Abstract
Over the last decade, it has been increasingly demonstrated that the genomes of many species are pervasively transcribed, resulting in the production of numerous long noncoding RNAs (lncRNAs). At the same time, it is now appreciated that many types of DNA regulatory elements, such as enhancers and promoters, regularly initiate bidirectional transcription. Thus, discerning functional noncoding transcripts from a vast transcriptome is a paramount priority, and challenge, for the lncRNA field. In this review, we aim to provide a conceptual and experimental framework for classifying and elucidating lncRNA function. We categorize lncRNA loci into those that regulate gene expression in cis versus those that perform functions in trans, and propose an experimental approach to dissect lncRNA activity based on these classifications. These strategies to further understand lncRNAs promise to reveal new and unanticipated biology, with great potential to advance our understanding of normal physiology and disease.
Introduction
Much of the non-protein coding portion of the human genome has historically been regarded as junk DNA. Over the last decade, however, the development of high-throughput technologies, such as next-generation sequencing, have allowed an in-depth examination of the noncoding genome with unprecedented resolution and scale. Such studies have surprisingly revealed that, although less than 2 percent of the human genome encodes proteins, the majority of all nucleotides are detectably transcribed under some conditions (Djebali et al., 2012). Among the various types of non-protein coding transcripts, a class referred to as long noncoding RNAs (lncRNAs) have attracted increasing attention. lncRNAs are defined as transcripts of more than 200 nucleotides that are not translated into proteins. They comprise a heterogeneous class of intergenic transcripts, enhancer RNAs (eRNAs), and sense or antisense transcripts that overlap other genes (Derrien et al., 2012). lncRNAs have been proposed to carry out diverse functions including transcriptional regulation in cis or trans, organization of nuclear domains, and regulation of proteins or RNA molecules (Ulitsky and Bartel, 2013). It has also been demonstrated that some transcripts that are annotated as lncRNAs actually encode for small proteins (Anderson et al., 2015; Matsumoto et al., 2017).
There are a broad range of estimates for the number of lncRNA genes in mammals, ranging from less than 20,000 to over 100,000 in humans (Harrow et al., 2012; Zhao et al., 2016). Nevertheless, the function and biological relevance of the vast majority of lncRNAs remain enigmatic. Given that transcriptional regulatory elements such as enhancers and promoters are now known to initiate transcription bi-directionally (Core et al., 2008; Li et al., 2016; Seila et al., 2008), it is likely that many lncRNAs, if not the majority, actually represent RNAs that initiate at enhancers or promoters, but do not perform sequence-specific functions. This conclusion is further suggested by the fact that many lncRNAs are localized to the nucleus with low expression levels and little primary sequence conservation (Derrien et al., 2012; Djebali et al., 2012). Recent reports of local gene regulation by lncRNA loci reinforce this notion and suggest that, in many cases, the act of transcription or DNA elements within the lncRNA locus are more likely to be the source of regulatory activity than the actual lncRNA itself (Anderson et al., 2016; Engreitz et al., 2016; Groff et al., 2016). Given these observations, it is clear that the mere existence or production of an RNA does not automatically imply its functionality. Indeed, we must assume until proven otherwise that of the tens of thousands of annotated lncRNAs, those that function independently of the DNA sequence from which they are transcribed represent a small minority. Nevertheless, even if a small percentage of lncRNAs are functional, they would still constitute a major gene class with hundreds or possibly thousands of members.
Here we will review our current understanding of lncRNA function, classifying these transcripts broadly into those that regulate local chromatin structure and/or gene expression in cis versus those that leave the site of transcription and perform cellular functions in trans. Mechanistic principles will be derived by highlighting select lncRNAs whose functions have been rigorously evaluated using the most robust available methods. Based on lessons learned from these examples, we propose an experimental framework for dissecting a lncRNA locus and elucidating its roles in molecular and cellular biology.
Functions of lncRNA loci in local gene regulation
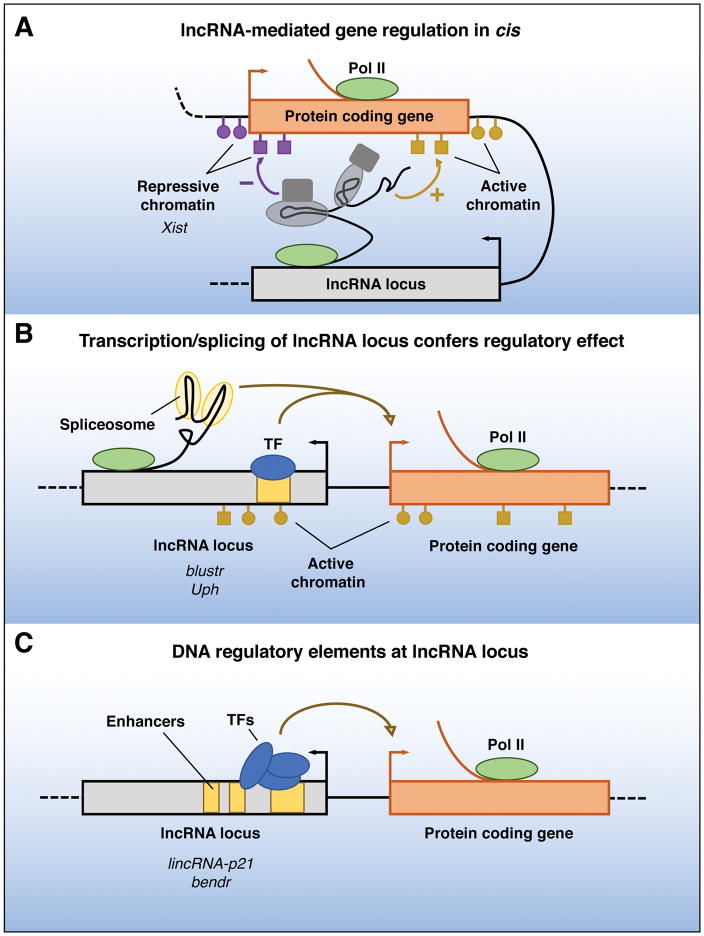
As an initial framework for understanding lncRNA function, we can broadly classify lncRNAs into those that act in cis, influencing the expression and/or chromatin state of nearby genes, and those that execute an array of functions throughout the cell in trans. For cis-acting lncRNAs, it is particularly challenging to distinguish functions of the RNA molecule itself from the DNA from which it is transcribed. One can envision at least three potential mechanisms through which a lncRNA locus can locally regulate chromatin or gene expression (Figure 1): I) the lncRNA transcript itself regulates the expression of neighboring genes through its ability to recruit regulatory factors to the locus and/or modulate their function; II) the process of transcription and/or splicing of the lncRNA confers a gene-regulation functionality that is independent of the sequence of the RNA transcript; or III) regulation in cis depends solely on DNA elements within the lncRNA promoter or gene locus and is completely independent of the encoded RNA or its production. These modes of action are discussed in turn below.
Figure 1. Functions of lncRNA loci in local gene regulation.
The ability of a lncRNA locus to regulate the expression of nearby genes in cis may be attributable to sequence-specific functions of the mature lncRNA transcript (A), may require transcription or splicing of an RNA, but the lncRNA itself is not functional (B), or may be due to DNA elements within the lncRNA promoter or gene body that function independently of the transcribed RNA (C). Pol II, RNA polymerase II; TF, transcription factor.
Sequence-dependent lncRNA regulation in cis
Among the most prominent proposed functions for lncRNAs is the establishment of repressive or activating chromatin cis (Figure 1A). The most famous and well-established example of a cis-acting lncRNA which functions in this manner is the X-inactive specific transcript Xist. During early embryonic development in female mammals, one of the two X chromosomes is transcriptionally silenced for dosage compensation. This critical process depends upon transcription of Xist from only one X chromosome, which will later become the inactive X (Xi). Following its induction, Xist spreads across the entire Xi and initiates a series of events that result in re-localization of the chromosome to the nuclear periphery, deposition of repressive chromatin marks, and eventual transcriptional silencing of almost the entire chromosome. The mechanisms of Xist-mediated X inactivation have been reviewed extensively elsewhere (Cerase et al., 2015; da Rocha and Heard, 2017) and therefore are only briefly described here. The ~17 kb Xist transcript consists of six repeat domains (A–F) that contribute to its silencing activity. While genetic studies have effectively dissected the sequence-specific requirements of Xist domains for X-inactivation, we now appreciate that early biochemical studies of the Xist ribonucleoprotein (RNP) complex that mediates silencing were confounded by the high rate of false positive interactions detected by in vitro binding studies and standard RNA immunoprecipitation (RIP) assays (Mili and Steitz, 2004). Fortunately, recently developed approaches that rely upon crosslinking of RNA-protein interactions followed by purification under denaturing conditions as well as forward genetic approaches have significantly improved our knowledge of the Xist-protein interactome and its mechanism of action (Chu et al., 2015; McHugh et al., 2015; Minajigi et al., 2015; Moindrot et al., 2015; Monfort et al., 2015). While aspects of the Xist-silencing mechanism are still controversial, such as whether the RNA directly recruits polycomb repressive complex 2 (PRC2) to establish repressive chromatin across Xi, multiple studies have documented that SMART/HDAC1-associated repressor protein (SHARP, also known as SPEN) is directly recruited to Xi through interaction with the Xist A-repeat. This leads to recruitment of the SHARP/SPEN interacting protein silencing mediator for retinoid and thyroid hormone receptors (SMRT) and its interactor histone deacetylase 3 (HDAC3), ultimately leading to X chromosome histone deacetylation, one of the earliest events in X inactivation. The fact that the Xist lncRNA has now been studied for over 25 years (Brockdorff et al., 1991; Brown et al., 1991), yet aspects of its molecular function still remain highly debated, emphasizes the difficulty in clearly establishing the mechanism of action of this class of cis-acting regulatory RNAs. At the same time, recent advances in our understanding of this transcript illustrate how classic genetics coupled with state-of-the-art molecular and biochemical methodologies can be applied to dissect lncRNA function.
Transcription or splicing-dependent regulation in cis by lncRNA loci
In some cases, production of an RNA from a lncRNA locus is critical for local gene regulation, yet the activity of the locus is independent of the mature lncRNA transcript or its sequence. A classic example of this type of regulation occurs at the mammalian imprinted Igf2r gene. An antisense lncRNA, Airn (antisense Igfr2 RNA noncoding), overlaps the Igfr2 gene body and promoter and is essential for silencing the paternal allele. Through an exhaustive series of polyadenylation (polyA) site insertions which truncated the Airn transcript before or after it spanned the Igf2r promoter, Barlow and colleagues definitively demonstrated that Airn silencing activity is solely dependent on antisense transcription through the Igf2r promoter, which likely produces transcriptional interference, and is independent of the Airn sequence itself (Latos et al., 2012).
Transcription of lncRNA loci that do not overlap with other transcription units may also alter RNA polymerase II occupancy on nearby promoters and gene bodies, as well as influence local chromatin states and transcription factor binding on promoter and enhancer regions (Figure 1B). (Anderson et al., 2016; Engreitz et al., 2016). This mechanism is exemplified by linc1319 or Blustr [bivalent locus (Sfmbt2) is upregulated by splicing and transcribing an RNA]. Promoter deletion, insertion of polyA signals which prematurely terminate transcription, or mutation of the first 5′ splice site of Blustr resulted in a significant reduction of expression of the neighboring gene Sfmbt2 (Engreitz et al., 2016). The cis-activating effect was correlated with the length of transcription, but appeared to be independent of specific Blustr sequences, as sequential deletions of downstream exons and introns of the lncRNA had no effect on Sfmbt2 expression. It should be noted, however, that the first exon of Blustr was not independently deleted or otherwise mutated in these experiments, leaving open the formal possibility that this portion of the transcript exhibits functionality at the RNA level. Impaired transcription or splicing of Blustr changed the chromatin state of the Sfmbt2 promoter, reducing histone 3 lysine 4 trimethylation (H3K4me3) and expanding histone 3 lysine 27 trimethylation (H3K27me3) as well as decreasing RNA polymerase occupancy at the transcription start site and within the gene body of Sfmbt2. Thus, transcription and splicing of the Blustr RNA is necessary to license expression of the neighboring gene.
Upperhand (Uph) presents another example where transcription, but most likely not the transcript itself, is responsible for local gene regulation (Anderson et al., 2016). Uph is transcribed from a bi-directional promoter that also produces Hand2, a transcription factor that is crucial for heart development. While the Uph sequence is not well conserved in different species, production of a transcript from this region, which spans multiple well-conserved Hand2 enhancers, is common across mammals. Premature termination of Uph transcription, through the insertion of a polyA signal early in the transcript, impaired Hand2 expression in cis, resulting in embryonic lethality in mice. Insertion of the tdTomato coding sequence at the same site had no effect on Hand2 expression. Moreover, knockdown of the mature Uph transcript using antisense oligonucleotides (ASOs) in cell lines did not influence Hand2 levels. While this result argues against a function of the mature Uph transcript, it remains formally possible that the nascent lncRNA transcript, which may not be efficiently targeted by the ASOs, performs an RNA-mediated function necessary for appropriate Hand2 regulation. This caveat notwithstanding, premature termination of Uph transcription in mice diminished the enhancer chromatin signature—histone 3 lysine 4 monomethylation (H3K4me1) and histone 3 lysine 27 acetylation (H3K27ac)—within the Uph gene body and prevented binding of the transcription factor GATA4 to a Hand2 enhancer in this region, resulting in reduced Hand2 expression. Notably, the mature Uph transcript was found to localize predominantly to the cytoplasm. While this finding may suggest additional functions for Uph, it also serves as a warning that care must be exercised when inferring a molecular function of a lncRNA based solely on its cellular localization.
The production of RNA from DNA regulatory elements has been proposed to function more generally in facilitating transcription factor (TF) binding. In light of pervasive bi-directional transcription of promoters and enhancers, Sigova et al. postulated that simultaneous binding of TFs to DNA and RNA might reinforce their association with transcribed elements. Indeed, the ubiquitously-expressed TF Yin-Yang 1 (YY1) was shown to bind both DNA and RNA at proximal promoters and enhancers (Sigova et al., 2015). Inhibiting transcription decreased occupancy of YY1, whereas Cas9-mediated tethering of an RNA sequence derived from a YY1 target gene promoter increased YY1 occupancy. While the generality of this “transcription factor trapping by RNA” model remains to be documented for TFs other than YY1, it provides a compelling mechanism to explain how RNA production from a regulatory DNA element could establish a positive feedback loop that reinforces TF association. Most likely, the promiscuous affinity of certain TFs for RNA, rather than sequence-specific RNA-protein interactions, allows them to take advantage of this mechanism. Notably, promiscuous interactions of the histone methyltransferase PRC2 with RNA has also been observed, suggesting that association of this complex with target loci might similarly be influenced by RNA production (Davidovich et al., 2015).
Functional DNA elements within lncRNA loci
The possibility that the cis-regulatory activity of a lncRNA locus is due to DNA elements within it that function independently of the production or sequence of the transcribed RNA must always be considered (Figure 1C). lincRNA-p21 provides an instructive example of this type of locus in which the regulatory functions can largely be ascribed to conventional cis-acting DNA elements embedded within the lncRNA gene body. lincRNA-p21 is a nuclear-localized transcript produced from an intergenic locus that neighbors the CDKN1A gene in human and mouse. An initial study reported that lincRNA-p21 functions as a p53-dependent trans-acting lncRNA that, when induced by DNA damage, mediates apoptosis by contributing to the repression of a gene network regulated by p53 (Huarte et al., 2010). These findings stimulated numerous additional studies that proposed additional trans-acting functions of lincRNA-p21. Careful genetic analyses of the lincRNA-p21 locus, however, later uncovered strong evidence that this lncRNA locus actually functions to regulate Cdkn1a in cis (Dimitrova et al., 2014). Downstream effects of lincRNA-p21 loss-of-function on global gene expression could be attributed to a secondary consequence of reduced levels of the Cdkn1a-encoded protein p21. This mechanism is further supported by the short half-life (less than 2 hours) and low copy number (8 copies per cell) of lincRNA-p21 (Dimitrova et al., 2014), which argues against a role as a trans-acting lncRNA with broad gene regulatory functions. Most recently, further genetic dissection of lincRNA-p21 provided evidence that this locus contains functional cis-regulatory DNA elements that modulate gene expression of neighboring genes, even in tissues in which lincRNA-p21 is not expressed (Groff et al., 2016). These findings are in line with another report that classified lincRNA-p21 as an enhancer RNA that originates from a p53 binding site associated with regulation of CDKN1A (Allen et al., 2014). Thus, there is emerging consensus that the lincRNA-p21 locus regulates neighboring gene expression in cis, and, at least in some contexts, DNA elements within this region appear to perform this function independently of production of the lncRNA. However, it is important to note that Dimitrova et al. convincingly showed that knockdown of lincRNA-p21 with ASOs was sufficient to cause downregulation of Cdkn1a in cultured murine fibroblasts, suggesting that in some cell contexts, the transcript—or at least the act of transcription—may be necessary for the regulatory activity of this locus.
The presence of DNA elements that locally regulate gene expression independently of transcription appears to be a common attribute of lncRNA loci. To assess whether lncRNA loci frequently regulate nearby genes in cis and whether such effects are mediated by DNA elements or the transcribed lncRNA, a recent study used an elegant genome editing approach in hybrid mouse ES cells that allowed the monitoring of expression of neighboring genes in an allele specific manner (Engreitz et al., 2016). Deletion of five out of 12 lncRNA promoters (and four out of six protein coding promoters) affected gene expression of neighboring genes on the same allele. By introducing early polyA signals to terminate transcription, it was further shown that in all but one case, transcription of the lncRNA or protein coding locus beyond the first 1–3 exons was not necessary for these cis-regulatory effects. For example, deletion of the promoter of linc1536, renamed Bendr (Bend4-regulating effects not dependent on the RNA), significantly decreased the expression and transcription of the adjacent protein coding gene Bend4 in cis. However, Bend4 expression was not affected by inserting a polyA signal into the first intron of Bendr, which fully abolished downstream transcription of the lncRNA. These findings suggest that the Bendr locus, and most of the other cis-acting lncRNA loci studied in this report, regulate local gene expression through enhancer-like elements, which function independently of the production of a noncoding transcript. Nevertheless, in order to completely rule out the possibility of RNA functionality within the earliest exons of these lncRNAs, further experiments in which the sequences upstream of the polyA site insertions are replaced with unrelated sequences will be necessary.
Functions of lncRNAs in trans
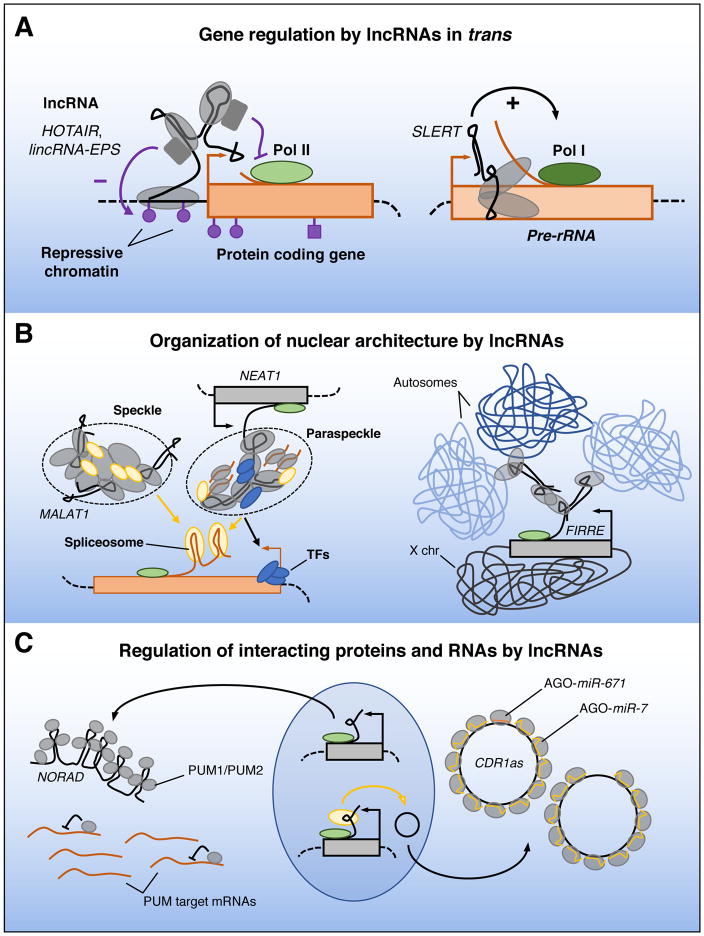
In addition to those that regulate gene expression and chromatin states in cis, there is an increasing number of examples of lncRNAs that leave the site of transcription and operate in trans. These lncRNAs can be categorized into at least three major subgroups (Figure 2): I) lncRNAs that regulate chromatin states and gene expression at regions distant from their transcription site; II) lncRNAs that influence nuclear structure and organization; and III) lncRNAs that interact with and regulate the behavior of proteins and/or other RNA molecules.
Figure 2. Functions of lncRNAs in trans.
(A) lncRNAs may regulate the expression of unlinked genes by interacting with promoters and enhancers, or other proteins bound to these sites, and influencing chromatin states and RNA polymerase activity. (B) Some lncRNAs are components of dynamic subcellular structures, such as MALAT1 in nuclear speckles and NEAT1 in paraspeckles, and may help to position these structures near actively transcribed genes to facilitate delivery of splicing and transcription factors. lncRNA FIRRE organizes interchromosomal interactions within the nucleus. C) Transacting lncRNAs may regulate the activity of interacting proteins and RNAs in a stoichiometric manner. For example, NORAD binds many molecules of PUMILIO (PUM) proteins, limiting the availability of these post-transcriptional regulators to interact with other targets. CDR1as binds and stabilizes AGO:miR-7 complexes, whereas AGO:miR-671 cleaves the lncRNA. Pol I, RNA polymerase I; X chr, X chromosome.
Gene regulation by lncRNAs in trans
Among the first lncRNAs reported to regulate gene expression in trans was the HOX antisense intergenic RNA HOTAIR, a ~2.2 kb spliced and polyadenylated mammalian transcript that is expressed from the HOXC locus. HOX transcription factors produced from four HOX gene clusters (HOXA, HOXB, HOXC, and HOXD) are deeply conserved developmental regulators of positional identity and differentiation. While noncoding RNAs had previously been shown to regulate Hox gene expression in cis in Drosophila (Petruk et al., 2006), Rinn et al. made the surprising discovery that HOTAIR is required to maintain repressive chromatin marks at the distant HOXD locus (Rinn et al., 2007). A later study using chromatin isolation by RNA purification (ChIRP), a technique to map genome-wide lncRNA-chromatin interactions, detected a HOTAIR binding site within the HOXD cluster (Chu et al., 2011). Depletion of HOTAIR with siRNAs resulted in transcriptional activation of HOXD genes with an associated decrease in the repressive chromatin mark H3K27me3 (Rinn et al., 2007). RIP and biotinylated RNA pull-down assays detected interactions between HOTAIR and components of PRC2, the complex that catalyzes H3K27me3 modification. These findings suggested a model (Figure 2A) in which HOTAIR acts as a scaffold that coordinates the recruitment of chromatin modifying complexes to the HOXD locus to establish a repressed chromatin state. Since these early studies, however, our understanding of the caveats associated with studying RNA-protein interactions have evolved, raising questions about some aspects of this model. As mentioned earlier in this review, in vitro binding and RIP experiments can yield false-positive interactions and PRC2 in particular is believed to interact non-specifically with RNA (Davidovich et al., 2015; Mili and Steitz, 2004). Moreover, in a recent study (Portoso et al., 2017), overexpression of HOTAIR in human breast cancer cells was found to result in the PRC2-independent repression of a limited number of target genes. Tethering experiments in which HOTAIR was guided to a luciferase reporter locus confirmed that recruitment of this lncRNA was sufficient to result in transcriptional repression, but again this effect was independent of PRC2. These recent findings are notable because the initial work on HOTAIR (Rinn et al., 2007; Tsai et al., 2010) and an early mechanistic study of Xist (Zhao et al., 2008) led to the still widely cited hypothesis that recruitment of chromatin modifying complexes to target gene loci is a predominant lncRNA function. Nevertheless, it is now clear that many questions remain regarding the mechanism through which HOTAIR and potentially other lncRNAs regulate target gene expression. The application of newer methods that can more reliably detect RNA-protein interactions will likely help resolve these uncertainties (Chu et al., 2015; McHugh et al., 2015; Minajigi et al., 2015).
Since the discovery of HOTAIR, many additional lncRNAs have been reported to regulate transcription in trans. We will highlight two further examples here that illustrate the diverse mechanisms through which this class of lncRNAs can function. The first of these is lincRNA-EPS (lincRNA erythroid prosurvival, annotated as Ttc39aos1), a 2.5 kb spliced and polyadenylated lncRNA that was originally discovered due to its induction during mouse erythropoiesis (Hu et al., 2011). Knockdown of lincRNA-EPS in erythroid progenitors resulted in apoptosis in vitro. Importantly, heterologous expression of the lncRNA at physiologic levels repressed apoptosis, demonstrating its ability to function in trans. The anti-apoptotic function of lincRNA-EPS was attributed, at least in part, to its ability to suppress expression of the pro-apoptotic gene Pycard, although the molecular mechanism of Pycard regulation was not determined. In a subsequent study, lincRNA-EPS was found to be downregulated in macrophages exposed to inflammatory stimuli such as LPS. Genetic deletion of the entire 4 kb lncRNA locus in mice resulted in the upregulation of many immune response genes (IRGs) in macrophages and conferred hypersensitivity to endotoxic shock induced by LPS administration (Atianand et al., 2016). Two lines of evidence strongly pointed to a trans-acting function of this lncRNA underlying these effects. First, deletion of lincRNA-EPS did not affect the expression of neighboring genes, arguing against cis-regulatory activity. Second, retroviral expression of lincRNA-EPS at physiologic levels in knockout macrophages reversed the induction of IRGs that occurs upon loss of the lncRNA. Mechanistically, lincRNA-EPS appears to interact directly with the promoters of the genes it represses (Figure 2A), as detected by RNA antisense purification (RAP), a stringent method that captures RNA-chromatin interactions (Engreitz et al., 2013). Recruitment of hnRNP L to these promoters through a direct interaction with lincRNA-EPS was proposed to play an important role in subsequent transcriptional repression. It is notable that lincRNA-EPS is expressed at only ~11 copies per cell in macrophages, suggesting that it has the capacity to interact with only a limited number of direct target genes which may indirectly be responsible for broader changes in IRG expression. Furthermore, how the specificity of lincRNA-EPS-target promoter interactions is determined remains unknown. A final note about lincRNA-EPS is that while a transcript is produced from the syntenic position in the human genome, it exhibits limited to no sequence similarity to the mouse lncRNA. This raises the question of whether lincRNA-EPS acquired gene-regulatory activity specifically within the rodent lineage and whether a yet-to-be identified lncRNA performs similar functions in human cells.
The regulation of gene expression in trans by lncRNAs is not limited to RNA polymerase II (pol II)-transcribed target genes. The recent discovery of SLERT (snoRNA-ended lncRNA enhances pre-ribosomal RNA transcription) demonstrates that lncRNAs can also influence transcription by RNA polymerase I (pol I) (Xing et al., 2017) (Figure 2A). SLERT represents a structurally distinct class of lncRNAs that are processed from introns of pol II transcripts and are stabilized by the presence of snoRNA precursor sequences at the lncRNA 5′ and 3′ ends. In the case of SLERT, the mature lncRNA is a 694 nucleotide (nt) RNA flanked by two box H/ACA snoRNAs that is processed from the human TBRG4 gene. Careful CRISPR knockout experiments that preserved expression of the TBRG4 host transcript as well as rescue experiments using heterologous expression plasmids established that SLERT enhances transcription of pre-rRNA by pol I in trans. Accordingly SLERT is highly enriched within nucleoli, the site of pre-rRNA transcription. Proteomics experiments revealed interaction between SLERT and DDX21, a protein that binds to pol I subunits and represses pol I transcription. Remarkably, super-resolution microscopy revealed that DDX21 forms ring-like structures around pol I within nucleoli, which presumably function to limit pre-rRNA production. SLERT localizes to these DDX21 rings and antagonizes DDX21-pol I interactions, a mechanism that plausibly explains the stimulatory effect of SLERT on pol I transcription.
Organization of nuclear architecture by lncRNAs
Some lncRNAs appear to influence nuclear architecture to orchestrate transcription, RNA processing, and other steps in gene expression, and to impart spatial regulation upon these processes. This role for lncRNAs was first suggested by studies of the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1, also called NEAT2), a ~8 kb highly abundant lncRNA that is conserved across vertebrates and localizes to nuclear speckles (Hutchinson et al., 2007; Ji et al., 2003; Pauli et al., 2012; Ulitsky et al., 2011). Nuclear speckles are dynamic compartments that contain many components of the splicing machinery such as spliceosomal subunits, small nuclear ribonucleoproteins (snRNPs), and serine/arginine-rich (SR) proteins (Spector and Lamond, 2011). MALAT1 is recruited to speckles through direct interactions with multiple splicing-associated proteins, although it is not necessary for speckle formation per se (Bernard et al., 2010; Miyagawa et al., 2012; Tripathi et al., 2010). In addition, capture hybridization analysis of RNA targets (CHART) (West et al., 2014) and RNA antisense purification for mapping RNA-chromatin interactions (RAP-DNA) and RNA-RNA interactions (RAP-RNA) (Engreitz et al., 2014) demonstrated that MALAT1 associates with actively transcribed gene bodies, most likely via interactions with proteins that indirectly guide the lncRNA to nascent pre-mRNAs. It has therefore been suggested that MALAT1 acts as a linker or scaffold that facilitates the positioning of nuclear speckles at active gene loci (Figure 2B). While it is possible that this serves to enhance delivery of the splicing machinery to nascent transcripts, this has not been demonstrated for endogenous genes and Malat1-deficient mice do not exhibit measureable splicing abnormalities. Thus, the precise molecular function of MALAT1 remains unclear.
Malat1 is not essential for mouse development or viability (Eissmann et al., 2012; Nakagawa et al., 2012; Zhang et al., 2012), although Malat1−/− mice display delayed neonatal retinal vascularization due to reduced proliferation of endothelial cells (Michalik et al., 2014). Additionally, high expression of MALAT1 has been associated with poor prognosis and metastasis in lung cancer and other tumor types (Gutschner et al., 2013a; Ji et al., 2003). Knockout studies in human cell lines and mice have corroborated the pro-metastatic activity of Malat1 in vivo (Arun et al., 2016; Gutschner et al., 2013b). Furthermore, therapeutic administration of ASOs targeting Malat1 in a mouse breast cancer model resulted in slower growing, more differentiated tumors and a significant reduction in metastases without overt toxic effects (Arun et al., 2016). It is presently unclear whether these anti-cancer effects can be attributed to disruption of the aforementioned role of MALAT1 within nuclear speckles or whether other additional, currently unrecognized, MALAT1 functions are responsible. Further interrogation of this question is warranted since a better understanding of the molecular basis for MALAT1 oncogenic activity may reveal new approaches for targeted anti-metastatic therapeutics.
The nuclear enriched abundant transcript 1 (NEAT1, also known as MEN ε/β) is another well-characterized example of a lncRNA that organizes nuclear structure. NEAT1 is a conserved single exon transcript that, in human cells, is alternatively processed to produce 3.7 kb (NEAT1_1) and 22.7 kb isoforms (NEAT1_2) (Hutchinson et al., 2007). Interestingly, NEAT1 and MALAT1 are expressed from adjacent gene loci, yet they localize to distinct subnuclear bodies. While MALAT1 is enriched in nuclear speckles, NEAT1 is localized to paraspeckles, which are dynamic nuclear compartments that contain proteins involved in transcription and RNA processing (Fox and Lamond, 2010). NEAT1 interacts with several paraspeckle proteins including p54nrb/NONO, PSPC1/PSP1, and PSF/SFPQ, and is essential for the formation and maintenance of these nuclear domains (Clemson et al., 2009; Mao et al., 2011; Sasaki et al., 2009; Sunwoo et al., 2009; West et al., 2014). The recent finding that NEAT1, like MALAT1, associates with actively transcribed gene loci (West et al., 2014) suggests that both of these lncRNAs may similarly function to localize their respective subnuclear bodies in proximity to nascent transcripts (Figure 2B).
Like Malat1, Neat1 is dispensable for overtly normal development in mice (Nakagawa et al., 2011). In fact, Neat1 and paraspeckles are only detectable in a subpopulation of cells in mouse tissues at baseline. Two such tissues are the corpus luteum, which forms in the ovary after ovulation and produces progesterone, and the developing mammary gland. Accordingly, these tissues are defective in Neat1−/− mice, resulting in reduced fecundity and impaired lactation (Nakagawa et al., 2014; Standaert et al., 2014). A role for NEAT1 in the p53 tumor suppressor pathway has also been uncovered as NEAT1 is a direct target of p53 and is induced under a variety of stresses including DNA damage and oncogene-induced replication stress (Adriaens et al., 2016; Blume et al., 2015; Botcheva et al., 2011; Mello et al., 2017). Neat1-deficient mouse fibroblasts are prone to transformation and loss of Neat1 enhances the formation of pancreatic cancer precursor lesions in Kras mutant mice (Mello et al., 2017). On the other hand, Neat1−/− mice are resistant to chemically-induced skin cancer (Adriaens et al., 2016). As is the case with Malat1, it is currently unclear how Neat1 specifically, and paraspeckles more generally, function to regulate tissue development and tumorigenesis in these contexts.
Firre (functional intergenic repeating RNA element) represents a distinct class of lncRNAs that contribute to nuclear organization. First identified in a screen for mouse lncRNAs that regulate adipogenesis (Sun et al., 2013), Firre was subsequently found to be a nuclear localized, conserved lncRNA containing many repeats of a 156 nt sequence that binds to hnRNP U (Hacisuleyman et al., 2014). While Firre is transcribed from the X-chromosome, it escapes X-inactivation and remains associated with its site of transcription in human and mouse cells. Intriguingly, genome-wide mapping of Firre-chromatin interactions in mouse embryonic stem (mES) cells using RAP (Engreitz et al., 2013) revealed that, in addition to its transcription site, Firre could be detected at five additional unlinked autosomal loci (Figure 2B). Trans-chromosomal interactions between the Firre locus on X with these sites was confirmed using fluorescent in situ hybridization (FISH). Deletion of Firre or knockdown of hnRNP U abolished these interchromosomal contacts, suggesting that the Firre RNA functions as hub to co-localize distant genomic regions at the site of Firre transcription. While deletion of this lncRNA did not affect expression of genes at these trans-chromosomal contact sites in mES cells, it remains possible that this remarkable RNA-directed organization provides a regulatory function in other settings. More generally, these findings highlight a broader role for lncRNAs as organizers of chromatin localization within the nucleus. Indeed, another prominent example of this mechanism is provided by Xist, which directly interacts with the lamin B receptor (LBR), a transmembrane constituent of the inner nuclear membrane, to tether the inactive X chromosome to a silencing domain at the nuclear periphery (Chen et al., 2016).
Regulation of interacting proteins and RNAs by lncRNAs
Trans-acting lncRNAs may also function by modulating the activity or abundance of proteins or RNAs to which they directly bind. A key feature of these lncRNAs is that they often require stoichiometric interaction with their target molecules in order to exert measureable regulatory effects. Therefore, when studying this class of transcripts, it is essential to carefully quantify the cellular copy number of the lncRNA and its target(s) to establish the plausibility of this mechanism. While in principle these regulatory interactions can take place in either the nucleus or cytoplasm, we focus on cytoplasmic cases here to complement the many examples of nuclear lncRNAs highlighted above.
The lncRNA NORAD (noncoding RNA activated by DNA damage), functions as a molecular decoy for the RNA binding proteins PUMILIO1 (PUM1) and PUMILIO2 (PUM2) (Lee et al., 2016; Tichon et al., 2016) (Figure 2C). PUMILIO proteins belong to a deeply conserved family of RNA binding proteins that specifically bind to the sequence UGUANAUA, termed the PUMILIO response element (PRE), which is generally found in the 3′ UTR of selected mRNAs. Binding of PUM1/PUM2 triggers accelerated mRNA decay and translational inhibition of these mRNA targets (Miller and Olivas, 2011; Wickens et al., 2002). NORAD is an abundant 5.3 kb unspliced polyadenylated transcript that localizes predominantly to the cytoplasm. The sequence of NORAD contains five repeated units of ~500 nt each, termed NORAD domains (Lee et al., 2016), and can be further divided into 12 shorter repeating sequences (Tichon et al., 2016). Enriched in these repeats are 15 perfect PREs, far more than expected by chance in a transcript of this length. Quantitative analysis of the number of NORAD and PUMILIO molecules per cell revealed that this lncRNA provides sufficient PREs to occupy the complete cellular pool of PUM1 and PUM2. Moreover, photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) experiments demonstrated that NORAD is the preferred RNA binding partner of PUM2 in human cells (Hafner et al., 2010; Lee et al., 2016). These observations suggested that NORAD functions as a negative regulator of PUM1/PUM2, limiting the availability of these proteins to interact with mRNA targets. Indeed, inactivation of NORAD through insertion of a polyA signal at its 5′ end (Lee et al., 2016) or siRNA-mediated knockdown (Tichon et al., 2016), resulted in PUMILIO hyperactivity as indicated by the broad downregulation of PUMILIO target transcripts. PUMILIO targets are highly enriched for regulators of mitosis and their repression results in chromosomal instability and aneuploidy in NORAD knockout cells (Lee et al., 2016). Further study of the NORAD-PUMILIO RNP will be important to illuminate how this lncRNA is able to successfully compete against the complete transcriptome-wide pool of PUMILIO targets to potently inhibit these RNA binding proteins.
While PUMILIO proteins are broadly conserved across vertebrates and invertebrates, NORAD orthologs appear to be limited to mammalian species, raising the question of why mammals use this mechanism to regulate PUMILIO activity. Interestingly, either overexpression or deletion of PUM1/PUM2 in mammalian cell lines resulted in aneuploidy, indicating that PUMILIO activity must be maintained within a narrow range to avoid deleterious consequences. This concept is further supported by the finding that Pum1 haploinsufficiency in mice causes neurodegeneration (Gennarino et al., 2015). Thus, it is likely that one important function of NORAD is to act as a buffer that tightly regulates the pool of free PUM1/PUM2 that is available to interact with target transcripts. In addition, NORAD expression is upregulated in response to diverse cellular stresses including DNA damage (Lee et al., 2016) and hypoxia (Michalik et al., 2014), pointing to a role for this lncRNA in the dynamic regulation of PUMILIO activity under some conditions. Further investigation of the regulation of the NORAD-PUMILIO axis and its physiologic role in vivo remain important areas for future research.
In addition to RNA binding proteins, lncRNAs also have the ability to regulate the abundance or activity of other RNAs to which they bind through base-pairing interactions. Prominent among this class are noncoding RNAs that regulate microRNA (miRNA) activity, a category of transcripts termed competing endogenous RNAs (ceRNAs). ceRNAs have been suggested to broadly titrate miRNA availability (Tay et al., 2014), although this proposal has remained controversial because the ceRNA:miRNA stoichiometry may not be adequate to support this mechanism in the majority of cases (Bosson et al., 2014; Denzler et al., 2014). Nevertheless, studies of the natural antisense transcript of cerebellar degeneration-related protein 1 (CDR1as, also called ciRS-7) clearly show that exceptional cases of miRNA regulation by noncoding RNAs do exist. CDR1as is one of many recently described circular RNAs (circRNAs), a class of transcripts that form from back-splicing of a 5′ splice site to an upstream 3′ splice site (circRNA biogenesis and function have been reviewed extensively elsewhere) (Chen, 2016). CDR1as, an abundant cytoplasmic circRNA, is broadly conserved in mammals and highly expressed in the brain. A major clue for understanding the function of CDR1as came from its highly unique sequence, containing over 70 conserved seed matches for miR-7 and one nearly perfectly complementary binding site for miR-671 (Figure 2C) (Hansen et al., 2013; Hansen et al., 2011; Memczak et al., 2013). CLIP experiments in human cells as well as human and mouse brains documented extensive binding of miR-7:Argonaute (AGO) complexes to CDR1as (Hansen et al., 2013; Memczak et al., 2013; Piwecka et al., 2017). Due to its circular structure, CDR1as lacks a 5′ cap and polyA tail, rendering it resistant to miRNA-induced deadenylation and decay. Thus, CDR1as has the capacity to bind and sequester free miR-7, resulting in de-repression of miR-7 targets when the circRNA is heterologously expressed in human cell lines in vitro and in zebrafish embryos in vivo (Hansen et al., 2013; Memczak et al., 2013). On the other hand, the circRNA can be readily sliced by miR-671 (Hansen et al., 2011), a mechanism that was proposed to release miR-7 from inhibition by this transcript. Despite the ability of CDR1as to inhibit miR-7 in experimental models, however, recent results obtained from the study of Cdr1as knockout mice revealed a more complex relationship between this circRNA and miR-7 activity in vivo (Piwecka et al., 2017). Surprisingly, deletion of Cdr1as resulted in a significant reduction of miR-7 levels and a consequent upregulation of many miR-7 target genes in the brain, causing defects in synaptic neurotransmission and phenotypes associated with neuropsychiatric disorders. Thus, Cdr1as appears to function as a positive regulator of miR-7 in vivo. Single molecule FISH in cultured neurons revealed localization of Cdr1as to both soma and neuronal processes, suggesting that the circRNA may transport miR-7 to designated subcellular locations such as synapses where it may be released by cleavage of Cdr1as by miR-671. In this model, binding and transport of miR-7 by Cdr1as is essential for stable expression of the miRNA. Further study will be necessary to substantiate this model and elucidate precisely how Cdr1as orchestrates miRNA activity in the mammalian brain.
Experimental dissection of lncRNA loci
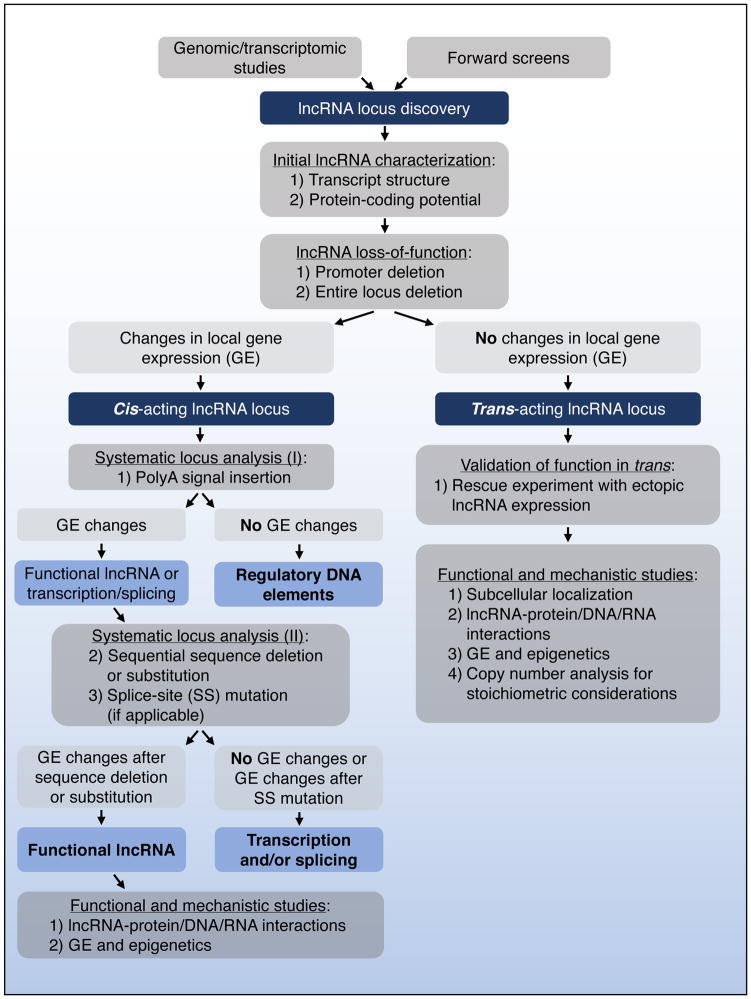
The classification of lncRNAs into those that regulate gene expression in cis versus those that act in trans not only provides a conceptual framework for understanding lncRNA function, but also suggests a logical experimental approach to interrogate lncRNA activity, which we propose below (Figure 3).
Figure 3. Experimental dissection of lncRNA loci.
A proposed experimental approach for classifying lncRNAs into those that function in cis or trans, followed by investigation of the underlying mechanisms of action.
Identification and initial characterization of lncRNA loci
The selection of a lncRNA for in-depth study often follows from the association of a given lncRNA locus with a specific developmental, physiologic, or pathophysiologic state. For example, many lncRNAs exhibit dynamic expression during development or cellular differentiation (Hu et al., 2012; Sauvageau et al., 2013). Numerous lncRNAs that are subject to copy number alteration or altered expression in diseases such as cancer have been reported (Yan et al., 2015), as have lncRNAs whose expression is differentially regulated under diverse stress conditions or after activation of signaling pathways (Atianand et al., 2016; Lee et al., 2016; Michalik et al., 2014).
Forward genetic screens using multiple CRISPR technologies have recently emerged as a complementary approach to identify lncRNA loci that influence phenotypes of interest, including cell proliferation and drug resistance (Joung et al., 2017; Liu et al., 2017; Zhu et al., 2016). Importantly, targeting Cas9 to lncRNA loci with individual single guide RNAs (sgRNAs) is unlikely to reliably cause loss of function since the small insertions-deletions (indels) induced by this strategy will often fail to result in functional inactivation of the lncRNA. To more consistently achieve lncRNA loss of function, libraries of paired sgRNAs that generate larger deletions at lncRNA loci have been employed (Zhu et al., 2016). Alternatively, fusion of Cas9 to domains that inhibit transcription (CRISPR interference or CRISPRi) or to domains that activate transcription (CRISPR activation or CRISPRa) have enabled genome-wide lncRNA loss- and gain-of-function screens, respectively (Joung et al., 2017; Liu et al., 2017). When interpreting the results of screens such as these, it is important to recognize that each of these technologies can equivalently inactivate or hyperactivate bona fide lncRNA-producing genes as well as DNA regulatory elements such as enhancers (Fulco et al., 2016; Klann et al., 2017). Thus, the candidates that emerge from these systematic approaches must be studied in detail to establish whether they are, in fact, functional RNAs and, if so, their mechanisms of action.
Once a lncRNA locus of interest has been identified, a thorough structural characterization of the transcript in the cell type or tissue of interest should be carried out. These studies could include rapid amplification of cDNA ends (RACE) or related methods to define the transcript 5′ and 3′ ends, coupled with reverse-transcriptase (RT)-PCR to document splicing patterns. Alternatively, analysis of publicly-available high-throughput sequencing datasets produced from consortia such as ENCODE (https://www.encodeproject.org) and FANTOM (http://fantom.gsc.riken.jp), if available for the relevant cell type or tissue, may be sufficient to structurally characterize lncRNAs of interest. These data include cap analysis of gene expression (CAGE), RNA-seq, and polyA site sequencing (PAS-seq), which can define transcript 5′ ends, splicing patterns, and 3′ ends, respectively. If possible, northern blotting provides a reliable method to confirm expression of transcript(s) of the expected size. These types of experiments are advisable since available annotations, often derived from automated analysis of high-throughput datasets, may be inaccurate or incomplete, especially if the transcript was identified initially in a distinct cell type. The cellular copy number of a lncRNA transcript should also be determined as this information will ultimately be important when formulating plausible hypotheses regarding the lncRNA mechanism of action. Since an increasing number of annotated lncRNAs have been demonstrated to encode small peptides (Anderson et al., 2015; Matsumoto et al., 2017), it is critical to rule out the protein-coding potential of any putative noncoding transcript. Analysis of the evolutionary pattern of codon substitution in a sequence of interest using algorithms such as PhyloCSF (Lin et al., 2011) provides one powerful predictor of the likelihood that it encodes a protein. This approach takes advantage of the fact that coding transcripts will be enriched for phylogenetic sequence variation that introduces synonymous codon substitutions or conservative amino acid changes, and will be relatively depleted of substitutions that create translation termination codons. The similarity of in silico translated sequence to known protein family (Pfam) domains (Finn et al., 2016) also increases the likelihood that it encodes a protein. Finally, direct observation of ribosome footprints over a putative open reading frame using ribosome profiling can often discriminate protein-coding transcripts from noncoding RNAs (Guttman et al., 2013; Ingolia et al., 2014).
Functional interrogation of cis-acting lncRNA loci
Once a putative lncRNA of interest is identified, the first step towards understanding its function is determining whether it acts locally to regulate gene expression in cis or whether it leaves the site of transcription and performs a function in trans. This discrimination can best be achieved by performing loss- and gain-of-function experiments. To assess the consequences of lncRNA loss of function, straightforward genome-editing approaches such as CRISPR/Cas9-mediated deletion of the lncRNA promoter or the entire lncRNA locus can initially be employed. For large multi-exonic lncRNA genes, it is preferable to delete the promoter region rather than the entire locus to reduce the chance of removing regulatory DNA elements embedded in the lncRNA gene body. Once clean loss-of-function is achieved, transcription and steady-state expression of nearby genes should be determined using sequencing based technologies such as RNA-seq and global run-on sequencing (GRO-seq) (Core et al., 2008) and/or quantitative RT-PCR (qRT-PCR). Altered expression of nearby genes following inactivation of the lncRNA strongly points to a cis-acting function. The inability of heterologous expression of the lncRNA to rescue altered gene expression in lncRNA knockout cells further establishes that the locus under study functions to regulate local gene expression in cis.
As described above, lncRNA loci that regulate local gene expression may do so via DNA regulatory elements embedded within the lncRNA promoter or gene body that may or may not require transcription for their function. In other cases, the transcribed RNA itself may be the functional entity (Figure 1). To begin to distinguish between these possibilities, insertion of a polyA signal near the 5′ end of the lncRNA locus will often be informative. This manipulation will prematurely terminate transcription while keeping all DNA elements in the promoter and gene body otherwise intact. Thus, if the expression of nearby genes is affected by this manipulation, transcription of the lncRNA locus is necessary for its activity, arguing against a role for traditional DNA regulatory elements as the sole functional entity. One caveat of this approach is that insertion of a long polyA site-containing sequence can increase the distance between DNA regulatory elements, thereby potentially altering their activity. Insertion of a control sequence at the same site, such as a mutated polyA site, can mitigate this concern. In cases where a polyA site insertion abolishes the regulatory activity of the lncRNA locus, the act of transcription or splicing of an RNA produced from the locus may be the key event or the RNA itself may perform a regulatory function in a sequence-specific manner. The systematic introduction of targeted mutations that disrupt splice sites and sequentially delete or replace sequences of the lncRNA (such as exons) will usually allow further discrimination between these potential mechanisms. For example, a lncRNA that functions in a sequence-specific manner will be sensitive to removal of exons whereas loci in which transcription and splicing are the essential events will often tolerate sequence substitutions, but will be inactivated by polyA site insertions and splice-site mutations. As an important aside, ASO- or siRNA-mediated knockdown is frequently used to assess whether a lncRNA locus produces a functional RNA. While these methods can clearly reduce the expression of mature lncRNA transcripts, it is currently unknown to what extent these reagents promote cleavage of nascent RNAs. This is a critical issue since nascent transcript cleavage by the polyadenylation machinery or through other mechanisms is a natural trigger for transcription termination (Ghazal et al., 2009; Kim et al., 2004; Wagschal et al., 2012; West et al., 2004). Upon cleavage of nascent transcripts, the newly formed free 5′ end becomes a substrate for 5′-to-3′ degradation by the exoribonuclease XRN2 whose activity induces disassembly of elongating Pol II complexes. Therefore, if ASOs or siRNAs can act upon nascent RNAs, they too may trigger transcription termination through this mechanism. In light of this caveat, ASO- or siRNA-mediated knockdown cannot, based on our present understanding, discriminate whether transcription or production of a functional RNA are the key activities of a lncRNA locus.
For cases where the lncRNA locus produces a functional RNA, the identification of interacting proteins and nucleic acids is an important next step. Methods that employ crosslinking and recover interactions under stringent or even denaturing conditions are highly preferred over methods that rely on native RIP or in vitro pull-down assays. As reviewed extensively elsewhere (McHugh et al., 2014), recently developed methodologies have greatly improved the specificity of de novo identification of the interaction partners of endogenous RNAs, including proteins, chromatin, and other RNAs (Chu et al., 2011; Chu et al., 2015; Engreitz et al., 2013; Engreitz et al., 2014; McHugh et al., 2015; Minajigi et al., 2015). Since cis-acting lncRNAs are often expressed at low levels, however, it may be difficult to apply these methods initially. If needed, candidate protein interaction partners of lncRNAs may be identified by in vitro pull-down of RNA in cellular extracts. In these cases, it is crucial to carefully validate any putative interactions among the endogenous molecules using crosslinking-based methods such as CLIP (Hafner et al., 2010; Konig et al., 2010; Licatalosi et al., 2008; Van Nostrand et al., 2016). The identification of protein or nucleic acid binding partners or sites of chromatin localization will often allow formulation of plausible hypotheses to test in further mechanistic studies of the cis-acting lncRNA.
Investigation of trans-acting lncRNAs
In cases where lncRNA loss of function does not affect local gene expression, a function in trans becomes more likely. Functions in trans can be unambiguously confirmed by rescue experiments in which lncRNA expression is ectopically restored in knockout cells, resulting in the reversal of the phenotypes observed upon lncRNA loss of function. As described above, lncRNAs that act in trans may perform diverse functions (Figure 2). An initial analysis of the subcellular localization of the lncRNA using cell fractionation and fluorescent in situ hybridization will often be informative. The identification of proteins or nucleic acids that interact with the RNA and/or its localization to specific regions of chromatin are an integral part of the functional interrogation of a trans-acting lncRNA and, as discussed above, should incorporate robust crosslinking-based methodologies that reduce false-positive interactions. Finally, it is particularly important when studying this class of lncRNAs to carefully quantify their expression levels and ensure that any proposed mechanistic models incorporate this information. If a lncRNA is believed to regulate a large number of target genes or sequester an abundant protein, plausible stoichiometry must exist between regulator and target.
Concluding remarks
Over the last decade, the study of lncRNAs has stimulated vigorous debate over the question of whether noncoding RNAs represent “transcriptional noise” or truly functional biomolecules. Clearly, there is no unifying answer – meaningful understanding of lncRNA function (or lack thereof) can only be achieved from detailed study on a case-by-case basis. Importantly, our evolving understanding of the prevalence of genomic elements that produce noncoding transcripts, such as enhancers, has mandated that we approach the experimental evaluation of a lncRNA locus with an agnostic view regarding whether the produced RNA is functional. As Occam’s razor dictates, the simplest hypothesis, in this case that the production of a lncRNA most likely marks the presence of a regulatory DNA element, is often the correct one. To substantiate a more complex model, sequential experimentation is needed to establish that transcription is necessary for activity, then that the transcript itself performs a function, followed finally by the possibility that the lncRNA leaves the site of transcription and performs a function elsewhere in the cell. Fortunately, technical advances in genome editing and biochemical analyses of RNP composition have significantly advanced our ability to definitively characterize lncRNA function and elucidate molecular mechanisms of action. The examples highlighted here show that the application of these rigorous approaches can convincingly identify functional lncRNAs and reveal their molecular activities. We are optimistic, therefore, that the continued study of the noncoding transcriptome holds outstanding promise to reveal new and unanticipated biology, with great potential to impact our understanding of normal physiology and disease.
Acknowledgments
We thank Asha Acharya, Mathew Augustine, Ryan Golden, Jaeil Han, Kathryn O’Donnell, Anu Thomas, and Xiaoqiang Zhu for helpful comments on the manuscript. J.T.M. is an Investigator of the Howard Hughes Medical Institute and is also supported by the Cancer Prevention and Research Institute of Texas (RP160249) and the National Institutes of Health (R35CA197311). F.K. is supported by the Leopoldina Fellowship Program (LPDS 2014-12) from the German National Academy of Sciences Leopoldina. We apologize to those whose work we were unable to cite due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume CJ, Hotz-Wagenblatt A, Hullein J, Sellner L, Jethwa A, Stolz T, Slabicki M, Lee K, Sharathchandra A, Benner A, et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29:2015–2023. doi: 10.1038/leu.2015.119. [DOI] [PubMed] [Google Scholar]
- Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botcheva K, McCorkle SR, McCombie WR, Dunn JJ, Anderson CW. Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle. 2011;10:4237–4249. doi: 10.4161/cc.10.24.18383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Cerase A, Pintacuda G, Tattermusch A, Avner P. Xist localization and function: new insights from multiple levels. Genome Biol. 2015;16:166. doi: 10.1186/s13059-015-0733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, Chow A, Cerase A, McDonel P, Guttman M. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Heard E. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol. 2017;24:197–204. doi: 10.1038/nsmb.3370. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Singh RK, White JJ, De Maio A, Han K, Kim JY, Jafar-Nejad P, di Ronza A, Kang H, Sayegh LS, et al. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell. 2015;160:1087–1098. doi: 10.1016/j.cell.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazal G, Gagnon J, Jacques PE, Landry JR, Robert F, Elela SA. Yeast RNase III triggers polyadenylation-independent transcription termination. Mol Cell. 2009;36:99–109. doi: 10.1016/j.molcel.2009.07.029. [DOI] [PubMed] [Google Scholar]
- Groff AF, Sanchez-Gomez DB, Soruco MM, Gerhardinger C, Barutcu AR, Li E, Elcavage L, Plana O, Sanchez LV, Lee JC, et al. In Vivo Characterization of Linc-p21 Reveals Functional cis-Regulatory DNA Elements. Cell Rep. 2016;16:2178–2186. doi: 10.1016/j.celrep.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013a;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013b;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Crawford GE, Reddy TE, Gersbach CA. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol. 2017;35:561–568. doi: 10.1038/nbt.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355 doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Russell P, Guttman M. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014;15:203. doi: 10.1186/gb4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello SS, Sinow C, Raj N, Mazur PK, Bieging-Rolett K, Broz DK, Imam JFC, Vogel H, Wood LD, Sage J, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31:1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA. 2011;2:471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
- Minajigi A, Froberg J, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349 doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18:738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep. 2015;12:562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells. Cell Rep. 2015;12:554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC, et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357 doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- Portoso M, Ragazzini R, Brencic Z, Moiani A, Michaud A, Vassilev I, Wassef M, Servant N, Sargueil B, Margueron R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017;36:981–994. doi: 10.15252/embj.201695335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20:1844–1849. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichon A, Gil N, Lubelsky Y, Havkin Solomon T, Lemze D, Itzkovitz S, Stern-Ginossar N, Ulitsky I. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun. 2016;7:12209. doi: 10.1038/ncomms12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150:1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ. Human 5′ --> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, Dong R, Yang L, Chen LL. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell. 2017;169:664–678. e616. doi: 10.1016/j.cell.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, Li Z, Bu D, Sun N, Zhang MQ, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–1286. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]