‐5 coding region and a reverse primer from the 5′ end of the nos terminator (Table S1). Transgene genomic copy number was estimated in the T0 and/or T1 progenies of selected transgenic lines by quantitative real‐time PCR (Q‐PCR), based on the 2−ΔΔCt method (Kovalchuk et al., 2013; Li et al., 2004). Nos terminator primers and a specific DNA probe (Yadav et al., 2015) were used for transgene amplification, and endogenous Puroindoline‐b (Pin‐b) gene primers and probe (Li et al., 2004; Yadav et al., 2015) were used for template loading normalization (Table S1). T1 lines with a single copy number were selected for further analysis. The selections of homozygous T1 lines were conducted in pilot experiments using the T2 progeny of transgenic T1 lines with two copies of transgene or T3 progeny of T2 lines. In these experiments, transgene integration was assessed by PCR in twelve seedlings of each line. The line was considered as homozygous if the expected PCR product was observed for all twelve plants. Seeds of homozygous T1 (or T2) lines from each construct were selected and used for phenotyping and stress tolerance tests.
Appendix S1 contain the descriptions of gene expression by quantitative real‐time PCR, cloning of promoters and the identification of ABA‐responsive cis‐elements, analysis of evolutionary relationships, in‐yeast activation assay, construction of 3D models, analysis of transgenic plants, drought tolerance tests and survival rates of seedlings under terminal drought, and frost tolerance tests.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Soil water tension monitored at 10 and 30 cm depths in large containers used for plant growth under well‐watered conditions or increasing drought.
Figure S2 Details of frost tolerance experiments.
Figure S3 Alignments of TdHDZipI‐5A and TdHDZipI‐5B promoter sequences and sequences of corresponding genes of Triticum aestivum cv. Chinese Spring, identified in the Whole Genome Reference Assembly Pseudomolecules v1.0 databases of the International Wheat Genome Sequencing Consortium, using the BLAST software (Altschul et al., 1997).
Figure S4 Alignment of TdHDZipI‐5B (5B) and TdHDZipI‐5A (5A) promoters. LALIGN (Huang and Miller, 1991) was used to find the best local alignments.
Figure S5 (a) Transgene copy numbers in T1 transgenic plants estimated by Q‐PCR. Plants seeds used in analyses are indicated by arrows. (b) Examples of selection of homozygous lines by PCR using transgene‐specific primers.
Figure S6 Growth characteristics and yield components of control wild‐type (WT) and transgenic T1 wheat (Triticum aestivum cv. Gladius) plants transformed with pUbi‐TaHDZipI‐5.
Figure S7 Expression levels of three stress‐inducible LEA (Late Embryogenesis Abundant)/COR (cold‐responsive)/DHN (dehydrin) genes (TaWZY2, GenBank: EU395844; TaCOR14B, GenBank: AF207546; TaRab15, GenBank: X59133) and the TaDREB3 (GenBank: DQ353853) regulatory gene, in leaves of unstressed control WT plants and T3 sublines of tree independent transgenic lines.
Table S1 List of PCR primers and DNA probes used in this study.
Table S2 A sequence alignment of 14 entries (with GenBank accession numbers) used to generate a phylogenetic tree displaying the evolutionary relationships of HD‐Zip I γ‐clade TFs from Arabidopsis and selected monocots, shown in Figure 1.
Table S3 Hydrogen bonds of homo‐dimeric TaHDZipI‐3 and TaHDZipI‐5, and hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5 with HDZ1 (5′‐CAATCATTGC‐3′/5′‐GCAATGATTG‐3′).
Table S4 Characteristics of the T2/T3 progenies of TaHDZipI‐5 transgenic lines analysed in large containers under well‐watered or mild drought condition.
Appendix S1 Materials and methods.
Acknowledgements
We acknowledge contributions of William Chew and Ainur Ismagul in plant transformation and growth experiments. We also thank Ursula Langridge, Yuriy Onyskiv, Alex Kovalchuk and Yongle Li for technical support; Margaret Pallotta for the assistance with the BAC library screen, Mario Fruzangohar for bioinformatic analyses, and Julie Hayes for critically reading the manuscript. YY is thankful to the University of Adelaide for a postgraduate scholarship. Contributions of SH, MR and YL were supported by the Australian Research Council Industrial Transforming Research Hub (IH130200027). SLu was funded by Dupont‐Pioneer. This work was supported by the Australian Research Council Linkage Project (LP120100201 to MH and SLo), the Australian Grains Research and Development Corporation, the Government of South Australia, and Dupont‐Pioneer.
Accession numbers: TdHDZipI‐5A promoter‐MF406150; TdHDZipI‐5B promoter‐MF406151.
References
- Agalou, A. , Purwantomo, S. , Overnas, E. , Johannesson, H. , Zhu, X. , Estiati, A. , Kam, R.J.D. et al. (2008) A genome‐wide survey of HD‐Zip genes in rice and analysis of drought‐responsive family members. Plant Mol. Biol. 66, 87–103. [DOI] [PubMed] [Google Scholar]
- Agarwal, P.K. , Gupta, K. , Lopato, S. and Agarwal, P. (2017) Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 68, 2135–2148. [DOI] [PubMed] [Google Scholar]
- Aharoni, A. , Dixit, S. , Jetter, R. , Thoenes, E. , Arkel, G.V. and Pereira, A. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis . Plant Cell, 16, 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel, F.D. , Manavella, P.A. , Dezar, C.A. and Chan, R.L. (2007) The true story of the HD‐Zip family. Trends Plant Sci. 12, 419–426. [DOI] [PubMed] [Google Scholar]
- Baranowskij, N. , Frohberg, C. , Prat, S. and Willmitzer, L. (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 13, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee, A. , Khurana, J.P. and Jain, M. (2016) Characterization of rice homeobox genes, OsHOX22 and OsHOX24, and over‐expression of OsHOX24 in transgenic Arabidopsis suggest their role in abiotic stress response. Front. Plant Sci. 7, 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee, A. , Sharma, R. and Jain, M. (2017) Over‐expression of OsHOX24 confers enhanced susceptibility to abiotic stresses in transgenic rice via modulating stress‐responsive gene expression. Front. Plant Sci. 8, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, H. , Luang, S. , Li, Y. , Bazanova, N. , Morran, S. , Song, Z. , Perera, M.A. et al. (2016) Identification and characterization of wheat drought‐responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 67, 5363–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, H. , Luang, S. , Li, Y. , Bazanova, N. , Borisjuk, N. , Hrmova, M. and Lopato, S. (2017) Wheat drought‐responsive WXPL transcription factors regulate cuticle biosynthesis genes. Plant Mol. Biol. 94, 15–32. [DOI] [PubMed] [Google Scholar]
- Borisjuk, N. , Hrmova, M. and Lopato, S. (2014) Transcriptional regulation of cuticle biosynthesis. Biotechnol. Adv. 32, 526–540. [DOI] [PubMed] [Google Scholar]
- Braam, J. , Sistrunk, M.L. , Polisensky, D.H. , Xu, W. , Purugganan, M.M. , Antosiewicz, D.M. , Campbell, P. et al. (1997) Plant responses to environmental stress: regulation and functions of the Arabidopsis TCH genes. Planta, 203, S35–S41. [DOI] [PubMed] [Google Scholar]
- Brandt, R. , Cabedo, M. , Xie, Y. and Wenkel, S. (2014) Homeodomain leucine‐zipper proteins and their role in synchronizing growth and development with the environment. J. Integr. Plant Biol. 56, 518–526. [DOI] [PubMed] [Google Scholar]
- Bray, E.A. (1997) Plant responses to water deficit. Trends Plant Sci. 2, 48–54. [Google Scholar]
- Cabello, J.V. and Chan, R.L. (2012) The homologous homeodomain–leucine zipper transcription factors HaHB1 and AtHB13 confer tolerance to drought and salinity stresses via the induction of proteins that stabilize membranes. Plant Biotechnol. J. 10, 815–825. [DOI] [PubMed] [Google Scholar]
- Cabello, J.V. , Arce, A.L. and Chan, R.L. (2012) The homologous HD‐Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis‐related and glucanase proteins. Plant J. 69, 141–153. [DOI] [PubMed] [Google Scholar]
- Cenci, A. , Chantret, N. , Kong, X. , Gu, Y. , Anderson, O.D. , Fahima, T. , Distelfeld, A. et al. (2003) Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp durum). Theor. Appl. Genet. 107, 931–939. [DOI] [PubMed] [Google Scholar]
- Chan, R.L. , Gago, G.M. , Palena, C.M. and Gonzalez, D.H. (1998) Homeoboxes in plant development. Biochim. Biophys. Acta, 1442, 1–19. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Fan, X. , Qian, K. , Zhang, Y. , Song, M. , Liu, Y. , Xu, G. et al. (2017) pOsNAR2.1: OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol. J. 15, 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew, W. , Hrmova, M. and Lopato, S. (2013) Role of homeodomain leucine zipper (HD‐Zip) IV transcription factors in plant development and plant protection from deleterious environmental factors. Int. J. Mol. Sci. 14, 8122–8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Phillips, J. , Meijer, A.H. , Salamini, F. and Bartels, D. (2002) Characterization of five novel dehydration‐responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum . Plant Mol. Biol. 49, 601–610. [DOI] [PubMed] [Google Scholar]
- Dezar, C.A. , Fedrigo, G.V. and Chan, R.L. (2005a) The promoter of the sunflower HD‐Zip protein gene Hahb4 directs tissue‐specific expression and is inducible by water stress, high salt concentrations and ABA. Plant Sci. 169, 447–456. [Google Scholar]
- Dezar, C.A. , Gago, G.M. , González, D.H. and Chan, R.L. (2005b) Hahb‐4, a sunflower homeobox‐leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 14, 429–440. [DOI] [PubMed] [Google Scholar]
- Eini, O. , Yang, N. , Pyvovarenko, T. , Pillman, K. , Bazanova, N. , Tikhomirov, N. , Eliby, S. et al. (2013) Complex regulation by Apetala2 domain‐containing transcription factors revealed through analysis of the stress‐responsive TdCor410b promoter from durum wheat. PLoS ONE, 8, e58713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, W. , Phillips, J. , Salamini, F. and Bartels, D. (1998) Two dehydration‐inducible transcripts from the resurrection plant Craterostigma plantagineum encode interacting homeodomain‐leucine zipper proteins. Plant J. 15, 413–421. [DOI] [PubMed] [Google Scholar]
- Gahlaut, V. , Jaiswal, V. , Kumar, A. and Gupta, P.K. (2016) Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Genet. 129, 2019–2042. [DOI] [PubMed] [Google Scholar]
- Gehring, W.J. , Müller, M. , Affolter, M. , Percival‐Smith, A. , Billeter, M. , Qian, Y.Q. , Otting, G. et al. (1990) The structure of the homeodomain and its functional implications. Trends Genet. 6, 323–329. [DOI] [PubMed] [Google Scholar]
- Harris, J.C. , Hrmova, M. , Lopato, S. and Langridge, P. (2011) Modulation of plant growth by HD‐Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 190, 823–837. [DOI] [PubMed] [Google Scholar]
- Harris, J.C. , Sornaraj, P. , Taylor, M. , Bazanova, N. , Baumann, U. , Lovell, B. , Langridge, P. et al. (2016) Molecular interactions of the γ‐clade homeodomain‐leucine zipper class I transcription factors during the wheat response to water deficit. Plant Mol. Biol. 90, 435–452. [DOI] [PubMed] [Google Scholar]
- Henriksson, E. , Olsson, A.S. , Johannesson, H. , Johansson, H. , Hanson, J. , Engström, P. and Söderman, E. (2005) Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 139, 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. and Korenaga, T. (1999) Plant cis‐acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova, M. and Lopato, S. (2014) Enhancing abiotic stress tolerance in plants by modulating properties of stress responsive transcription factors. In Genomics of Plant Genetic Resources ( Tuberosa, R. , Graner, A. and Frison, E. , eds), pp. 291–316. Dordrecht, the Netherlands: Springer. [Google Scholar]
- Huang, X. and Miller, W. (1991) A time‐efficient, linear‐space local similarity algorithm. Adv. Appl. Math. 12, 337–357. [Google Scholar]
- Hwang, I. , Chen, H.C. and Sheen, J. (2002) Two‐component signal transduction pathways in Arabidopsis . Plant Physiol. 129, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismagul, A. , Iskakova, G. , Harris, J.C. and Eliby, S. (2014) Biolistic transformation of wheat with centrophenoxine as a synthetic auxin. Biolistic transformation of wheat with centrophenoxine as a synthetic auxin. Methods Mol. Biol. 1145, 191–202. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. and Deyholos, M.K. (2009) Functional characterization of Arabidopsis NaCl‐inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, N. , Smith, J. , Pallotta, M. , Singh, R. , Ismagul, A. , Eliby, S. , Bazanova, N. et al. (2009) Characterization of the wheat endosperm transfer cell‐specific protein TaPR60. Plant Mol. Biol. 71, 81–98. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, N. , Jia, W. , Eini, O. , Morran, S. , Pyvovarenko, T. , Fletcher, S. , Bazanova, N. et al. (2013) Optimization of TaDREB3 gene expression in transgenic barley using cold‐inducible promoters. Plant Biotechnol. J. 11, 659–670. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, N. , Chew, W. , Sornaraj, P. , Borisjuk, N. , Yang, N. , Singh, R. , Bazanova, N. et al. (2016) The homeodomain transcription factor TaHDZipI‐2 from wheat regulates frost tolerance, flowering time and spike development in transgenic barley. New Phytol. 211, 671–687. [DOI] [PubMed] [Google Scholar]
- Laughon, A. and Scott, M.P. (1984) Sequence of a Drosophila segmentation gene: protein structure homology with DNA‐binding proteins. Nature, 310, 25–31. [DOI] [PubMed] [Google Scholar]
- Li, Z.W. , Hansen, J.L. , Liu, Y. , Zemetra, R.S. and Berger, P.H. (2004) Using real‐time PCR to determine transgene copy number in wheat. Plant Mol. Biol. Rep. 22, 179–188. [Google Scholar]
- Lopato, S. , Bazanova, N. , Morran, S. , Milligan, A.S. , Shirley, N. and Langridge, P. (2006) Isolation of plant transcription factors using a modified yeast one‐hybrid system. Plant Methods, 2, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J. , Söderman, E. , Svenson, M. , Borkird, C. and Engström, P. (1992) A new homeobox‐leucine zipper gene from Arabidopsis thaliana . Plant Mol. Biol. 18, 1019–1022. [DOI] [PubMed] [Google Scholar]
- Meijer, A.H. , Scarpella, E. , Dijk, E.L. , Qin, L. , Taal, A.J. , Rueb, S. , Harrington, S.E. et al. (1997) Transcriptional repression by Oshox1, a novel homeodomain leucine zipper protein from rice. Plant J. 11, 263–276. [DOI] [PubMed] [Google Scholar]
- Mercy, I.S. , Meeley, R.B. , Nichols, S.E. and Olsen, O.A. (2003) Zea mays ZmMybst1 cDNA, encodes a single Myb‐repeat protein with the VASHAQKYF motif. J. Exp. Bot. 54, 1117–1119. [DOI] [PubMed] [Google Scholar]
- Morran, S. , Eini, O. , Pyvovarenko, T. , Parent, B. , Singh, R. , Ismagul, A. , Eliby, S. et al. (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. [DOI] [PubMed] [Google Scholar]
- Mukherjee, K. and Bürglin, T.R. (2006) MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain‐leucine zipper III proteins. Plant Physiol. 140, 1142–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, A. , Engström, P. and Söderman, E. (2004) The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis . Plant Mol. Biol. 55, 663–677. [DOI] [PubMed] [Google Scholar]
- Otting, G. , Qian, Y.Q. , Billeter, M. , Müller, M. , Affolter, M. , Gehring, W.J. and Wüthrich, K. (1990) Protein‐DNA contacts in the structure of a homeodomain‐DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 9, 3085–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palena, C.M. and Gonzalez, D.H. (1999) A monomer–dimer equilibrium modulates the interaction of the sunflower homeodomain leucine‐zipper protein Hahb‐4 with DNA. Biochem. J. 341, 81–87. [PMC free article] [PubMed] [Google Scholar]
- Pearce, R.S. (1999) Molecular analysis of acclimation to cold. Plant Growth Regul. 29, 47–76. [Google Scholar]
- Peng, X. , Wu, Q. , Teng, L. , Tang, F. , Pi, Z. and Shen, S. (2015) Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 15, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotti, M.F. , Ribone, P.A. and Chan, R.L. (2017) Plant transcription factors from the homeodomain‐leucine zipper family I. Role in development and stress responses. IUBMB Life, 69, 280–289. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P. and Aravind, L. (1999) START: a lipid‐binding domain in StAR, HD‐ZIP and signalling proteins. Trends Biochem. Sci. 24, 130–132. [DOI] [PubMed] [Google Scholar]
- Ren, X. , Chen, Z. , Liu, Y. , Zhang, H. , Zhang, M. , Liu, Q. , Hong, X. et al. (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti, I. , Sessa, G. , Lucchetti, S. and Morelli, G. (1991) A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 10, 1787–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali, A. and Blundell, T. (1993) Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Schena, M. and Davis, R.W. (1992) HD‐Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc. Natl Acad. Sci. USA, 89, 3894–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, M. and Davis, R.W. (1994) Structure of homeobox‐leucine zipper genes suggests a model for the evolution of gene families. Proc. Natl Acad. Sci. USA, 91, 8393–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick, K. , Nguyen, D. , Karlowski, W.M. and Mayer, K.F. (2004) START lipid/sterol‐binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz, J.W. , Rousseau, F. , Martins, I.C. , Ferkinghoff‐Borg, J. , Stricher, F. and Serrano, L. (2005) Prediction of water and metal binding sites and their affinities by using the Fold‐X force field. Proc. Natl Acad. Sci. USA, 102, 10147–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, P.J. , Lee, S.B. , Suh, M.C. , Park, M.J. , Go, Y.S. and Park, C.M. (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis . Plant Cell, 23, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa, G. , Morelli, G. and Ruberti, I. (1993) The Athb‐1 and‐2 HD‐Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 12, 3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavrukov, Y. , Gupta, N.K. , Miyazaki, J. , Baho, M.N. , Chalmers, K.J. , Tester, M. , Langridge, P. et al. (2010) HvNax3 – a locus controlling shoot sodium exclusion derived from wild barley (Hordeum vulgare ssp. spontaneum). Funct. Integr. Genomics, 10, 277–291. [DOI] [PubMed] [Google Scholar]
- Shepherd, J.C. , McGinnis, W. , Carrasco, A.E. , De Robertis, E.M. and Gehring, W.J. (1984) Fly and frog homoeo domains show homologies with yeast mating type regulatory proteins. Nature, 310, 70–71. [DOI] [PubMed] [Google Scholar]
- Söderman, E. , Mattsson, J. and Engström, P. (1996) The Arabidopsis homeobox gene ATHB‐7 is induced by water deficit and by abscisic acid. Plant J. 10, 375–381. [DOI] [PubMed] [Google Scholar]
- Szilák, L. , Moitra, J. and Vinson, C. (1997) Design of a leucine zipper coiled coil stabilized 1.4 kcal mol−1 by phosphorylation of a serine in the e position. Protein Sci. 6, 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y. , Li, M. , Yang, Y. , Sun, X. , Wang, N. , Liang, B. and Ma, F. (2017) Overexpression of MpCYS4, a phytocystatin gene from Malus prunifolia (Willd.) Borkh., enhances stomatal closure to confer drought tolerance in transgenic Arabidopsis and apple. Front. Plant Sci. 8, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena, G. , Asai, T. , Chiu, W.L. and Sheen, J. (2001) Plant mitogen‐activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Zhou, W. , Gong, X. and Cheng, B. (2016) Expression of ZmHDZ4, a maize homeodomain‐leucine zipper I gene, confers tolerance to drought stress in transgenic rice. Plant Mol. Biol. Rep. 34, 845–853. [Google Scholar]
- Yadav, D. , Shavrukov, Y. , Bazanova, N. , Chirkova, L. , Borisjuk, N. , Kovalchuk, N. , Ismagul, A. et al. (2015) Constitutive overexpression of the TaNF‐YB4 gene in transgenic wheat significantly improves grain yield. J. Exp. Bot. 66, 6635–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Haider, I. , Kohlen, W. , Jiang, L. , Bouwmeester, H. , Meijer, A.H. , Schluepmann, H. et al. (2012) Function of the HD‐Zip I gene Oshox22 in ABA‐mediated drought and salt tolerances in rice. Plant Mol. Biol. 80, 571–585. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Chen, X. , Guan, X. , Liu, Y. , Chen, H. , Wang, T. , Mouekouba, L.D. et al. (2014) A genome‐wide survey of homeodomain‐leucine zipper genes and analysis of cold‐responsive HD‐Zip I members’ expression in tomato. Biosci. Biotechnol. Biochem. 78, 1337–1349. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Zhou, Y. , Jiang, H. , Li, X. , Gan, D. , Peng, X. , Zhu, S. et al. (2011) Systematic analysis of sequences and expression patterns of drought‐responsive members of the HD‐Zip gene family in maize. PLoS ONE, 6, e28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Ma, Q. , Jin, X. , Peng, X. , Liu, J. , Deng, L. , Yan, H. et al. (2014) A novel maize homeodomain–leucine zipper (HD‐Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis . Plant Cell Physiol. 55, 1142–1156. [DOI] [PubMed] [Google Scholar]
- Zhu, J.K. (2016) Abiotic stress signaling and responses in plants. Cell, 167, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Soil water tension monitored at 10 and 30 cm depths in large containers used for plant growth under well‐watered conditions or increasing drought.
Figure S2 Details of frost tolerance experiments.
Figure S3 Alignments of TdHDZipI‐5A and TdHDZipI‐5B promoter sequences and sequences of corresponding genes of Triticum aestivum cv. Chinese Spring, identified in the Whole Genome Reference Assembly Pseudomolecules v1.0 databases of the International Wheat Genome Sequencing Consortium, using the BLAST software (Altschul et al., 1997).
Figure S4 Alignment of TdHDZipI‐5B (5B) and TdHDZipI‐5A (5A) promoters. LALIGN (Huang and Miller, 1991) was used to find the best local alignments.
Figure S5 (a) Transgene copy numbers in T1 transgenic plants estimated by Q‐PCR. Plants seeds used in analyses are indicated by arrows. (b) Examples of selection of homozygous lines by PCR using transgene‐specific primers.
Figure S6 Growth characteristics and yield components of control wild‐type (WT) and transgenic T1 wheat (Triticum aestivum cv. Gladius) plants transformed with pUbi‐TaHDZipI‐5.
Figure S7 Expression levels of three stress‐inducible LEA (Late Embryogenesis Abundant)/COR (cold‐responsive)/DHN (dehydrin) genes (TaWZY2, GenBank: EU395844; TaCOR14B, GenBank: AF207546; TaRab15, GenBank: X59133) and the TaDREB3 (GenBank: DQ353853) regulatory gene, in leaves of unstressed control WT plants and T3 sublines of tree independent transgenic lines.
Table S1 List of PCR primers and DNA probes used in this study.
Table S2 A sequence alignment of 14 entries (with GenBank accession numbers) used to generate a phylogenetic tree displaying the evolutionary relationships of HD‐Zip I γ‐clade TFs from Arabidopsis and selected monocots, shown in Figure 1.
Table S3 Hydrogen bonds of homo‐dimeric TaHDZipI‐3 and TaHDZipI‐5, and hetero‐dimeric TaHDZipI‐3/TaHDZipI‐5 with HDZ1 (5′‐CAATCATTGC‐3′/5′‐GCAATGATTG‐3′).
Table S4 Characteristics of the T2/T3 progenies of TaHDZipI‐5 transgenic lines analysed in large containers under well‐watered or mild drought condition.
Appendix S1 Materials and methods.


