Abstract
During development, the embryo transitions from a metabolism favoring glycolysis to a metabolism favoring mitochondrial respiration. How metabolic shifts regulate developmental processes, or how developmental processes regulate metabolic shifts, remains unclear. To test the requirement of mitochondrial function in developing endoderm-derived tissues, we genetically inactivated the mitochondrial transcription factor, Tfam, using the Shh-Cre driver. Tfam mutants did not survive postnatally, exhibiting defects in lung development. In the developing intestine, TFAM-loss was tolerated until late fetal development, during which the process of villus elongation was compromised. While progenitor cell populations appeared unperturbed, markers of enterocyte maturation were diminished and villi were blunted. Loss of TFAM was also tested in the adult intestinal epithelium, where enterocyte maturation was similarly dependent upon the mitochondrial transcription factor. While progenitor cells in the transit amplifying zone of the adult intestine remained proliferative, intestinal stem cell renewal was dependent upon TFAM, as indicated by molecular profiling and intestinal organoid formation assays. Taken together, these studies point to critical roles for the mitochondrial regulator TFAM for multiple aspects of intestinal development and maturation, and highlight the importance of mitochondrial regulators in tissue development and homeostasis.
Keywords: TFAM, mitochondria, intestinal development, intestinal stem cells
1. Introduction
Shifts in cellular metabolic state accompany distinct developmental transitions. The acquisition of induced pluripotency depends upon a metabolic shift from oxidative phosphorylation to glycolysis (Folmes et al., 2011; Shyh-Chang et al., 2013; Zhou et al., 2012). Glycolysis is the preferred biochemical pathway during early stages of embryonic development; proliferating cells are dependent upon glycolysis and the production of glycolytic intermediates to increase biomass. Conversely, differentiated cells produce energy more efficiently without glycolytic intermediates via oxidative phosphorylation (Vander Heiden et al., 2009). Links between metabolic and developmental processes are providing new insights into developmental mechanisms. Many advances linking metabolic state to cellular differentiation leverage the opportunities provided by in vitro study in the fields of regenerative medicine (Wanet et al., 2015), and while fewer in number, in vivo demonstrations of a coupling between metabolic activity and developmental processes are increasing. Metabolic shifts have been implicated in differentiation of mesenchymal, hematopoietic, and muscle stem cells (Bracha et al., 2010; Chen et al., 2008; Qian et al., 2016; Shyh-Chang et al., 2013; Takubo et al., 2013; Tormos et al., 2011), and upon activation of macrophages (Tannahill et al., 2013).
A key hallmark of intestinal development involves a morphological restructuring of the early fetal gut, which transitions from a highly proliferative, pseudostratified epithelium to a columnar epithelium that projects post-mitotic villus structures into the lumen (Walton et al., 2016a). Interspersed between villi, proliferative cellular pockets continue to expand the cellular content of the expanding gut. As the gut matures, these proliferative zones will give rise to Crypts of Lieberkühn that house an active intestinal stem cell population and proliferating progenitor cells in the adult (Chin et al., 2017). Adult villi harbor differentiated cells of absorptive (enterocyte) or secretory (goblet and enteroendocrine) lineages. Previous studies have implicated a variety of transcription factors and signaling molecules in the process of embryonic villus formation (Choi et al., 2006; Kaestner et al., 1997; Karlsson et al., 2000; Kim et al., 2007; Lepourcelet et al., 2005; Madison et al., 2005; McLin et al., 2009; Ormestad et al., 2006; Shyer et al., 2013; Spence et al., 2011; van den Brink, 2007; Walker et al., 2014; Walton et al., 2012; Walton et al., 2016), but the role of metabolic regulators in villus formation is less appreciated. In the developing intestine, genes encoding components of the electron transport chain, which mediate mitochondrial Ox-Phos, show elevated expression coinciding with the onset of villus morphogenesis in fetal stages (Kumar et al., 2016), and pharmacological inhibition of the electron transport chain compromises villus maturation in an ex vivo culture system (Kumar et al., 2016). However, the genetic disruption of a direct mitochondrial regulator has not been conducted in the context of intestinal development.
In adult mice, there is also a shift from glycolysis to oxidative phosphorylation that correlates with cell position along the crypt-villus axis. Stem cells located at the bottoms of the crypts show increased glycolysis, whereas differentiated cells near the villus tips exhibit increased oxidative phosphorylation (Stringari et al., 2012). This gradient is presumed to make glycolytic intermediates available for increased biomass in the proliferative crypt, while in the differentiated villus, efficient oxidation of glucose via the electron transport chain maximizes ATP production to fuel the enzymatic activities driving digestion. It has been suggested that this glycolysis-Ox-Phos gradient along the crypt-villus axis can be exploited for regulatory function. Luminal butyrate is consumed via aerobic respiration in colonocytes at the luminal surface, and thus bioavailability of butyrate is diminished at the crypt base, where stem cells are spared the growth-inhibitory function of butyrate (Kaiko et al., 2016). Stem cell renewal itself is subject to ROS signaling and may be influenced by mitochondrial-generated ROS, as Ox-Phos inhibitors block stem cells from functioning in an organoid assay (Rodriguez-Colman et al., 2017), and ROS promotes stem cell signature gene expression in a Rac1-dependent manner, particularly upon APC inactivation in colon polyps (Myant et al., 2013). Despite the important advances, regulators of the metabolic gradient along the crypt-villus axis in the adult tissue or in the embryonic transition to villus formation are incompletely understood. Knockout models that disrupt mitochondrial function, such as YY1, trigger stem cell loss (Berger et al., 2016; Perekatt et al., 2014), but may also disrupt other cellular processes (Blattler et al., 2012; Jeong et al., 2017). Genetic inactivation of a factor specific for mitochondrial function would clarify the importance of mitochondrial function in intestinal development and homeostasis.
TFAM (transcription factor A, mitochondrial) is a nuclear gene critical for transcription and replication of the mitochondrial genome. (reviewed in (Campbell et al., 2012)). TFAM regulates thirteen genes in the mitochondrial genome that code for critical components of the electron transport chain, making TFAM a vital factor for energy production via oxidative phosphorylation (Shi et al., 2012). Thus, genetic inactivation of TFAM provides a precise tool to interrogate the role of mitochondrial metabolism in biological processes. TFAM inactivation early in embryonic development with the β-Actin-Cre driver leads to enlarged mitochondria with abnormal cristae, and embryonic lethality before E10.5 (Larsson et al., 1998). However, prior to E8.5, no stark phenotype was observed, suggesting that mitochondrial activity is dispensable for early embryo growth and proliferation. In developmental processes, TFAM was shown to be important for differentiation of the epidermis, as differentiation of keratinocytes is compromised upon conditional inactivation of TFAM, while epidermal progenitor proliferation was unperturbed (Hamanaka et al., 2013). In the intestine, TFAM heterozygous mice were observed to be more susceptible to APCmin-driven polyp formation, possibly owing to increased oxidative stress and DNA damage (Woo et al., 2012), and truncating mutations of TFAM have been observed in colon cancers (Guo et al., 2011). However, homozygous loss of TFAM in the adult intestine or in the developing intestine has not been explored.
In the current study, we use both developmental and adult-onset intestinal epithelial Cre drivers to specifically test the requirements for TFAM in the developing and adult intestinal epithelium. We find that TFAM loss has no consequence for early intestinal growth and expansion, but is required for proper villus maturation. Similarly, we find that ablation of TFAM in the adult intestinal epithelium compromises maturation of enterocytes on intestinal villi, leading to weight loss and death. Interestingly, we also find that TFAM-loss disrupts intestinal stem cell homeostasis in the adult niche, with loss of resident stem cells and an inability of TFAM-deficient crypts to self-renew in organoid cultures. Taken together, these studies point to critical roles for the mitochondrial regulator TFAM for multiple aspects of intestinal development and adult functions, and highlight the importance of mitochondrial regulators in tissue development and homeostasis.
2. Results
TFAM is required for proper lung development
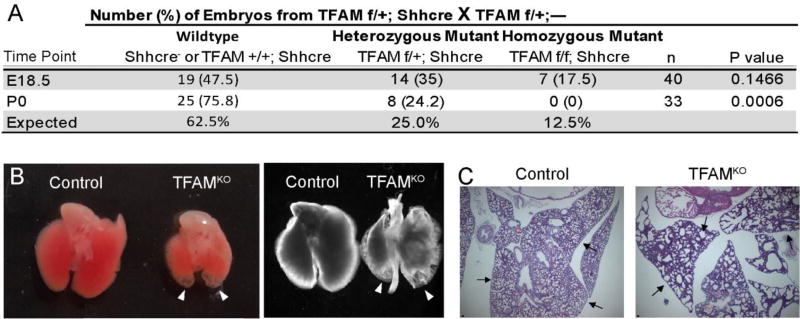
We created a genetic model employing the Shh-Cre driver (Harfe et al., 2004) to recombine Tfamf/f alleles (Hamanaka et al., 2013; Larsson et al., 1998). The Shh-Cre allele drives Cre expression, beginning at approximately E9.5, throughout the epithelium of several endoderm-derived organs, including the intestinal epithelium, but also in the stomach and lungs. The Shh-Cre; Tfamf/f mutation was neonatal lethal, as Mendelian ratios were observed at E18.5, but no viable pups were observed on or after postnatal day 1 (Fig. 1A). We examined the lungs for a potential cause of an acute, neonatal lethal phenotype. Indeed, the lungs in E18.5 embryos exhibited transparent cysts in the distal lobes (Fig. 1B–C), a characteristic of branching morphological defects and incomplete development of the peripheral airway (Cardoso and Lu, 2006; Mucenski et al., 2003). Thus, TFAM is essential for proper lung development and postnatal survival.
Figure 1. Loss of TFAM in the developing endoderm-derivatives is neonatal lethal with compromised lung development.
(A) Mice lacking TFAM in developing endoderm tissues were generated using the Shh-Cre driver. No surviving pups were observed after birth, whereas pups were present at expected Mendelian ratios one day prior to birth (Chi-squared test). (B) Whole mount images of E18.5 lungs from TFAM mutant or littermate controls (either front-lit or back-lit). Arrowheads point to transparent tips at the distal regions, which correspond to under-developed cysts upon histological examination (C). Scale bar = 50µm. Arrows point to peripheral airway differences.
TFAM loss results in an immature neonatal gut, particularly affecting villus elongation
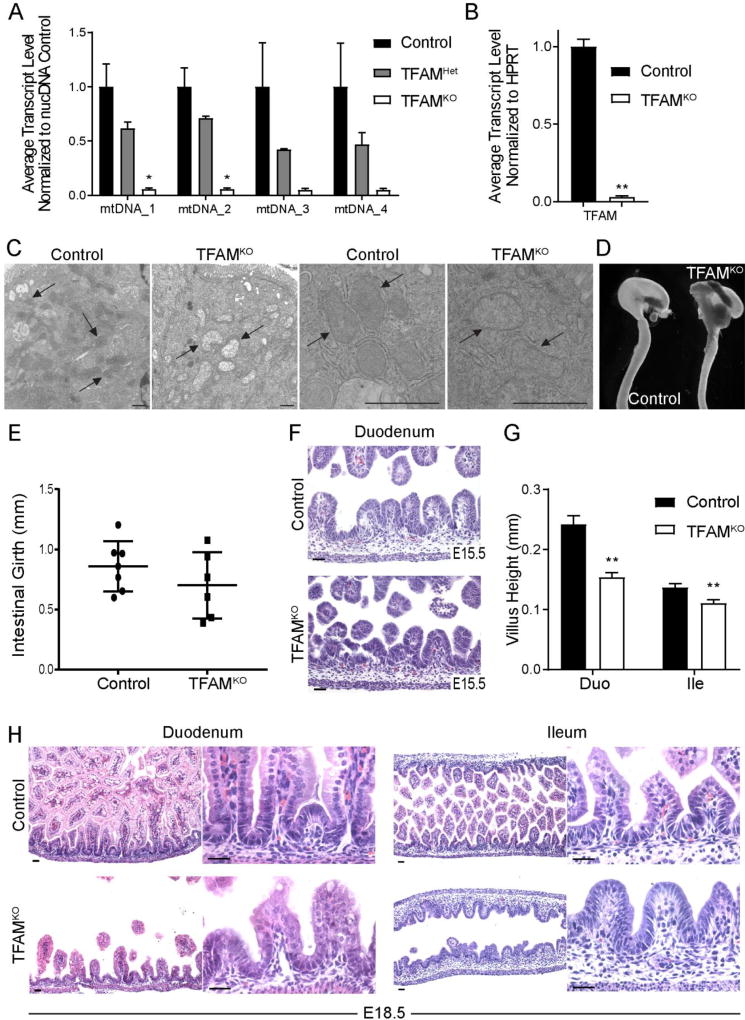
To determine whether TFAM-driven control of mitochondrial function was also important for intestinal development, we characterized Tfam mutant intestines versus littermate controls. mtDNA was diminished in the Tfam mutant epithelium, as were transcripts encoding Tfam, reflecting the loss of the mitochondrial transcription factor in our model (Fig. 2A–B). Severe reduction in Tfam transcripts were observed in both the proximal and distal gut epithelium, and, consistent with loss of TFAM function, transcripts of genes residing in the mitochondrial genome were similarly compromised (Supplemental Figure 1A–B). Loss of TFAM function also corresponded to disrupted mitochondrial architecture (Figure 2C, Supplemental Figure 1C), with transmission electron micrographs indicating that the mitochondria in mutant cells lack inner mitochondrial membranes and lose electron density. At E18.5, Tfam mutant intestines were similar in length to their littermate controls (Supplemental Figure 1D–E), but exhibited diminished girth (Fig. 2D–E). We then examined the consequence of Tfam loss across a timecourse of intestinal development. At E15.5 no morphological differences were observed, however by E18.5, mutant embryos exhibited shorter, stunted villi (Fig. 2F–H). Reduced villus height was observed in both the duodenum (p = 2.376E-06) and, to a lesser degree, in the ileum where villus maturation is developmentally slower to progress (p = 0.00365) (Fig. 2G–H). Thus, while Tfam appears dispensable for embryonic gut elongation and initiation of villus morphogenesis, it is required for fetal maturation of the gut epithelium and proper villus elongation.
Figure 2. TFAM is required for developmental maturation of the intestinal epithelium.
(A) Mitochondrial DNA levels are diminished in Shh-Cre; Tfamf/+, and nearly lacking in Shh-Cre; Tfamf/f gut epithelial cells at E18.5, consistent with reduced TFAM function (qPCR, Students t-test, n=4, p< 0.05), and (B) loss of Tfam mRNA in these mutants (qRT-PCR, students t-test, n=4). (C) TEM indicates that mitochondrial ultrastructure is compromised in the jejunum of E18.5 mutants (representative of 2 knockout and 3 control samples). (D) Whole mount images of representative guts dissected from TFAMKO or littermate control mice at E18.5. While length of the gut was unaffected by TFAM-loss (Supplemental Figure 1D–E), (E) the girth of the gut trended smaller in the mutants. (F) At E15.5, no differences were observed in the morphology of the developing intestine, suggesting that villus formation initiates normally. (G–H) However, by E18.5, villi were noticeably shorter, particularly in the duodenum, indicating that TFAM is required for villus maturation and extension into the developing gut lumen (t-test, n= 4). Scale bars = 50µm.
Enterocyte differentiation is compromised upon loss of TFAM
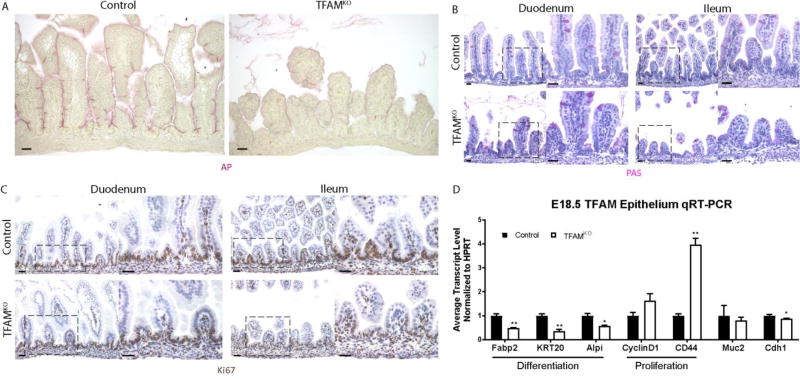
We next investigated the role of Tfam in promoting the development of specific epithelial cell lineages. Alkaline phosphatase activity, a marker of mature enterocytes, was dramatically reduced in Tfam mutants whereas goblet cells, marked by Periodic-acid-Schiff staining, exhibited no obvious differences (Fig. 3A–B, Supplemental Figure 1F). These results indicated that enterocyte differentiation is particularly sensitive to loss of TFAM. Conversely, cell proliferation, which occurs at intervillus domains of the epithelium in the fetal gut, remained unchanged in epithelium lacking TFAM (as defined by Ki67 immunostain, Fig. 3C, Supplemental Figure 1G). These findings were observed in both the proximal and distal neonatal intestine. Thus, while maturation of the enterocyte lineage and villus elongation appeared dependent upon TFAM, proliferation proceeded normally. To corroborate these observations, we measured transcript levels from the isolated intestinal epithelia of E18.5 TFAM-deficient embryos and littermate controls. Transcript levels of enterocyte differentiation markers such as Fabp2, Krt20, and Alpi are significantly reduced in TFAM mutants, whereas proliferation markers such as Ccnd1 and Cd44 remain relatively unchanged or even increase (Fig 3D, Supplemental Figure 1H). Muc2 and Tff3, markers for mucin-secreting goblet cells, remained unchanged, mirroring histological findings, and Cdh1 levels, a marker of epithelial cells, was modestly affected. Taken together, these data provide strong evidence that the mitochondrial regulator TFAM is required for enterocyte maturation during intestinal development.
Figure 3. Enterocyte maturation is particularly susceptible to TFAM-loss.
(A) Alkaline phosphatase activity is diminished in the duodenum of TFAMKO embryos compared to littermate controls at E18.5, whereas goblet cells appear unaffected (B). (C) Proliferation in the intervillus regions appears similar between control and TFAM-mutant embryos at E18.5. (D) qRT-PCR analysis of transcripts isolated from the E18.5 whole gut epithelium indicate that markers of enterocyte or proliferating cells are expressed in a manner consistent with histological analysis. These data point to a specific requirement for TFAM in enterocyte maturation, whereas TFAM function appears dispensable for cellular proliferation. (t-test, n = 4) Scale bars = 50µm.
TFAM is required in the mature intestinal epithelium for terminal differentiation
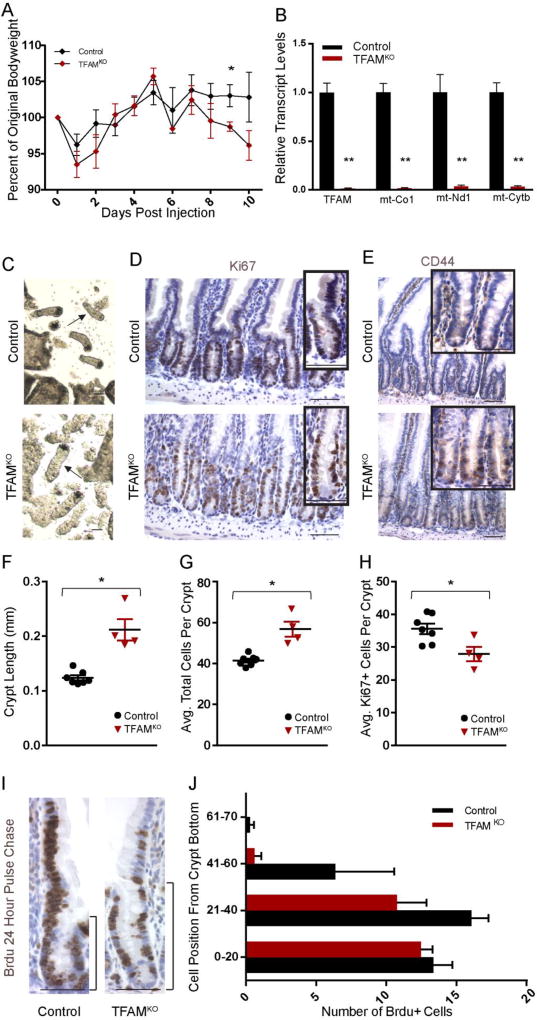
Since TFAM loss in embryos results in a disruption of fetal differentiation, we wondered whether TFAM may play a similar role in mediating differentiation in the adult intestinal epithelium. To conditionally inactivate TFAM in the adult intestinal epithelium, we employed the intestine-specific, tamoxifen inducible Villin-CreERT2 driver. Tamoxifen treatment of Tfamf/f; Villin-CreERT2 mice had no immediate consequence on the animals, however by 9 days post initiation of treatment, the mutants exhibited significantly decreased bodyweight compared to littermate controls (Fig. 4A), appeared moribund, and required humane euthanasia the following day as their bodyweight continued to decline. Dissection of the mutants revealed fluid-filled, distended intestines. Efficient Tfam inactivation, and compromised mitochondrial function was indicated by precipitous drops in mitochondrial-encoded transcripts such as mt-Co1, mt-Nd1, and mt-Cytb (Fig. 4B). These observations were corroborated by immunoblotting for the mitochondrial complex 1 gene NDUFS3 (Supplemental Figure 2A). Upon epithelial isolation of the jejunum, it was immediately observed that the crypts were significantly elongated in the mutants, and this was confirmed in histological sections of the duodenal epithelium (Fig. 4C–E). More cells were observed per crypt in the mutant mice, and the crypts were longer (Fig. 4F–G), with a broader zone of Ki67+ staining and more robust expression of the transit amplifying cell marker CD44 (Fig. 4 D–E). Paradoxically, there was a slight decrease in the number of proliferating cells (Fig. 4 H), and there were no differences in apoptosis to explain differences in crypt length, as assayed by cleaved caspase-3 staining (Supplemental figure 2B). Longer crypts without increased numbers of proliferating cells suggested the possibility that Tfam mutant crypt cells failed to mature and were slow to transition to the villus, which was consistent with 24-hour BrdU pulse chase analysis, indicating that proliferating cells in the mutant crypts were slower to transit onto the villus (Fig. 4I–J). Taken together, these results suggest that loss of Tfam leads to elongated crypts via a slower crypt cell transit time and a prolonged period of crypt cell identity, which may come at the expense of cellular differentiation.
Figure 4. TFAM mutant epithelia harbor elongated crypts.
(A) Adult-onset loss of TFAM, using Villin-CreERT2; Tfamf/f mice, results in weight loss and death within 11 days after the initial tamoxifen treatment. (t-test, n= 7 controls, 5 mutants). (B) Consistent with efficient loss of TFAM, transcripts of Tfam and targets of TFAM in the mitochondrial genome are almost undetectable in the mutants (t-test, n=7 controls, 4 mutants). (C) Crypt lengths are noticeably longer in adult mice 10 days after TFAM inactivation, as observed in whole mount images, and histologically, longer crypts are also observed (D–E). Longer crypt length corresponds to an increase in total cell numbers in the crypts (F–G), and (E) elevated immunoreactivity for CD44, a crypt cell marker. Surprisingly longer crypts are not accompanied by an increased number of proliferating cells (D, H). BrdU pulse-chase analysis at 24 hours post-treatment suggests there is reduced migration rates of BrdU-labeled cells onto the villus of Tfam mutants (t-test, n= 3 controls, 3 mutants) (I–J). Taken together, these results suggest increased numbers of cells with crypt identity upon TFAM-loss. Histology is imaged and quantified from the duodenum. Scale bars = 50µm.
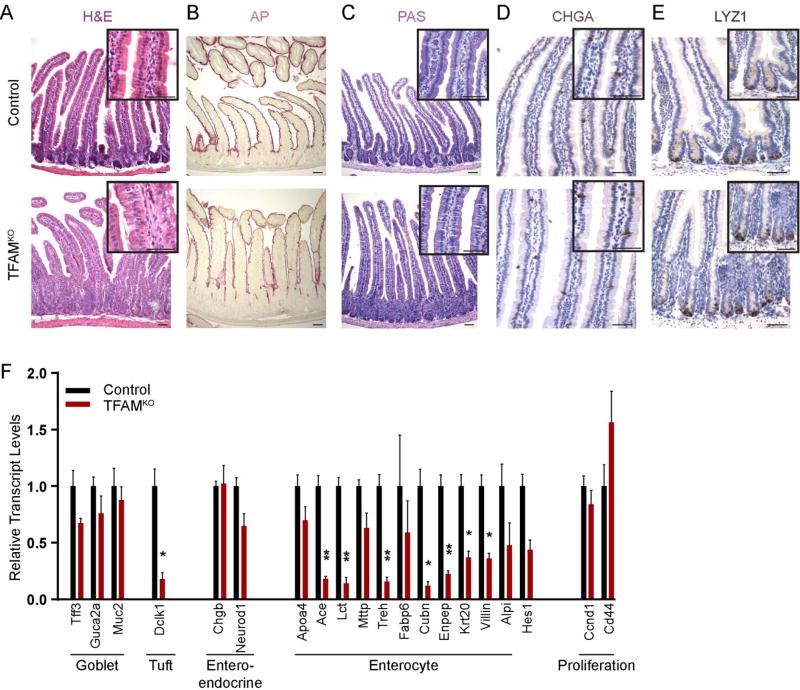
We next investigated epithelial differentiation in the adult Tfam mutants. Mutant villi harbored a poorly organized epithelium, with reduced apical hematoxylin staining and diminished alkaline phosphatase activity (Fig. 5A–B). Conversely, no differences between control and mutant mice were observed in PAS-stained samples, CHGA immunostaining, or LYZ1 immunostaining, suggesting that goblet, enteroendocrine, and Paneth cell maturation proceeded normally in the absence of TFAM (Fig. 5C–E, Supplemental Figure 2C–E). Quantification of tuft cells, marked by DCLK1 immunostaining also did not reveal significant changes (Supplemental Figure 2F–G). These findings suggested that enterocyte maturation may be more specifically compromised upon adult-onset loss of TFAM, similar to what was observed upon TFAM loss in the embryo (Figs. 2–3). Indeed, transcript levels characteristic of enterocyte function were reduced in the Tfam mutants, while markers of goblet cell function, enteroendocrine cell function, and cellular proliferation (Ccnd1 and Cd44) were unchanged (Fig. 5F). The tuft cell marker Dclk1 was significantly reduced, which was surprising given that there were no differences in the numbers of cells marked by DCLK1 expression (Supplemental Figure 2F). Taken together, these results indicate that in the absence of TFAM, cells fail to undergo terminal differentiation, leading to an expanded crypt cell population. This echoes the Tfam mutant phenotype observed in fetal development, where cells are competent to proliferate, but fail to undergo terminal differentiation, with enterocyte differentiation appearing particularly susceptible to TFAM loss.
Figure 5. Enterocyte differentiation is incomplete upon loss of TFAM.
(A–E) Villus epithelial cells lack differentiated features characteristic of enterocytes (Alkaline phosphatase activity, B), but do not exhibit appreciable differences in goblet (PAS, C), enteroendocrine (CHGA, D), or Paneth (LYZ1, E) cell lineages. (F) These findings are corroborated by qRT-PCR analysis of markers of differentiated intestinal cell lineages, with markers of the enterocyte lineage particularly affected by TFAM loss, relative to proliferative cell markers or markers of other differentiated cell lineages. (t-test, n=7 controls, 4 mutants). All histology is imaged from the duodenum. Scale bars = 50µm.
TFAM loss compromises intestinal stem cell renewal
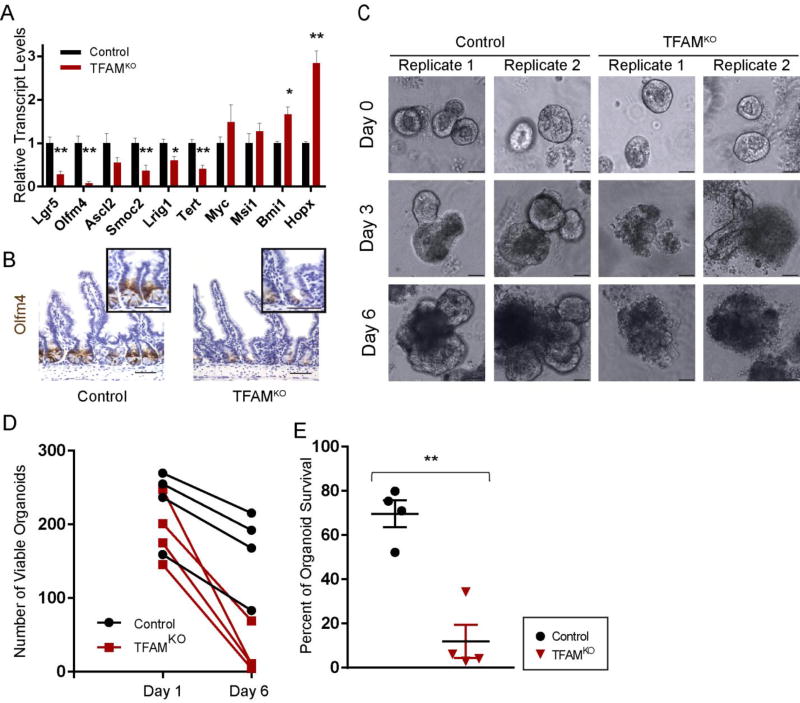
Molecular profiling of the TFAM-deficient epithelium also pointed to a disruption in the intestinal stem cell population, as markers for the crypt-base-columnar cell population were significantly reduced in the TFAM mutants (Fig. 6A). Echoing these findings, immunoreactivity for the crypt-base-columnar stem cell marker OLFM4 was notably diminished in crypts lacking TFAM compared to littermate controls (Fig. 6B). To directly assay stem cell activity, we tested the ability of TFAM-deficient crypt epithelial cells to propagate in organoid cultures. Unlike crypts isolated from Cre-negative littermate controls, Tfamf/f;Vil-CreERT2 crypts isolated 4 days after the onset of tamoxifen treatment rapidly atrophied over the course of 6 days, despite initiating organoid structures (Fig. 6C–E). Taken together, these results indicate that TFAM has dual functions in the mature intestinal epithelium; it maintains an active stem cell population, and promotes enterocyte maturation. These results underscore a growing appreciation for the role of the metabolic state in regulation of tissue homeostasis, and suggest that stem cells and enterocytes are particularly sensitive to disruptions in mitochondrial regulators, whereas transit amplifying cells and goblet cells are more tolerant to loss of the mitochondrial transcription factor, TFAM.
Figure 6. TFAM is required for intestinal stem cell maintenance.
(A) Transcript levels of genes known to express in the crypt-base-columnar stem cells are dramatically reduced after TFAM-loss in the adult epithelium (t-test, n= 7 controls, 4 mutants). (B) Immunohistochemistry for the crypt-base-columnar stem cell marker OLFM4 shows that this protein is greatly reduced in Tfam mutants. (C) Crypt cultures were performed to assay stem cell activity in control and TFAM-mutant mice. Crypts from mice 4 days after treatment to induce TFAM loss gave rise to initial organoid structures but could not sustain their growth, consistent with a lack of stem cell activity. Control crypts gave rise to viable organoids. (Images correspond to 2 independent biological replicates. Six biological replicates exhibited similar results.) (D–E) Quantification of organoid forming efficiency and survival to 6 days after plating duodenal crypts (t-test, n= 4 controls, 4 mutants). Histology is imaged from the ileum, but representative of the entire small intestine. Scale bars = 50µm.
3. Discussion
Warburg’s observation that cancer cells, even when situated in an oxygen-rich environment, prefer to ferment glucose (Warburg O., 1924) foreshadowed links between the fields of cancer and metabolism through which exciting therapeutic opportunities are being pursued. Links between metabolism and developmental processes are likely to be just as important. Developmental tissues, depending upon whether their cells reside in an actively proliferative or a more quiescent or post-mitotic state are expected to have different energy demands. The early embryo is rapidly expanding and thus is dependent on glycolytic intermediates to increase biomass (Vander Heiden et al., 2009). In the developing gut and in the transit amplifying zone, cells are highly proliferative, and, based upon our results, appear less dependent on mitochondria. It has been shown that proliferative cells in the crypts predominantly undergo glycolytic metabolism compared to the differentiated cells in the villus, which mainly rely on oxidative phosphorylation (Kaiko et al., 2016; Stringari et al., 2012). Our results are consistent with these findings, and provide genetic evidence based upon differential susceptibility of these cell lineages to loss of the mitochondrial transcription factor, TFAM (Figure 7). Cellular differentiation is particularly susceptible to TFAM loss, as mitochondria-dependent oxidative phosphorylation is likely required to provide ATP concentrations necessary to fuel the uptake, processing, and transport of dietary nutrients by enterocytes. We speculate that goblet cells would exhibit less energy demands, and thus goblet cell differentiation is less dependent upon TFAM function. Additionally, more cell divisions occur in the development of the enterocyte lineage (as transit amplifying cells) than in the development of the secretory lineages (Goblet, Paneth, enteroendocrine), which are direct descendants of the stem cells. The increased number of cell cycles that precede enterocyte differentiation could put additional stress on the mitochondrial populations of enterocytes. Single cell analyses will be important to address these questions in the future.
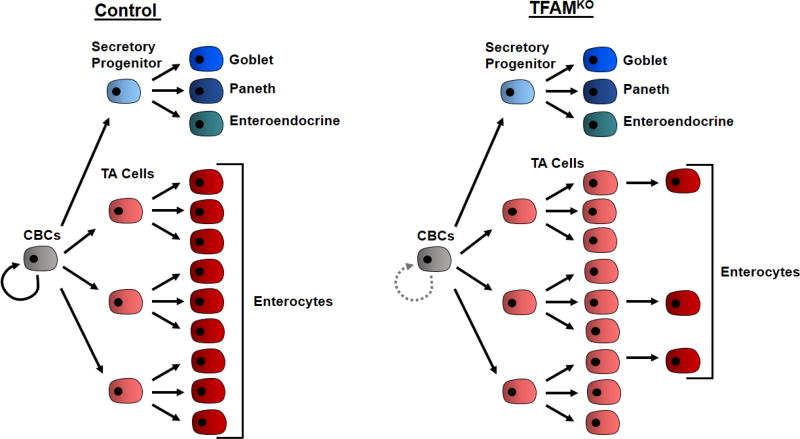
Figure 7. Overall effect of TFAM loss.
Upon loss of TFAM, stem cells are deficient in self-renewal. Tfam-mutant stem cells are still able to differentiate into transit amplifying progenitor cells, and secretory progenitor cells. However, while development of secretory lineage progeny (goblet cells, Paneth cells and enteroendocrine cells) appears normal, transit amplifying cells do not transition efficiently to their enterocyte progeny in TFAMKO epithelium.
Intestinal stem cells are dependent upon the mitochondrial regulator TFAM, whereas their proliferating progeny in the crypt appear tolerant of TFAM loss. This may reflect the nature of stem cells being less mitotically active than transit amplifying cells, and thus being less obliged to rely upon glycolysis, and more dependent upon oxidative phosphorylation (Rodriguez-Colman et al., 2017). However, the intestinal stem cell loss observed in TFAM mutants may also reflect sensitivity of intestinal stem cells to ROS levels, which can be elevated upon disruption of mitochondrial function (Asano et al., 2017; Kumar et al., 2016; Myant et al., 2013; Xu et al., 2017).
It is unclear whether developmental signaling processes dictate the cellular metabolic state, or if the opposite occurs, where shifts in cell metabolism regulate developmental processes. Loss of differentiation-promoting transcriptional networks in the intestine lead to downregulation of genes functioning in oxidation/reduction (San Roman et al., 2015), suggesting that developmental signaling pathways instruct metabolic state. Conversely, inactivation of TFAM, or pharmacological block of the electron transport chain, blocks enterocyte maturation (this study and (Kumar et al., 2016)), demonstrating that the metabolic state can function epistatically to developmental regulators. Whether the differentiation deficit observed upon TFAM loss has origins in alterations in CBC cell function also remains an open question. Most likely, developmental signaling pathways and metabolic states function in a complex and collaborative process to achieve proper tissue development and function. The regulatory relationship between developmental signaling pathways and metabolism is an important frontier for both developmental biology and regenerative medicine.
4. Materials and Methods
Mice
Tfamf/f mice (Hamanaka et al., 2013; Larsson et al., 1998), and Shh-cre mice (Harfe et al., 2004) were obtained from Jackson Labs. Villin-CreERT2 mice (el Marjou et al., 2004) were kindly shared by Dr. Sylvie Robine. All experiments were approved by the Rutgers University IACUC.
Tissue Preparation
Mouse embryos and adult intestinal tissue were dissected, and after embedding the tissue sections into paraffin blocks, 5µm sections were taken for histology. Regions from the small intestine (duodenum and ileum or the proximal region including the stomach and distal region including the caecum) were left overnight in 4% paraformaldehyde fixative at 4°C. The tissues were then washed with Phosphate-Buffered Saline (PBS) and dehydrated through an increasing concentration of ethanol for 1 hour in each solution, and then put into Xylene for 1 hour, twice. After the sections were completely dehydrated, they were placed in a heated Xylene/paraffin mixture for 1 hour, and then transferred to pure liquid paraffin for an additional hour. The tissue sections were then embedded into wax and left overnight at −20°C to harden until needed for sectioning. For epithelial cell isolation, embryos were collected at E18.5, and the embryonic intestine was opened with forceps and incubated in 3mM EDTA for 40 minutes at 4°C on a rotator. Adult jejunal epithelium was collected in a similar manner. In the case of tissue prepared for BrdU immunohistochemistry, the mice were injected with 1 mg BrdU, 30 minutes or 24 h before euthanasia.
Antibodies and Staining Conditions
Sections were processed for immunostaining with different antibodies using the Vectastain ABC (avidin biotin complex) Kit by Vector Laboratories (PK6101) and counterstained with hematoxylin. A one hour antigen retrieval step in 10mM sodium citrate solution (pH 6) under 15 psi pressure was used for all immunostains and then the slides were placed in 0.5% H2O2 in water for 20 minutes for a peroxidase quenching step. After washes in PBS, the slides were left in 0.5% Triton X-100 in PBS for 5 minutes. The slides were then blocked with 1.5% Normal Goat Serum (NGS) or Fetal Bovine Serum (FBS) in PBS for 1 hour and then were incubated in primary antibody overnight at 4°C in a humid environment. The next day they were incubated in secondary antibody and ABC solution (in the dark) each for 1 hour in a humid chamber. The slides were then placed into 0.1M Tris solution (pH 7.4) before being developed with freshly DAB solution (1:20 dilution of 1% DAB and 1:20 dilution of 0.3% H2O2 in 0.1M Tris pH 7.4).
The primary antibodies and dilutions used for staining are as follows: BrdU (1:500, AbD Sterotec, MCA2060GA), CC3 (1:200, Cell Signaling, 9661), CD44 (1:200, BD Pharmingen, 558739), CHGA (1:300, Santa Cruz, sc-1488), DCLK1 (1:300, Abcam, ab37994), Ki67 (1:300, Abcam, ab16667), OLFM4 (1:1000, Cell Signaling, 39141), LYZ1 (1:250, AR-024-5R). For Cleaved Caspase 3, the primary antibody was diluted in the Signal Stain Ab Diluent (Cell Signaling, 8112S). For a Periodic acid-Schiff (PAS) stain, slides were incubated in 0.5% periodic acid and stained with Schiff ’s reagent (J612171, Alfa Aesar). For alkaline phosphatase staining, 1-Step NBT/BCIP (34042, Thermo Scientific) was used. Before developing the alkaline phosphatase, the slides were incubated in a 0.1M Tris-HCL buffer of pH 8.
Images were taken using a Retiga 1300CCD (Q-Imaging) camera and a Nikon Eclipse E800 microscope with the QC-Capture imaging software. Oil was used as the medium for 60×/1.4 N.A. magnification and air as the medium for 10×/0.45 N.A., 20×/0.75 N.A., and 40×/0.75 N.A.
Transmission EM
Intestinal tissues were freshly collected, flushed in cold PBS, dissected into 1–3 mm fragments, and fixed overnight at 4 °C in 0.1 M sodium cacodylate buffer (pH 7.4) containing 2.5% glutaraldehyde and 2.0% (vol/vol) para-formaldehyde. Fixed tissues were washed briefly with 0.1 M sodium cacodylate buffer and postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 1 h at 4 °C. Tissues were washed with excess distilled water and En bloc-stained with 1% aqueous uranyl acetate for 30 min in the dark. Tissues were then washed with distilled water and dehydrated through a graded series of ethanol and propylene oxide. Tissues were then transferred into EMBed 812 (Electron Microscopy Sciences 14120) and propylene oxide (mixed at 1:1) overnight at room temperature in tightly capped vials on a shaker. Tissues were then transferred into 100% EMBed 812 overnight at room temperature and then placed into embedding molds at 65°C for 48 hours. Ultrathin sections (~70 nm) were cut; grids were stained with uranyl acetate and lead citrate and observed using FEI Tecnai 12 transmission electron microscope and micrographs were recorded using a Gatan OneView 16-megapixel camera. Multiple grids were analyzed for each tissue sample.
Histological Quantification
Image-J software was used to perform calculations using specific pixel/mm ratios. In order to get pixel aspect ratios used in calculating villus length and width, hemocytometer images were used at each magnification to calculate the number of pixels per 0.05mm. These ratios were then applied to each image, depending on the magnification, and were used to calculate the length and width of villi in mm. Villi that were measured for quantification were whole and protruded from the basement membrane. Intestinal length and duodenal girth was measured 10mm from the gastroduodenal junction. A straight line was drawn from one side of the gut tube to the other and the length was calculated on ImageJ using a scale individually set for each image (based on a ruler placed in whole mount images of the intestines). BrdU positive cells were quantified by counting in 20 cell intervals above the base of the crypts. The percentage of Ki67 positive cells per crypt was quantified by counting total Ki67 positive cells and dividing them by the total number of crypt cells. Similarly, goblet cells were quantified by counting goblet cells per villus and dividing it by the total number of villus cells. Tuft cells were quantified by counting the total number of tuft cells per villus. 10 counts were averaged per biological replicate for each quantification.
DNA/RNA Preparation and qPCR
Tissue was dissolved in Trizol and RNA was prepared using manufacturer’s protocols. The RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen, 18080051) to prepare cDNA. DNA was isolated from both E18.5 epithelial cell pellets and E18.5 luminal contents stored in 1× PBS by the QIAamp DNA Mini Kit (Qiagen, 51304). qPCR analysis was conducted using gene-specific primers and SYBR Green PCR Master Mix (Applied Biosystems, 4309155), with relative transcript levels normalized to Hprt levels in each sample for qRT-PCR. Mitochondrial DNA primer sequences are:
| Primer Pair name | Sequence |
|---|---|
| mtDNA_1 | 5’-GGATCCGAGCATCTTATCCA-3’ |
| 5’- GGTGGTACTCCCGCTGTAAA-3’ | |
| mtDNA_2 | 5’-ATGGGTGTAATGCGGTGAAT-3’ |
| 5’- ACCAGCATTCCAGTCCTCAC-3’ | |
| mtDNA_3 | 5’- CCCAGCTACTACCATCATTCAAGT-3’ |
| 5’- GATGGTTTGGGAGATTGGTTGATGT-3’ | |
| mtDNA_4 | 5’- CCCATTCCACTTCTGATTACC-3’ |
| 5’- ATGATAGTAGAGTTGAGTAGCG-3’ |
Intestinal Organoid Culture
Primary organoid cultures were obtained from duodenal epithelium of Tfam mutant or littermate control mice 4 days after the onset of tamoxifen treatment to induce knockout. The intestine was washed in PBS, cut into 1 cm pieces, and then rotated for increments of 5, 10, and 25 minutes in 3 mM EDTA diluted in PBS. The tissue was then agitated by shaking approximately 30 times, and then filtered through a 70-um filter to isolate the crypts. The crypts were then washed with PBS and pelleted with low-speed centrifugation (170 × g) at 4 degrees Celsius. Crypts were then resuspended in matrix (BME-R1, Trevigen) and plated at a density of 250 crypts per 25 ul matrix.
The cultures were overlayed with Crypt culture media including primocin (InvivoGen), R-Spondin conditioned media, EGF (PMG8041), Noggin (250-38-25046), 0.5 M N-Acetyl Cysteine, B27 (Gibco 12587-010), and N2 supplement (Gibco 17502-048) suspended in Advanced DMEM/F12 (12634-028) supplemented with Glutamax (REF 35050-61), HEPES (15630-080), and Pen/Srep (15140-122), as previously described (Sato et al., 2009).
Statistics
In each case the statistical test is indicated in the figure legend. One asterisk: p<0.05, two asterisks: p<0.01.
Supplementary Material
(A–B) Transcript levels of Tfam and genes encoded in the mitochondrial genome are strongly reduced in both the proximal and distal fetal small intestinal epithelium (E18.5) (t-test, n=2). (C) Original images that were contrast-adjusted for use in Figure 2C. All other images in Figure 2C were not contrast adjusted. (D) Whole-mount preparation of E18.5 GI tract shows similar lengths of intestines of Control and TFAMKO mice, quantified in (E) (t-test, n= 7 control, 5 mutants). (F–G) Quantification of the number of cells in the E18.5 TFAM-mutant duodenal epithelium for PAS+ goblet cells (t-test, n= 4 control, 3 mutants) and Ki67+ cells (t-test, n= 3). (H) Additional markers of intestinal lineages were analyzed from RNA isolated from whole E18.5 control and TFAM-mutant small intestinal epithelial cells (t-test, n=2). Note that markers of the enterocyte lineage are particularly compromised upon TFAM loss.
(A) Loss of TFAM in the adult leads to loss of NDUFS3 expression. (B) No differences in cleaved caspase-3+ cells were observed between control and mutants. (C–F) Quantification of cell populations to accompany immunohistochemical analysis presented in Figure 5 and Figure S2G (t-test, n=6 control, 4 mutants C–D; n=7 control, 4 mutants E–F). (G) Immunostaining for DCLK1 is similar in control and mutant tissues. The duodenum is shown in these images, but the results are representative of the entire small intestine.
Highlights.
The mitochondrial transcription factor TFAM is required in the developing endoderm for proper lung development and post-natal survival
TFAM is dispensable early in intestinal development, but required in late fetal stages for maturation of the tissue and enterocyte differentiation
In the adult intestine, TFAM loss leads to expansion of cells in the crypt progenitor zone at the expense of differentiated villus cells.
Intestinal stem cell renewal also depends upon TFAM, highlighting a complex set of functions for the mitochondrial regulator in intestinal development and homeostasis.
Stem, progenitor, and differentiated intestinal epithelial cells exhibit unique dependencies on mitochondrial regulators, suggesting distinct metabolic requirements for each cell type.
Acknowledgments
This work was supported by the NIDDK of the NIH under grants R03DK099251 and U01DK103141 (MPV) and a startup grant from the Human Genetics Institute of New Jersey. Cancer Genetics Summer Undergraduate Research Fellowships supported BNW and NHT. MS was supported by Aresty fellowships. MS, BNW, and NHT also thank Drs. Namit Kumar, Kevin Tong, and Lei Chen for their mentorship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest
References
- Asano J, Sato T, Ichinose S, Kajita M, Onai N, Shimizu S, Ohteki T. Intrinsic Autophagy Is Required for the Maintenance of Intestinal Stem Cells and for Irradiation-Induced Intestinal Regeneration. Cell reports. 2017;20:1050–1060. doi: 10.1016/j.celrep.2017.07.019. [DOI] [PubMed] [Google Scholar]
- Berger E, Rath E, Yuan D, Waldschmitt N, Khaloian S, Allgauer M, Staszewski O, Lobner EM, Schottl T, Giesbertz P, Coleman OI, Prinz M, Weber A, Gerhard M, Klingenspor M, Janssen KP, Heikenwalder M, Haller D. Mitochondrial function controls intestinal epithelial stemness and proliferation. Nat Commun. 2016;7:13171. doi: 10.1038/ncomms13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler SM, Cunningham JT, Verdeguer F, Chim H, Haas W, Liu H, Romanino K, Ruegg MA, Gygi SP, Shi Y, Puigserver P. Yin Yang 1 deficiency in skeletal muscle protects against rapamycin-induced diabetic-like symptoms through activation of insulin/IGF signaling. Cell metabolism. 2012;15:505–517. doi: 10.1016/j.cmet.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha AL, Ramanathan A, Huang S, Ingber DE, Schreiber SL. Carbon metabolism-mediated myogenic differentiation. Nat Chem Biol. 2010;6:202–204. doi: 10.1038/nchembio.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012;1819:921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Cardoso WV, Lu J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–968. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- Chin AM, Hill DR, Aurora M, Spence JR. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin Cell Dev Biol. 2017;66:81–93. doi: 10.1016/j.semcdb.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MY, Romer AI, Hu M, Lepourcelet M, Mechoor A, Yesilaltay A, Krieger M, Gray PA, Shivdasani RA. A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development. 2006;133:4119–4129. doi: 10.1242/dev.02537. [DOI] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell metabolism. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zheng L, Liu W, Wang X, Wang Z, Wang Z, French AJ, Kang D, Chen L, Thibodeau SN, Liu W. Frequent truncating mutation of TFAM induces mitochondrial DNA depletion and apoptotic resistance in microsatellite-unstable colorectal cancer. Cancer Res. 2011;71:2978–2987. doi: 10.1158/0008-5472.CAN-10-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, Getsios S, Gottardi CJ, DeBerardinis RJ, Lavker RM, Chandel NS. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Jeong DE, Lee D, Hwang SY, Lee Y, Lee JE, Seo M, Hwang W, Seo K, Hwang AB, Artan M, Son HG, Jo JH, Baek H, Oh YM, Ryu Y, Kim HJ, Ha CM, Yoo JY, Lee SV. Mitochondrial chaperone HSP-60 regulates anti-bacterial immunity via p38 MAP kinase signaling. EMBO J. 2017;36:1046–1065. doi: 10.15252/embj.201694781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Silberg DG, Traber PG, Schutz G. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, Stappenbeck TS. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson L, Lindahl P, Heath JK, Betsholtz C. Abnormal gastrointestinal development in PDGFA and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development. 2000;127:3457–3466. doi: 10.1242/dev.127.16.3457. [DOI] [PubMed] [Google Scholar]
- Kim BM, Mao J, Taketo MM, Shivdasani RA. Phases of canonical Wnt signaling during the development of mouse intestinal epithelium. Gastroenterology. 2007;133:529–538. doi: 10.1053/j.gastro.2007.04.072. [DOI] [PubMed] [Google Scholar]
- Kumar N, Srivillibhuthur M, Joshi S, Walton KD, Zhou A, Faller WJ, Perekatt AO, Sansom OJ, Gumucio DL, Xing J, Bonder EM, Gao N, White E, Verzi MP. A YY1-dependent increase in aerobic metabolism is indispensable for intestinal organogenesis. Development. 2016;143:3711–3722. doi: 10.1242/dev.137992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lepourcelet M, Tou L, Cai L, Sawada J, Lazar AJ, Glickman JN, Williamson JA, Everett AD, Redston M, Fox EA, Nakatani Y, Shivdasani RA. Insights into developmental mechanisms and cancers in the mammalian intestine derived from serial analysis of gene expression and study of the hepatoma-derived growth factor (HDGF) Development. 2005;132:415–427. doi: 10.1242/dev.01579. [DOI] [PubMed] [Google Scholar]
- Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development. 2005;132:279–289. doi: 10.1242/dev.01576. [DOI] [PubMed] [Google Scholar]
- McLin VA, Henning SJ, Jamrich M. The role of the visceral mesoderm in the development of the gastrointestinal tract. Gastroenterology. 2009;136:2074–2091. doi: 10.1053/j.gastro.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, Wert SE, Nation JM, Loudy DE, Huelsken J, Birchmeier W, Morrisey EE, Whitsett JA. beta-Catenin is required for specification of proximal/distal cell fate during lung morphogenesis. J Biol Chem. 2003;278:40231–40238. doi: 10.1074/jbc.M305892200. [DOI] [PubMed] [Google Scholar]
- Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB, Schwitalla S, Kalna G, Ogg EL, Athineos D, Timpson P, Vidal M, Murray GI, Greten FR, Anderson KI, Sansom OJ. ROS production and NF-kappaB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell. 2013;12:761–773. doi: 10.1016/j.stem.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- Perekatt AO, Valdez MJ, Davila M, Hoffman A, Bonder EM, Gao N, Verzi MP. YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc Natl Acad Sci U S A. 2014;111:7695–7700. doi: 10.1073/pnas.1400128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P, He XC, Paulson A, Li Z, Tao F, Perry JM, Guo F, Zhao M, Zhi L, Venkatraman A, Haug JS, Parmely T, Li H, Dobrowsky RT, Ding WX, Kono T, Ferguson-Smith AC, Li L. The Dlk1-Gtl2 Locus Preserves LT-HSC Function by Inhibiting the PI3K-mTOR Pathway to Restrict Mitochondrial Metabolism. Cell Stem Cell. 2016;18:214–228. doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Colman MJ, Schewe M, Meerlo M, Stigter E, Gerrits J, Pras-Raves M, Sacchetti A, Hornsveld M, Oost KC, Snippert HJ, Verhoeven-Duif N, Fodde R, Burgering BM. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- San Roman AK, Aronson BE, Krasinski SD, Shivdasani RA, Verzi MP. Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J Biol Chem. 2015;290:1850–1860. doi: 10.1074/jbc.M114.620211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Shi Y, Dierckx A, Wanrooij PH, Wanrooij S, Larsson NG, Wilhelmsson LM, Falkenberg M, Gustafsson CM. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc Natl Acad Sci U S A. 2012;109:16510–16515. doi: 10.1073/pnas.1119738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer AE, Tallinen T, Nerurkar NL, Wei Z, Gil ES, Kaplan DL, Tabin CJ, Mahadevan L. Villification: how the gut gets its villi. Science. 2013;342:212–218. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development. 2013;140:2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Lauf R, Shroyer NF. Vertebrate intestinal endoderm development. Dev Dyn. 2011;240:501–520. doi: 10.1002/dvdy.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C, Edwards RA, Pate KT, Waterman ML, Donovan PJ, Gratton E. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Scientific reports. 2012;2:568. doi: 10.1038/srep00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, Soga T, Hirao A, Suematsu M, Suda T. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O'Neill LA. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormos KV, Anso E, Hamanaka RB, Eisenbart J, Joseph J, Kalyanaraman B, Chandel NS. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell metabolism. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink GR. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev. 2007;87:1343–1375. doi: 10.1152/physrev.00054.2006. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EM, Thompson CA, Battle MA. GATA4 and GATA6 regulate intestinal epithelial cytodifferentiation during development. Dev Biol. 2014;392:283–294. doi: 10.1016/j.ydbio.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Freddo AM, Wang S, Gumucio DL. Generation of intestinal surface: an absorbing tale. Development. 2016a;143:2261–2272. doi: 10.1242/dev.135400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Kolterud A, Czerwinski MJ, Bell MJ, Prakash A, Kushwaha J, Grosse AS, Schnell S, Gumucio DL. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc Natl Acad Sci U S A. 2012;109:15817–15822. doi: 10.1073/pnas.1205669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Whidden M, Kolterud A, Shoffner SK, Czerwinski MJ, Kushwaha J, Parmar N, Chandhrasekhar D, Freddo AM, Schnell S, Gumucio DL. Villification in the mouse: Bmp signals control intestinal villus patterning. Development. 2016b;143:427–436. doi: 10.1242/dev.130112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanet A, Arnould T, Najimi M, Renard P. Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev. 2015;24:1957–1971. doi: 10.1089/scd.2015.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo DK, Green PD, Santos JH, D'Souza AD, Walther Z, Martin WD, Christian BE, Chandel NS, Shadel GS. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. Am J Pathol. 2012;180:24–31. doi: 10.1016/j.ajpath.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Luo J, He L, Montell C, Perrimon N. Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca2+ signaling in the Drosophila midgut. Elife. 2017;6 doi: 10.7554/eLife.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, Ruohola-Baker H. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–B) Transcript levels of Tfam and genes encoded in the mitochondrial genome are strongly reduced in both the proximal and distal fetal small intestinal epithelium (E18.5) (t-test, n=2). (C) Original images that were contrast-adjusted for use in Figure 2C. All other images in Figure 2C were not contrast adjusted. (D) Whole-mount preparation of E18.5 GI tract shows similar lengths of intestines of Control and TFAMKO mice, quantified in (E) (t-test, n= 7 control, 5 mutants). (F–G) Quantification of the number of cells in the E18.5 TFAM-mutant duodenal epithelium for PAS+ goblet cells (t-test, n= 4 control, 3 mutants) and Ki67+ cells (t-test, n= 3). (H) Additional markers of intestinal lineages were analyzed from RNA isolated from whole E18.5 control and TFAM-mutant small intestinal epithelial cells (t-test, n=2). Note that markers of the enterocyte lineage are particularly compromised upon TFAM loss.
(A) Loss of TFAM in the adult leads to loss of NDUFS3 expression. (B) No differences in cleaved caspase-3+ cells were observed between control and mutants. (C–F) Quantification of cell populations to accompany immunohistochemical analysis presented in Figure 5 and Figure S2G (t-test, n=6 control, 4 mutants C–D; n=7 control, 4 mutants E–F). (G) Immunostaining for DCLK1 is similar in control and mutant tissues. The duodenum is shown in these images, but the results are representative of the entire small intestine.