Abstract
The receptor for advanced glycation end-products (RAGE) is a pattern recognition receptor and member of the immunoglobulin superfamily. RAGE is constitutively expressed in the distal lung where it co-localizes with the alveolar epithelium; RAGE expression is otherwise minimal or absent, except with disease. This suggests RAGE plays a role in lung physiology and pathology. We used proteomics to identify and characterize the effects of RAGE on rat alveolar epithelial (R3/1) cells. LC-MS/MS identified 177 differentially expressed proteins and the PANTHER Classification System further segregated proteins. Proteins involved in gene transcription (RNA and mRNA splicing, mRNA processing) and transport (protein, intracellular protein) were overrepresented; genes involved in a response to stimulus were underrepresented. Immune system processes and response to stimuli were downregulated with RAGE knockdown. Western blot confirmed RAGE-dependent changes in protein expression for NFκB and NLRP3 that was functionally supported by a reduction in IL-1β and phosphorylated p65. We also assessed RAGE’s effect on redox regulation and report that RAGE knockdown attenuated oxidant production, decreased protein oxidation, and increased reduced thiol pools. Collectively the data suggest that RAGE is a critical regulator of epithelial cell response and has implications for our understanding of lung disease, specifically acute lung injury.
Keywords: oxidative stress, ARDS, lung injury, redox regulation
Graphical Abstract
Introduction
The receptor for advanced glycation end-products (RAGE) is a pattern recognition receptor and member of the immunoglobulin superfamily whose function is to amplify and perpetuate the inflammatory response. [1] Much of our understanding of RAGE occurs within the context of diseases such as Diabetes Mellitus and Alzheimer’s disease as RAGE is a known contributor to their pathogenesis. In humans, RAGE expression is notably downregulated in the absence of disease, except along the basolateral surface of the alveolar epithelium where it predominantly co-localizes with the alveolar epithelial type 1 (T1) cell. [2, 3] T1 cells are large, flat squamous epithelial cells that cover 95%–98% of the alveolar surface area. T1 cells are a crucial barrier between the air filled alveolus and the blood filled capillary, and T1 cells contain a full complement of ion channels, pumps, and pores that regulate ion and water composition required for optimal gas-exchange. [4] This unique expression pattern suggests that RAGE may play an important, albeit unclear, role in the lung.
The active form of RAGE is a ~55 kDa transmembrane protein composed of a variable domain for ligand binding, two constant domains, a transmembrane domain, and a short cytoplasmic tail that is crucial for cell signaling. [5] Splice variants typically produce truncated forms of RAGE (of various size) that are either secreted into the extracellular space (sRAGE) or inserted into the plasma membrane (tRAGE). Both sRAGE and tRAGE serve as a decoy and reduce RAGE signaling. RAGE is a promiscuous receptor with many known ligands and as such RAGE functions along an axis where outcomes range from cell proliferation and regeneration to profound inflammation and injury.
Redox regulation, changes in oxidant balance that promote reversible modifications to proteins, is controlled through oxidant production, a balance between redox pairs, reduced thiol pools and reductases. [6] Redox regulation is crucial for cell signaling, and a disruption in redox regulation coupled with overwhelming oxidant production results in oxidative stress. Oxidative stress culminates in a loss cell in function and is a significant contributor to lung pathology. Targeting pathways that regulate oxidative stress during disease offer potential therapeutic points for intervention.
Acute respiratory distress syndrome (ARDS) is a severe form of lung injury that affects the alveolar epithelium, and ARDS is characterized by profound inflammation, pulmonary edema, and oxidative stress that culminate in respiratory failure. [7–9] Data from multiple studies converge to show that RAGE signaling is likely an important contributor to lung homeostasis and disease pathogenesis, particularly with lung fluid balance and inflammation. [2, 3, 5, 10–16] For example, we have shown that RAGE regulates lung fluid balance, and that higher levels of sRAGE correlate with increased volumes of epithelial ling fluid in humans. [17] Thus, suggesting that RAGE is a contributing pathway in the regulation of lung fluid balance. Furthermore studies describe inflammatory cytokine production through NFκB and NLRP3 signaling cascades following RAGE binding. [18–21] However, the role of RAGE in the alveolar epithelium, the site of highest constitutive expression, has not been conclusively shown.
Our objective was to obtain insight into the role of RAGE in the alveolar epithelium by using proteomics coupled with complementary biochemical and molecular studies to support identified functions. We identified 177 differentially expressed proteins that were further segregated using PANTHER classification system from which we report that RAGE contributes to alveolar epithelium’s response to stimuli. Using western blotting and ELISAs we found RAGE knockdown attenuated NF-κB and NLRP3 with an accompanying decrease in IL-1β, while decreasing protein ubiquitination. RAGE was also assessed for its role in redox regulation. RAGE knockdown decreased oxidant production, decreased agonist-derived protein oxidation, increased reduced (free) thiol pools, and attenuated levels of the disulfide reductase, Thioredoxin-1. Collectively, these data suggest that RAGE plays an important role the alveolar epithelium’s response during the evolution and resolution of lung injury. Moreover, our cellular and molecular biological studies validate important new findings obtained via high throughput mass spectrometry analysis of alveolar cells.
Materials and Methods
Reagents
Unless stated otherwise all reagents were obtained from Sigma-Millipore (Burlington, MA).
Cell Culture and siRNA
Rat alveolar epithelial (R3/1) cells were grown in 50:50 DMEM/Hams F12 media supplemented with 10% FBS and Penicillin/Streptomycin in a humidified 5% CO2 chamber. At 30% confluence, cells were transfected with 5pmole RAGE or scramble sequence siRNA (Santa Cruz, Dallas, TX) according to the manufacturer’s protocol. 48 hours after transfection, cells were treated with 2μL/mL advanced glycation end-products (AGEs) for 30 minutes. Confirmation of RAGE knockdown was determined through RAGE mRNA and protein expression.
ROS measurements
For superoxide generation, R3/1 cells were seeded to 96 well plates subjected to RAGE knockdown and then treated with AGEs (2μL/mL) for 30 minutes, followed by labeling with dihydroethedium (DHE, Thermo Fisher Scientific, San Jose, CA) (5μM for 30 minutes at 37°C in 5%CO2) before being rinsed with PBS and fluorescence intensity (Ex 510, Em 615) measured using a Tecan plate reader (Basil, Switzerland). For H2O2 measures, the cells were seeded to 6 well plates and subjected to knock down as described above ± AGEs and supernatants removed and assayed using Amplex Red (Thermo Fischer) per the manufacturer’s recommendations.
ELISA
Cleaved IL1-β concentrations were quantified from cell lysates using the IL1-β kit from R&D systems (Minneapolis, MN) per the manufacturer’s recommendations. Absorbance measures were taken using a Tecan plate reader at 450 nm.
Protein biochemistry
Following treatment, R3/1 cells were rinsed with ice-cold PBS and then lysed in RIPA buffer containing 1X protease and phosphatase inhibitors (Calbiochem, Burlington, MA). R3/1 cell lysates were then electrophoresed on acrylamide gels (8%, 12%, or 15% depending on molecular weight of target) and transferred to Protran nitrocellulose membranes (Scheicher Schuell) for Western blot analysis. Membranes were incubated with either goat polyclonal anti-RAGE (Santa Cruz, sc-8229, 1:1000), anti NFκB (Abcam, ab31481, 1:1000), anti-NLRP3 (Nova Biochemi, NBP212446, 1:1000), anti-phopho-p65 (Cell Signaling, 93H1, 1:1000) or anti-ubiquitin (Cell Signaling, 39365, 1:1000) overnight. A horseradish peroxidase conjugated secondary antibody (1:20,000) was applied and incubated for 1hr at room temperature. A Chemiluminescent signal was detected using Supersignal West Dura (Thermo Fischer Scientific) and exposed using a UVP chemiluminescent imaging station and compatible software.
PCR
Total RNA was extracted using an RNeasy isolation kit (Qiagen, Grand Island, NY) following the protocol of the manufacturer, and then treated with DNaseI and reverse-transcribed using Superscript II RNaseH-reverse transcriptase (Thermo Fischer Scientific). The level of RAGE mRNA expression was determined using the following primers: RAGE (forward) ACT ACC GAG TCC GAG TCT ACC, RAGE (reverse) GTA GCT TCC CTC AGA CAC ACA. Threshold levels of mRNA expression (ΔΔCt) were normalized to rat GAPDH levels, and values represent the mean of triplicate samples ±S.E. Data are representative of 3 independent studies.
OxyBlot
Detection of carbonyl groups in proteins can be used to quantify oxidative modification of proteins. The protein carbonyl contents in R3/1 cells following RAGE knockdown were detected by the OxyBlot protein oxidation kit (Thermo Fischer Scientific) per the manufacturer’s instructions. To derivatize the carbonyl group for Western blot detection, 10 μg protein was diluted with 12% SDS for a final concentration of 6% SDS, followed by the addition of 2 volumes of 20 nM 2,4-DNPH (dintrophenylhydrazine) in 10% trifluoracetic acid. The mixture was incubated at room temperature for 20 minutes, and a neutralization solution (1.5 volumes) from the OxyBlot kit was added to stop the reaction. Non-derivatized controls were run for each sample. A total of 10μg protein from each sample was loaded in a 4–20% gradient gel, and SDS-PAGE and western blotting were performed according to the manufacturer’s instructions. Rabbit polyclonal antibody raised against DNP (dinitrophenylhydrazone) was used at 1:150 dilution followed by an HRP-conjugated secondary antibody (1:300). Chemiluminescent signal was detected using Supersignal West Dura (Thermo Fischer Scientific) and exposed using a UVP chemiluminescent imaging station and compatible software.
Fluorescein 5-malemide labeling of reduced thiols
Fluoroscein-5-Maleimide (F5M; Invitrogen) was used to identify available reduced thiols with RAGE-ligand binding. R3/1 cells were treated as indicated and then 50 μL of 0.6 mM F5M (reconstituted in 20 mM Tris-HCL (pH 7.4), 0.1 mM MgCl2, and 1 mM MnCl2) was applied to 50 μL protein in standard RIPA buffer. Signal controls included R3/1 cells treated with 20 μM AAPA (R,R′-2-Acetylamino-3-[4-(2-acetylamino-2-carboxyethylsulfanylthiocarbonylamino) phenylthiocarbamoylsulfanyl]propionic acid hydrate, S, S′-[1,4-Phenylenebis (iminocarbonothioyl)]bis[N-acetyl-L-Cysteine] hydrate) to inhibit glutathione reductase or pre-labeling with N-ethyl malemide (NEM) to block free sulfhydryl before labeling with F5M. In all experiments, F5M reaction was quenched by adding equal volumes of 5X sample buffer containing 100 mM DTT. Samples were then boiled at 95°C for 10 min, spun down, and then electrophoresed on 10% acrylamide gel. F5M labeled bands were visualized by exposure to UV light on a trans-illuminator (UVP imaging station) and a UV filter. F5M signal was pseudo-colored and densimetric analysis performed using UVP Chemi-Doc-It compatible software.
Proteomic assay preparation and protein analysis
Following treatment with AGEs, R3/1 cells were lysed in RIPA buffer (Thermo Fischer Scientific). Cell lysates were diluted to 80% acetone and stored in −20°C overnight. The next day, lysates were centrifuged at 16k ×g for 10 min at 4°C, washed twice with ice-cold 100% acetone, centrifuged again and then placed under vacuum. The precipitated proteins were stored at −20°C until digested.
LC-MS/MS
Protein precipitates were solubilized in 100μL of 50mM Tris HCl/8M urea pH 8.0, vortexed, sonicated and spun at room temperature for 10 minutes at 16,000g. The supernatant was assayed for protein using the Pierce 660nm protein assay (Pierce Biotechnology, Rockford, IL). Four or 5 micrograms of protein were subsequently digested in 50mM ammonium bicarbonate pH 7.8, with 20:1 (w/w) trypsin (Pierce Biotechnology) for 3 hours at 37°C using ProteaseMax™ Surfactant trypsin enhancer following reduction and alkylation with dithiothreitol and iodoacetamide, respectively, according to the manufacturer’s instructions (Promega Corporation, Madison, WI). LC-MS/MS analysis was carried out using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) equipped with a nanoESI source, following 2μg capacity ZipTip (Millipore, Billeric, MA) C18 sample clean-up according to the manufacturer’s instructions. Peptides (500 ng digest material loaded) were eluted from an Acclaim Pepmap™ 100 precolumn (100-μm ID × 2 cm, Thermo Fischer Scientific) onto an Acclaim PepMap™ RSLC analytical column (75-μm ID × 15 cm, Thermo Fischer Scientific) using a 5% hold of solvent B (acetonitrile, 0.1% formic acid) for 15 minutes, followed by a 5–22% gradient of solvent B over 105 minutes, 22–32% solvent B over 15 minutes, 32–95% of solvent B over 10 minutes, 95% hold of solvent B for 10 minutes, and finally a return to 5% of solvent B in 0.1 minutes and another 14.9 minute hold of solvent B. All flow rates were 300 nL/min using a Dionex Ultimate 3000 RSLCnano System (Thermo Fischer Scientific). Solvent A consisted of water and 0.1% formic acid. Data dependent scanning was performed by the Xcalibur v 4.0.27.19 software [22] using a survey scan at 70,000 resolution scanning mass/charge (m/z) 400–1600 at an automatic gain control (AGC) target of 3e6 and a maximum injection time (IT) of 100 msec, followed by higher-energy collisional dissociation (HCD) tandem mass spectrometry (MS/MS) at 27nce (normalized collision energy), of the 10 most intense ions at a resolution of 17,500, an isolation width of 1.5 m/z, an AGC of 2e5 and a maximum IT of 50 msec. Dynamic exclusion was set to place any selected m/z on an exclusion list for 20 seconds after a single MS/MS. Ions of charge state +1, 7, 8, >8 and unassigned were excluded from MS/MS, as were isotopes. Tandem mass spectra were searched against the Rattus Norvegicus fasta protein database from UniprotKB downloaded on April 06, 2016, to which common contaminant proteins (e.g., human keratins obtained at ftp://ftp.thegpm.org/fasta/cRAP) were appended. All MS/MS spectra were searched using Thermo Proteome Discoverer 1.3 (Thermo Fisher Scientific) considering fully tryptic peptides with up to 2 missed cleavage sites. Variable modifications considered during the search included methionine oxidation (15.995 Da), and cysteine carbamidomethylation (57.021 Da). At the time of the search, the database contained 29,782 entries. Proteins were identified at 99% confidence with XCorr score cut-offs [23] as determined by a reversed database search. The protein and peptide identification results were also visualized with Scaffold v 4.3.4 (Proteome Software Inc., Portland OR), a program that relies on various search engine results (i.e.: Sequest, X!Tandem, MASCOT) and which uses Bayesian statistics to reliably identify more spectra. [24] Proteins were accepted that passed a minimum of two peptides identified at 0.1% peptide FDR and 90–99.9% protein confidence by the Protein Profit algorithm, within Scaffold. Three biological replicates were performed for each condition, and differentially expressed proteins were uploaded into the PANTHER Classification System (Version 12, released 2017-07-10) to enable high through put analysis.
Statistical analysis
All data are summarized as mean ± S.E. Statistical significance was set at p<0.05 and statistical analysis performed using SigmaStat. Single comparisons were performed using Student’s t-test. One-way analysis of variance was used for multiple comparisons followed by Bonferroni’s correction for post-hoc test for pair-wise comparisons. Bonferroni’s correction was also performed for multiple testing in the PANTHER Overrepresentation Test.
Results
Proteomics identify RAGE-dependent proteins in R3/1 cells
Using PANTHER we investigated relevant cellular proteins by comparison between RAGE knockdown and scramble (Figure 1). The analysis was conducted to determine differential proteins and pathways relevant to RAGE in alveolar epithelial cells. The Scaffold reference spectral library contained 890 proteins at a global false discovery rate of <1%. 177 proteins were found to be significantly different with RAGE knockdown (Supplemental Tables 1 and 2). All of these proteins were uploaded to PANTHER classification system (http://pantherdb.org) to classify the protein according to their biological process (Figure 2A) and molecular function (Figure 2B).
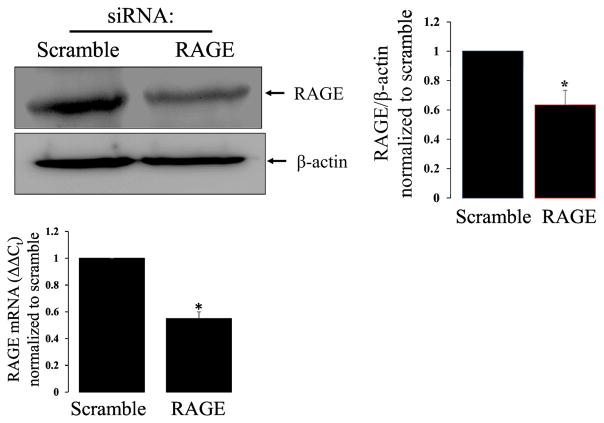
Figure 1. RAGE knockdown attenuates RAGE protein expression in R3/1 cells.
Representative western blot of RAGE knockdown and bar graph demonstrating ~40% reduction in RAGE protein expression with bar graph demonstrating ~50% reduction in RAGE mRNA with knockdown. N=4 *=p<0.05.
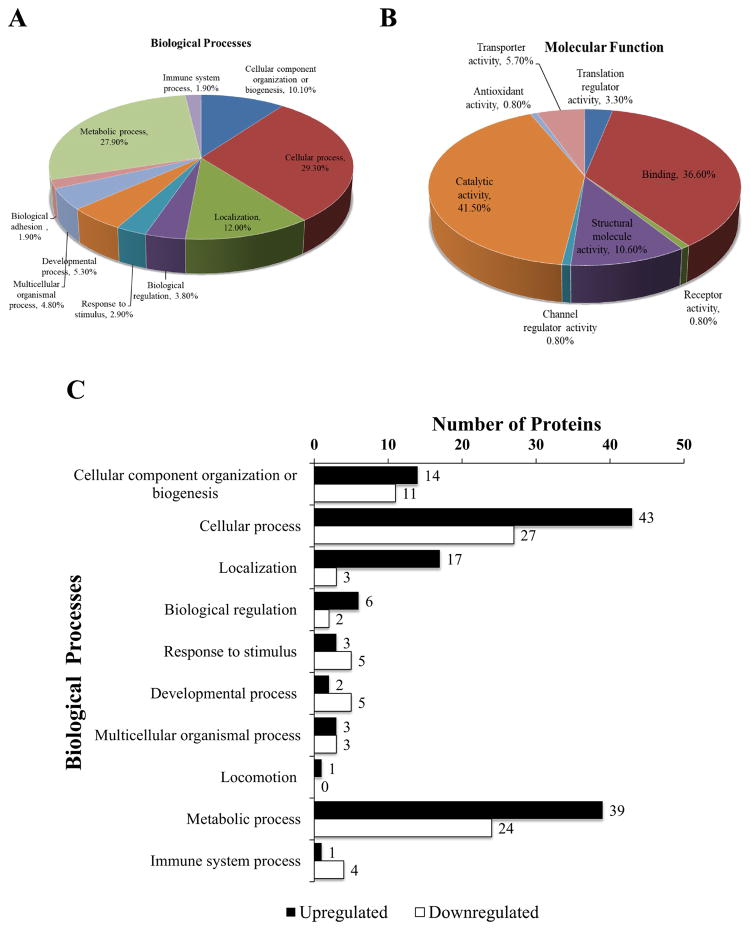
Figure 2. Classification of 177 differentially regulated proteins observed after LC-MS/MS according to their protein class using PANTHER Classification System.
Classification of differentially regulated proteins according to (A) biological processes and (B) molecular functions in which they are involved. (C) Bar graph demonstrating the frequency of biological processes that were upregulated or downregulated.
A comparison of the number of differentially regulated proteins (103 upregulated and 74 downregulated) based on their biological class was conducted and we found that proteins related to cellular and metabolic processes were well represented in the group of upregulated proteins (Figure 2C). Proteins related to response to stimulus, developmental processes, and immune systems responses were better represented in downregulated proteins. We also tested the biological processes overrepresented in this set of differentially regulated proteins. Table 1 shows a comparison of biological processes associated with this group of differentially expressed proteins, compared to all the processes present in the organism Rattus norvegicus (total 23,781) using PANTHER Overrepresentation Test. Bonferroi correction was used for multiple testing and the displayed results were significant (P<0.05). This analysis identified that many proteins involved in gene transcription (RNA and mRNA splicing, mRNA processing), transport (protein, intracellular protein), and response to stimuli were overrepresented in the differentially expressed proteins.
Table 1.
Biological processes of differentially expressed proteins
| PANTHER GO-Slim Biological Process | Number of proteins in Rattus norvegicus | Number of proteins (177) | Expected | Expression | Fold Enrichment | P-value |
|---|---|---|---|---|---|---|
| RNA splicing, via transesterification reactions | 148 | 8 | 0.84 | + | 9.49 | 5.97E−04 |
| mRNA splicing, via spliceosome | 188 | 8 | 1.07 | + | 7.47 | 3.35E−03 |
| mRNA processing | 249 | 8 | 1.42 | + | 5.64 | 2.38E−02 |
| Intracellular protein transport | 846 | 16 | 4.82 | + | 3.32 | 6.48E−03 |
| Protein transport | 869 | 16 | 4.95 | + | 3.23 | 8.92E−03 |
| Vesicle-mediated transport | 786 | 14 | 4.47 | + | 3.13 | 3.97E−02 |
| Transport | 1802 | 23 | 10.26 | + | 2.24 | 4.90E−02 |
| Unclassified | 9023 | 38 | 51.37 | − | 0.74 | 0.00E+00 |
| Response to stimulus | 3694 | 6 | 21.03 | − | 0.29 | 9.97E−03 |
RAGE-dependent changes in inflammation
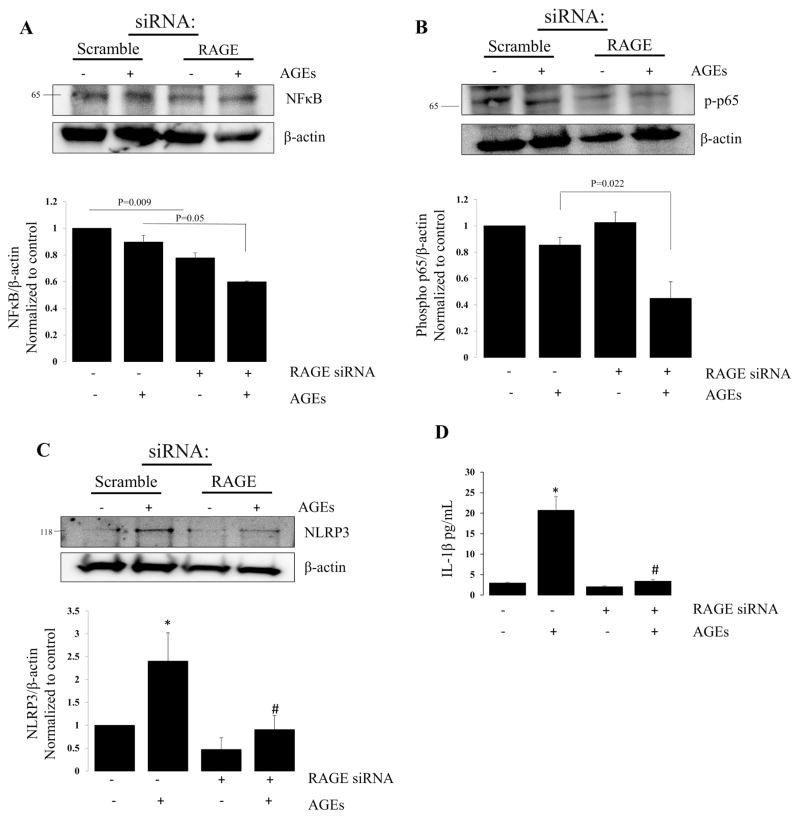
Because we observed a change in biological processes related to immune system response and a response to stimuli, both of which would be important to the alveolus during the evolution of lung injury, we assessed for the effects of - knock down on NFκB (Figure 3A), phosphorylated p65 (Figure 3B), and NLRP3 (Figure 3C) protein expression. RAGE knockdown with AGEs decreased expression of each compared to AGEs alone. AGEs increased cleaved IL-1β (Figure 3D) in cell lysates, an effect that was attenuated with RAGE knockdown (Scramble: 2.947 ± 0.213; Scramble + AGEs: 20.715 ± 3.175; RAGE siRNA: 2.066 ± 0.201; RAGE siRNA + AGEs: 3.397 ± 0.381).
Figure 3. RAGE knockdown attenuates inflammatory response genes.
Representative western blots for (A) NFκB, (B) phosphorylated NFκB (p65), (C) NLRP3 and respective bar graphs demonstrating a reduction in proteins. N=4,*=p<0.01 compared to scramble alone; #= P<0.05 compared to scramble with AGE treatment. (D) Bar graph of IL-1β following treatment with AGEs in scramble and RAGE knockdown groups. *=p<0.05 compared to scramble alone; #’ p<0.01 compared to scramble with AGEs.
RAGE-dependent changes in redox regulation
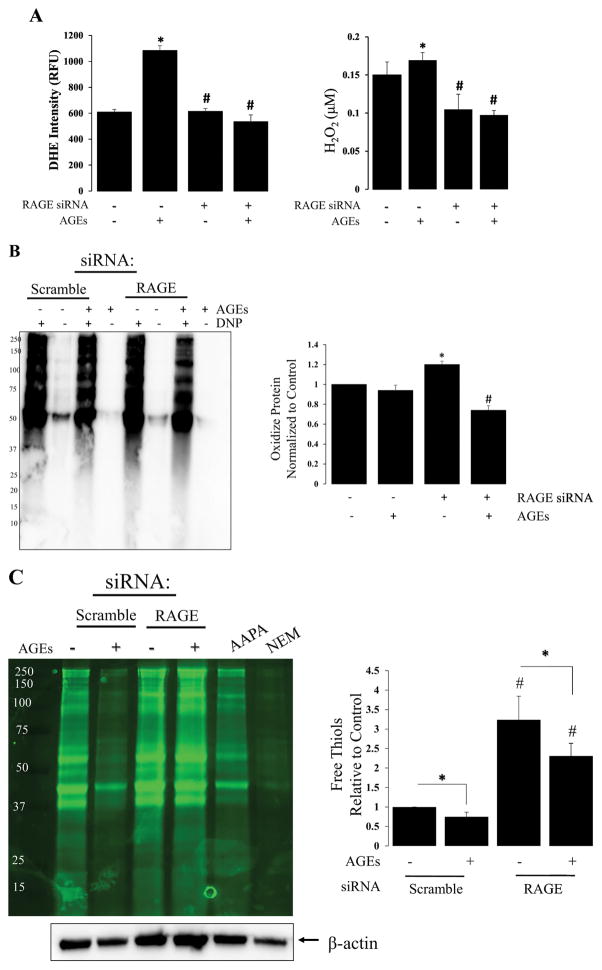
Cell signaling is vital for physiologic function, and redox regulation is an important mechanism of cell signaling that affects subsequent cellular responses. Key processes of redox regulation include: oxidant production leading to protein oxidation, post-translational modifications that occur in response to protein oxidation, and disulfide reductases that reduce oxidized (or post-translationally modified) proteins to their reduced forms. Therefore, we assessed the effect of RAGE on these key processes of redox regulation (Figure 4). Figure 4A shows an increase in RAGE-dependent superoxide (DHE) and H2O2 production in response to AGEs. RAGE-knockdown attenuated AGE-induced global protein oxidation (Figure 4B) as measured using OxyBlot. This novel observation indicates that RAGE plays a critically important role in the progression of lung injury. In line with this observation, RAGE knockdown also increased our ability to detect reduced thiols using fluorescently labeled maleimide reagent (F5M) in alveolar epithelial cells (Figure 4C). Together, Figures 4B and 4C provides evidence that RAGE signaling mediates cell function via post-translational modification of free thiols from the reduced to oxidized form(s), and contributes significantly to redox regulation of cellular responses. Specifically, the disulfide reductase, Thioredoxin-1 was down-regulated with RAGE knock-down (See Supplemental Table 1). Additional down-regulated proteins, known to play a role in maintaining redox balance, include: Cu2+-ZN2+ superoxide dismutase, Thioredoxin-like protein 1 and Thioredoxin domain-containing protein 17 (Supplemental Table 1). These proteins are highlighted because the SODs and Thioredoxins are known biological markers for lung injury. [25–28]
Figure 4. RAGE signaling is redox regulated.
(A) Bar graph demonstrating the effect of AGEs on superoxide and H2O2 production in R3/1 cells (± RAGE knockdown); N=3, *=p<0.05 compared, * = p<0.01 compared to scramble; # = P<0.05 compared to scramble with AGEs. (B) OxyBlot of RAGE dependent protein oxidation with associated bar graph demonstrating a ~30% reduction in protein oxidation with RAGE knockdown; N=4, *=p<0.05 compared to scramble; #= p<0.05 compared to scramble with AGEs. (C) Representative western blot of F5M labeled free thiols from scramble and RAGE knock-down groups ± AGEs; AAPA, R,R′-2-Acetylamino-3-[4-(2-acetylamino-2-carboxyethylsulfanylthiocarbonylamino)phenylthiocarbamoylsulfanyl]propionic acid hydrate, S, S′-[1,4-Phenylenebis(iminocarbonothioyl)]bis[N-acetyl-L-Cysteine] hydrate; NEM, N-ethyl malemide; Bar graph quantifying free thiols relative to β-actin; N=4, *= p<0.01 between groups, #= p<0.01 compared to control conditions.
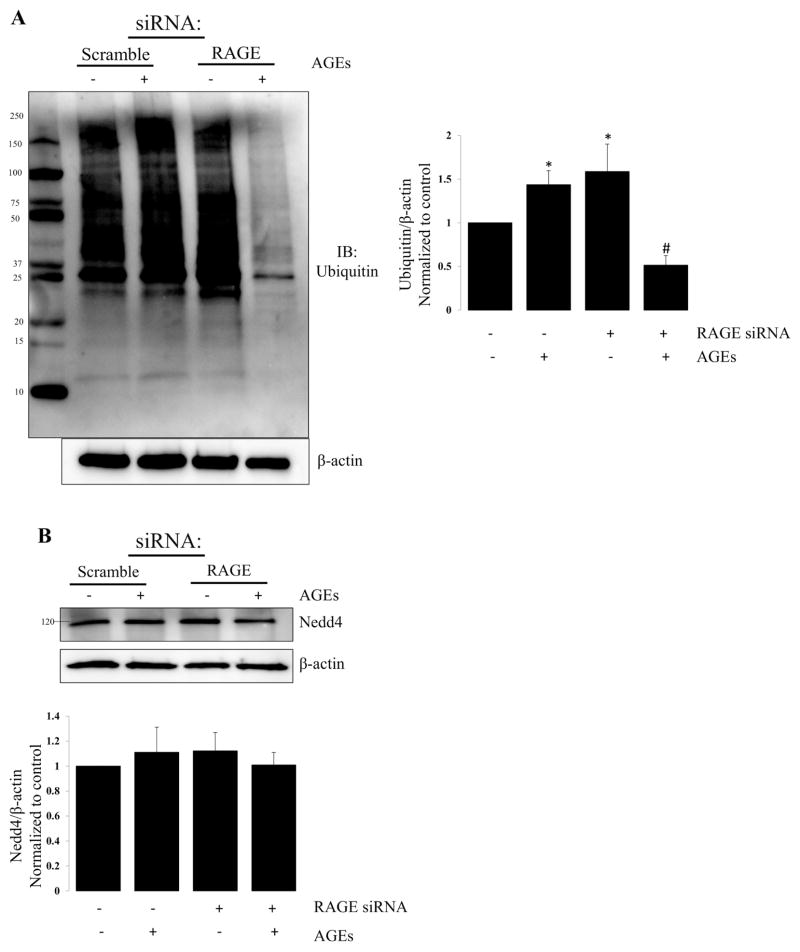
Protein oxidation can serves as an important signal for protein turnover. Therefore, we evaluated global protein ubiquitination in alveolar epithelial cells in which RAGE was knocked down using a siRNA approach. In Figure 5, we show the effects of RAGE knockdown on ubiquitination in alveolar epithelial cells. RAGE knockdown reduced protein ubiquitination by ~35%–40% suggesting that redox regulation of RAGE-ligand binding activates protein turnover.
Figure 5. RAGE knockdown attenuates ubiquitination.
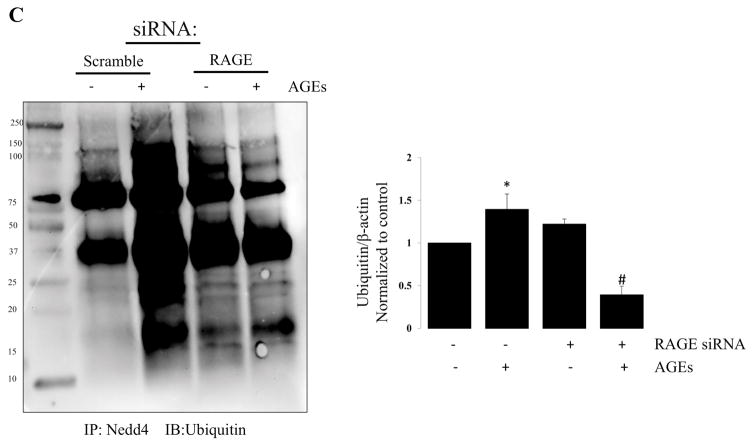
Representative western blot demonstrating the effects of RAGE knockdown on protein ubiquitination. (A) Representative western blot demonstrating changes in global protein ubiquitination in R3/1 cells following RAGE knockdown ± AGEs; Bar graph quantifying ubiquitin relative to β-actin. N=4; *=p<0.05 compared to control; #= p<0.01 compared to scramble with AGEs. (B) Representative western blot demonstrating no change in Nedd4 with RAGE knockdown ± AGEs; N=4. (C) Representative western blot of immune-precipitated Nedd 4 probed for Ubiquitin. Bar graph quantifying ubiquitinated Nedd4; N=4 *=p<0.05 compared to scramble alone; #=p<0.01 compared to scramble with AGEs.
Nedd4 is an E3 ubiquitin-protein ligase that targets proteins for ubiquitination, and plays a critical role in protein homeostasis. Nedd4 is widely expressed, and numerous proteins have been predicted or demonstrated to bind in vitro, and Need4 is involved in the regulation of a diverse range of processes. [29] We assessed the effect of RAGE knockdown with and without AGEs on Nedd4 expression in R3/1 cells (Figure 5B). Nedd4 levels were not affected; however, Nedd4-regulated ubiquitination is reduced with RAGE knockdown in the setting of AGEs, suggesting that an active RAGE receptor ultimately contributes to protein levels.
Discussion
Our objective was to obtain insight into the role of RAGE in the alveolar epithelium. We used proteomics coupled with complementary biochemical and molecular techniques to characterize RAGE function within the alveolar epithelium. We identified 177 differentially expressed proteins that were further classified based on biological processes and molecular function with overrepresentation testing showing an increased expression of genes related to RNA/mRNA splicing and processing and protein transport. Genes representing a response to stimuli were underrepresented. Collectively, these data demonstrate an active receptor when engaged.
Studies support a role for RAGE in the development of a variety of cancers, diabetes mellitus, and Alzheimer’s disease. [14, 20, 30–32] Canonical thinking suggests that RAGE functions along an axis where ligand-receptor binding produces pro-inflammatory and pro-growth stimuli leading to RAGE-dependent cell differentiation and cell survival. Indeed, RAGE-ligand signaling has been shown to promote cell differentiation in neonatal cells and may contribute to alveolarization. [33, 34] As the axis shifts, ligand-receptor binding leads to a more persistent feedforward signal culminating in persistent inflammation and inhibition of apoptosis. We observed a reduction in cleaved caspase-3 (data not shown) with RAGE knockdown and an accompanying decrease in protein ubiquitination suggesting that RAGE also contributes to cell and protein fate in the alveolar epithelium. During our optimization of siRNA we also found that concentrations of siRNA exceeding 5pmole led to significant cell death. PANTHER reported an overrepresentation of proteins involved in intracellular protein transport, protein transport, transport, and vesicle-mediated protein transport. Our previous work has also shown a role for RAGE in response to vesicle-mediated protein transport in that exosomes obtained from cells treated with H2O2 promote RAGE expression in alveolar epithelial cells. [35] This suggests that RAGE may play a role in directing the content and formation of exosomes in the alveolus.
Acute respiratory distress syndrome (ARDS) is a severe form of lung injury that is characterized by profound inflammation, oxidative stress, and marked hypoxemia that culminate in respiratory failure. Treatment for ARDS is merely supportive and mortality remains excessive. Therefore understanding the underlying pathology is crucial for the development of therapeutics. Recent studies suggest that RAGE may play an important role in ARDS, albeit evidence is peripheral as studies show elevated concentrations of IL-1β in the edema fluid of individuals with ARDS. [36] IL-1β is downstream of many pro-inflammatory signaling cascades, and we show that inhibition of RAGE reduced NF-κB and NLRP3 inflammasome signaling with an accompanying reduction in IL-1β in alveolar epithelial cells. Also, studies show a relationship between levels of sRAGE, a decoy, and edema fluid in persons with ARDS which argues for a relationship between RAGE signaling and fluid balance. [10, 16] Our previous work demonstrated a causal relationship between RAGE and activation of the epithelial sodium channel (ENaC), the rate limiting factor in the resolution of pulmonary edema; our work also demonstrated a correlation between AGEs and volume of the epithelial lining fluid in human subjects. [17] Collectively this suggests a role for RAGE in lung fluid balance.
Oxidative stress, an imbalance between oxidants and antioxidants in favor of oxidants that leads to a disruption in redox regulation or molecular damage, is common in lung disease and is a significant contributor to lung injury. [37–41] Redox regulation is a reversible processes in which oxidants oxidize free thiols (e.g.–SH groups on a given protein) leading to either disulfide formation or post-translational modifications that ultimately effects physiologic function. This process is reversed through activity of reductases, such as the disulfide reductase Thioredoxin-1, in which the disulfide bond is broken and the oxidized thiol is then reduced. The cycle of protein oxidation and subsequent reduction serves as a molecular switch to regulate physiologic function. Oxidative stress is impairment in this mechanism, that is, dysfunctional redox signaling.
Our data suggest that RAGE is a contributor to redox regulation in alveolar epithelial cells. Specifically, we demonstrate an increase in oxidants and the oxidation of proteins in response to RAGE-ligand binding, as well as an increase in free—SH groups with RAGE knock-down. Moreover, RAGE knockdown decreased Thioredoxin-1 (Trx-1) protein expression (See Supplemental Table 1). In addition, expression of antioxidant enzymes, such as Superoxide dismutase 1, Thioredoxin-like protein 1, and Thioredoxin domain containing protein 17 were also attenuated with RAGE knockdown. These data suggest that RAGE is an active contributor to redox regulation, and as such RAGE may play a significant role in the evolution and resolution of lung injury.
RAGE-ligand binding appears to affect protein ubiquitination (Figure 5). In Figure 5C we show a reduction in ubiquitination through Nedd4, an E3 ligase directly involved in the ubiquitination of numerous proteins. Collectively these data suggest that RAGE-ligand binding promotes protein turn-over. This is further supported by our proteomics data in Figure 3C showing an increase in cellular and metabolic processes that may require changes in protein expression to accommodate an increase in cellular and metabolic needs.
By using a proteomics approach the data presented here suggest that RAGE contributes to multiple functions in the alveolar epithelium. We have identified a unique function for RAGE in that the receptor contributes to intracellular redox state and redox-mediated signaling in cells that abundantly express the RAGE protein. This observation has implications for our understanding of RAGE within the context of known diseases, but is particularly relevant to the study of lung physiology and pathology, specifically acute lung injury where many RAGE ligands are present and where oxidative stress is a defining characteristic.
Supplementary Material
Significance statement.
In the present study, we undertook the first proteomic evaluation of RAGE-dependent processes in alveolar epithelial cells. The alveolar epithelium is a primary target during acute lung injury, and our data support a role for RAGE in gene transcription, protein transport, and response to stimuli. More over our data suggest that RAGE is a critical driver of redox regulation in the alveolar epithelium. The conclusions of the present work assist to unravel the molecular events that underlie the function of RAGE in alveolar epithelial cells and have implications for our understanding of RAGE signaling during lung injury. Our study was the first proteomic comparison showing the effects of RAGE activation from alveolar epithelial cells that constitutively express RAGE and these results can affect a wide field of lung biology, pulmonary therapeutics, and proteomics.
Highlights.
RAGE is constitutively expressed in the alveolar epithelium although its role is unclear.
RAGE contributes to alveolar epithelial cell response to stimuli.
RAGE is a significant contributor to redox regulation in the alveolar epithelium.
RAGE contributes to protein ubiquitination and inflammation in the alveolar epithelium.
Acknowledgments
Funding Source: Parker B. Francis Fellowship & University of Arizona Health Sciences Career Development Award awarded to C.A.D. Mass spectrometry and proteomics data were acquired by the University of Arizona Analytical and Biological Mass Spectrometry Facility supported by NIH/NCI grant CA023074 to the University of Arizona Cancer Center and by the BIO5 Institute of the University of Arizona.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kierdorf K, Fritz G. RAGE regulation and signaling in inflammation and beyond. J Leukoc Biol. 2013;94:55–68. doi: 10.1189/jlb.1012519. [DOI] [PubMed] [Google Scholar]
- 2.Frank JA, Briot R, Lee JW, Ishizaka A, et al. Physiological and biochemical markers of alveolar epithelial barrier dysfunction in perfused human lungs. Am J Physiol Lung Cell Mol Physiol. 2007;293:L52–59. doi: 10.1152/ajplung.00256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shirasawa M, Fujiwara N, Hirabayashi S, Ohno H, et al. Receptor for advanced glycation end-products is a marker of type I lung alveolar cells. Genes Cells. 2004;9:165–174. doi: 10.1111/j.1356-9597.2004.00712.x. [DOI] [PubMed] [Google Scholar]
- 4.Helms MN, Jain L, Self JL, Eaton DC. Redox regulation of epithelial sodium channels examined in alveolar type 1 and 2 cells patch-clamped in lung slice tissue. J Biol Chem. 2008;283:22875–22883. doi: 10.1074/jbc.M801363200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynolds PR, Kasteler SD, Cosio MG, Sturrock A, et al. RAGE: developmental expression and positive feedback regulation by Egr-1 during cigarette smoke exposure in pulmonary epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1094–1101. doi: 10.1152/ajplung.00318.2007. [DOI] [PubMed] [Google Scholar]
- 6.Go YM, Chandler JD, Jones DP. The cysteine proteome. Free Radic Biol Med. 2015;84:227–245. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esper A, Burnham EL, Moss M. The effect of alcohol abuse on ARDS and multiple organ dysfunction. Minerva Anestesiol. 2006;72:375–381. [PubMed] [Google Scholar]
- 8.Moss M, Steinberg KP, Guidot DM, Duhon GF, et al. The effect of chronic alcohol abuse on the incidence of ARDS and the severity of the multiple organ dysfunction syndrome in adults with septic shock: an interim and multivariate analysis. Chest. 1999;116:97s–98s. [PubMed] [Google Scholar]
- 9.Moss M, Burnham EL. Chronic alcohol abuse, acute respiratory distress syndrome, and multiple organ dysfunction. Crit Care Med. 2003;31:S207–212. doi: 10.1097/01.CCM.0000057845.77458.25. [DOI] [PubMed] [Google Scholar]
- 10.Jabaudon M, Blondonnet R, Roszyk L, Bouvier D, et al. Soluble Receptor for Advanced Glycation End-Products Predicts Impaired Alveolar Fluid Clearance in Acute Respiratory Distress Syndrome. Am J Respir Crit Care Med. 2015;192:191–199. doi: 10.1164/rccm.201501-0020OC. [DOI] [PubMed] [Google Scholar]
- 11.Kyung SY, Byun KH, Yoon JY, Kim YJ, et al. Advanced glycation end-products and receptor for advanced glycation end-products expression in patients with idiopathic pulmonary fibrosis and NSIP. Int J Clin Exp Pathol. 2014;7:221–228. [PMC free article] [PubMed] [Google Scholar]
- 12.Liliensiek B, Weigand MA, Bierhaus A, Nicklas W, et al. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J Clin Invest. 2004;113:1641–1650. doi: 10.1172/JCI18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutterloh EC, Opal SM, Pittman DD, Keith JC, Jr, et al. Inhibition of the RAGE products increases survival in experimental models of severe sepsis and systemic infection. Crit Care. 2007;11:R122. doi: 10.1186/cc6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt AM, Hofmann M, Taguchi A, Yan SD, Stern DM. RAGE: a multiligand receptor contributing to the cellular response in diabetic vasculopathy and inflammation. Semin Thromb Hemost. 2000;26:485–493. doi: 10.1055/s-2000-13204. [DOI] [PubMed] [Google Scholar]
- 15.Su X, Looney MR, Gupta N, Matthay MA. Receptor for advanced glycation end-products (RAGE) is an indicator of direct lung injury in models of experimental lung injury. Am J Physiol Lung Cell Mol Physiol. 2009;297:L1–5. doi: 10.1152/ajplung.90546.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uchida T, Shirasawa M, Ware LB, Kojima K, et al. Receptor for advanced glycation end-products is a marker of type I cell injury in acute lung injury. Am J Respir Crit Care Med. 2006;173:1008–1015. doi: 10.1164/rccm.200509-1477OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downs CA, Kreiner LH, Johnson NM, Brown LA, Helms MN. Receptor for advanced glycation end-products regulates lung fluid balance via protein kinase C-gp91(phox) signaling to epithelial sodium channels. Am J Respir Cell Mol Biol. 2015;52:75–87. doi: 10.1165/rcmb.2014-0002OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alves M, Calegari VC, Cunha DA, Saad MJ, et al. Increased expression of advanced glycation end-products and their receptor, and activation of nuclear factor kappa-B in lacrimal glands of diabetic rats. Diabetologia. 2005;48:2675–2681. doi: 10.1007/s00125-005-0010-9. [DOI] [PubMed] [Google Scholar]
- 19.Hammes HP, Hoerauf H, Alt A, Schleicher E, et al. N(epsilon)(carboxymethyl)lysin and the AGE receptor RAGE colocalize in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1999;40:1855–1859. [PubMed] [Google Scholar]
- 20.Pillai SS, Sugathan JK, Indira M. Selenium downregulates RAGE and NFkappaB expression in diabetic rats. Biol Trace Elem Res. 2012;149:71–77. doi: 10.1007/s12011-012-9401-1. [DOI] [PubMed] [Google Scholar]
- 21.Song Y, Wang Y, Zhang Y, Geng W, et al. Advanced glycation end products regulate anabolic and catabolic activities via NLRP3-inflammasome activation in human nucleus pulposus cells. J Cell Mol Med. 2017;21:1373–1387. doi: 10.1111/jcmm.13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andon NL, Hollingworth S, Koller A, Greenland AJ, et al. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2:1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Qian WJ, Liu T, Monroe ME, Strittmatter EF, et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 24.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 25.Wang G, Song Y, Feng W, Liu L, et al. Activation of AMPK attenuates LPS-induced acute lung injury by upregulation of PGC1alpha and SOD1. Exp Ther Med. 2016;12:1551–1555. doi: 10.3892/etm.2016.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen XJ, Zhang B, Hou SJ, Shi Y, et al. Osthole improves acute lung injury in mice by up-regulating Nrf-2/thioredoxin 1. Respir Physiol Neurobiol. 2013;188:214–222. doi: 10.1016/j.resp.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Yashiro M, Tsukahara H, Matsukawa A, Yamada M, et al. Redox-active protein thioredoxin-1 administration ameliorates influenza A virus (H1N1)-induced acute lung injury in mice. Crit Care Med. 2013;41:171–181. doi: 10.1097/CCM.0b013e3182676352. [DOI] [PubMed] [Google Scholar]
- 28.Yodoi J, Tian H, Masutani H, Nakamura H. Thiol redox barrier; local and systemic surveillance against stress and inflammatory diseases. Arch Biochem Biophys. 2016;595:88–93. doi: 10.1016/j.abb.2015.11.029. [DOI] [PubMed] [Google Scholar]
- 29.Cao XR, Lill NL, Boase N, Shi PP, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1:ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad S, Khan H, Siddiqui Z, Khan MY, et al. AGEs, RAGEs and s-RAGE; friend or foe for cancer. Semin Cancer Biol. 2017 doi: 10.1016/j.semcancer.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Azizan N, Suter MA, Liu Y, Logsdon CD. RAGE maintains high levels of NFkappaB and oncogenic Kras activity in pancreatic cancer. Biochem Biophys Res Commun. 2017;493:592–597. doi: 10.1016/j.bbrc.2017.08.147. [DOI] [PubMed] [Google Scholar]
- 32.Deane R, Singh I, Sagare AP, Bell RD, et al. A multimodal RAGE-specific inhibitor reduces amyloid beta-mediated brain disorder in a mouse model of Alzheimer disease. J Clin Invest. 2012;122:1377–1392. doi: 10.1172/JCI58642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasparitsch M, Arndt AK, Pawlitschek F, Oberle S, et al. RAGE-mediated interstitial fibrosis in neonatal obstructive nephropathy is independent of NF-kappaB activation. Kidney Int. 2013;84:911–919. doi: 10.1038/ki.2013.171. [DOI] [PubMed] [Google Scholar]
- 34.Tsoporis JN, Izhar S, Proteau G, Slaughter G, Parker TG. S100B-RAGE dependent VEGF secretion by cardiac myocytes induces myofibroblast proliferation. J Mol Cell Cardiol. 2012;52:464–473. doi: 10.1016/j.yjmcc.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 35.Downs CA, Dang VD, Johnson NM, Denslow ND, Alli AA. Hydrogen Peroxide Stimulates Exosomal Cathepsin B Regulation of the Receptor for Advanced Glycation End-Products (RAGE) J Cell Biochem. 2017 doi: 10.1002/jcb.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geiser T, Atabai K, Jarreau PH, Ware LB, et al. Pulmonary edema fluid from patients with acute lung injury augments in vitro alveolar epithelial repair by an IL-1beta-dependent mechanism. Am J Respir Crit Care Med. 2001;163:1384–1388. doi: 10.1164/ajrccm.163.6.2006131. [DOI] [PubMed] [Google Scholar]
- 37.Downs CA, Kreiner LH, Trac DQ, Helms MN. Acute effects of cigarette smoke extract on alveolar epithelial sodium channel activity and lung fluid clearance. Am J Respir Cell Mol Biol. 2013;49:251–259. doi: 10.1165/rcmb.2012-0234OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Downs CA, Trac DQ, Kreiner LH, Eaton AF, et al. Ethanol alters alveolar fluid balance via Nadph oxidase (NOX) signaling to epithelial sodium channels (ENaC) in the lung. PLoS One. 2013;8:e54750. doi: 10.1371/journal.pone.0054750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Downs CA, Kumar A, Kreiner LH, Johnson NM, Helms MN. H2O2 regulates lung epithelial sodium channel (ENaC) via ubiquitin-like protein Nedd8. J Biol Chem. 2013;288:8136–8145. doi: 10.1074/jbc.M112.389536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Downs CA, Helms MN. Regulation of ion transport by oxidants. Am J Physiol Lung Cell Mol Physiol. 2013;305:L595–603. doi: 10.1152/ajplung.00212.2013. [DOI] [PubMed] [Google Scholar]
- 41.Downs CA, Kreiner L, Zhao XM, Trac P, et al. Oxidized glutathione (GSSG) inhibits epithelial sodium channel activity in primary alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2015;308:L943–952. doi: 10.1152/ajplung.00213.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.