Abstract
Innate immune recognition is classically mediated by the interaction of host pattern-recognition receptors and pathogen-associated molecular patterns; this triggers a series of downstream signaling events that facilitate killing and elimination of invading pathogens. In this report, we provide the first evidence that peroxidasin (PXDN; also known as vascular peroxidase-1) directly binds to gram-negative bacteria and mediates bactericidal activity, thus, contributing to lung host defense. PXDN contains five leucine-rich repeats and four immunoglobulin domains, which allows for its interaction with lipopolysaccharide, a membrane component of gram-negative bacteria. Bactericidal activity of PXDN is mediated via its capacity to generate hypohalous acids. Deficiency of PXDN results in a failure to eradicate Pseudomonas aeruginosa and increased mortality in a murine model of Pseudomonas lung infection. These observations indicate that PXDN mediates previously unrecognized host defense functions against gram-negative bacterial pathogens.
Author summary
Multicellular organisms have evolved diversified host defense mechanisms for survival against invading pathogens. Of the mechanisms, the recognition of pathogens is classically mediated by the interaction of host and pathogen, which triggers a series of downstream responses to eliminate pathogens. Proteins that both selectively and directly interact with and kill pathogens are not well identified. In current study, we have determined the dual-function mechanisms of PXDN -mediated bacteria killing. We provide the first evidence for a novel role of PXDN in directly binding to gram-negative bacteria and mediating bactericidal activity. PXDN is highly expressed in the lung and secreted into epithelial lining fluid of the lung, and is induced by LPS and TNF-α. PXDN mutant mice reveal impaired lung host defense in acute lung infection model of P. aeruginosa. PXDN is a new class of bactericidal enzyme with dual function of recognizing and killing pathogens. This finding of an enzyme with dual function has important implications for new conceptual understanding of the innate immunity as well as for therapeutic development.
Introduction
The lung is exposed to a constant barrage of inhaled harmful agents and microorganisms. Several layers of defense in the normal lung help prevent infection from inhaled or aspirated microorganisms. These include the mechanical filtering of particles that occur in the nasal airway, the trapping of particles in mucus and mucociliary clearance. Respiratory epithelial cells also secrete surfactant proteins, antimicrobial peptides and complements; all of these secreted proteins are important in innate immunity [1, 2]. In addition, alveolar macrophages, neutrophils, lymphocytes and circulating antibodies participate in the clearance of microorganisms from the lung [3].
Innate immune responses are classically initiated by recognition of pathogens through host pattern-recognition receptors (PRRs) [4, 5]. The interaction between PRRs and the specific pathogen-associated ligands, named pathogen-associated molecular patterns (PAMPs) activates the downstream signaling events and host defense mechanisms to eliminate invading pathogens [4, 5]. Recognition of the PAMPs allows the host immune system to distinguish infectious pathogens from the host. Four families of PRRs have been identified, the Toll-like receptors (TLRs), nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs), retinoic acid inducible gene-1 (RIG-1) like receptors (RLRs) and C-type lectin receptors (CLRs) [6]. Members of these families contain at least one of nine highly conserved protein domains such as leucine-rich repeats (LRRs) and immunoglobulin-like (Ig) domains [6]. These domains are crucial for recognizing PAMPs within invading pathogens. Common bacterial PAMPs include lipopolysaccharide (LPS), peptidoglycan (PGN), bacterial flagellin and lipoteichoic acid [7].
The heme-containing peroxidase (hPx) family is known to participate in host defense [8, 9]. Myeloperoxidase (MPO), the proto-enzyme of hPx family, has been extensively investigated [8, 9]. MPO was thought to be the only peroxidase capable of generating HOCl under physiological conditions [8]. It has been proposed that MPO binds to pathogens as a result of its higher cationic surface charge (pI = 9.3) in the acidic environment of the phagosome [10, 11]; however, this interaction is non-specific and any molecule with anionic surface charge, including but not limited to pathogens, may potentially interact with MPO with its attendant risk of collateral damage. Lactoperoxidase (LPO) is secreted into some body fluids including milk, saliva and mucus of airway, but not plasma and alveolar lining fluid; its physiological role is to prevent microbial growth at these mucosal surfaces [12, 13]. However, the presence of hPx with bactericidal activity in the lower airways and alveolar regions has not been demonstrated.
Vascular peroxidase-1 (VPO1) is a newly-identified member of the hPx family in mammals [14]. The ortholog of VPO1 in Drosophila is known as peroxidasin (PXDN; as accepted by HUGO Gene Nomenclature Committee) [15]. In this article, the official name of PXDN is used instead of the alias, VPO1. PXDN catalyzes generation of hypohalous acids and kills bacteria in vitro [16, 17]. Homozygous mutation in PXDN causes developmental defects including congenital cataract, corneal opacity and glaucoma [18], abnormalities which are also present in PXDN mutant mice [19]. Unlike the classic hPx’es (MPO, eosinophil peroxidase, LPO and thyroid peroxidase), the expressions of which are restricted to specific cells or tissues, PXDN is more ubiquitously expressed [14]. PXDN is found at high circulating levels in human and mouse plasma, approximately 1.1 μM and 2.6 μM, respectively [20]. We previously reported that PXDN is the second mammalian hPx capable of catalyzing the oxidation of chloride in the presence of H2O2 to generate HOCl [16]. Interestingly, PXDN is unique among members of the hPx family [14]; it has additional domains in its N-terminus that include five LRRs and four Ig C2 domains (Fig 1A). LRRs and Ig domains are highly conserved protein domains and important in innate immune pattern recognition. However, the physiological function of PXDN in host defense is unclear. In the current study, we have identified a novel dual-function activity of PXDN, with its N-terminus recognizing LPS and its C-terminus mediating bactericidal killing, utilizing both in vitro and in vivo approaches. This is the first study, to our knowledge, demonstrating a critical role for PXDN in host defense of the lung.
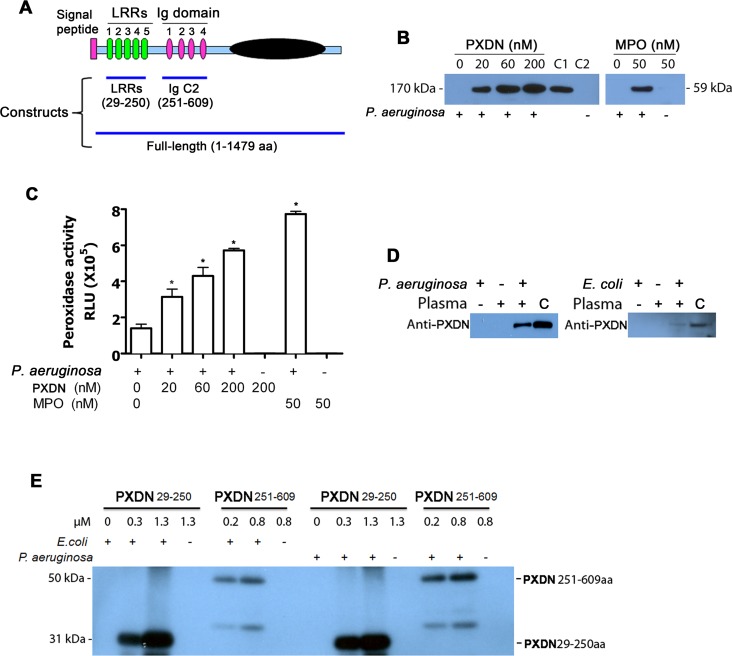
Fig 1. PXDN binds to GN bacteria.
(A) Diagram of human PXDN and recombinant proteins of PXDN used in the study. All constructs for preparation of recombinant proteins of PXDN were polyhistidine-tagged either cloned in plasmid pET30 or pcDNA3.1. The subcloned region and related domain structure are indicated. (B) P. aeruginosa binds to full-length PXDN. P. aeruginosa strain K was added to PBS containing PXDN or MPO as indicated. The mixtures were incubated at 37°C for 1 h. P. aeruginosa mixtures were centrifuged at 3099 x g for 5 min. The pellets were washed twice. One-half of the bacterial suspension was subjected to immunoblotting using anti-His antibody (left panel) or anti-MPO (right panel) antibody. C1 was PXDN positive control which was directly loaded. C2 was negative control in which 200 nM PXDN did not mixed with bacteria; it was centrifuged at 3099 x g for 5 min as experimental groups. (C) The remaining bacterial suspension in “B” was subjected to peroxidase activity assay using L-012 as chemiluminescent substrate. Note: MPO is known to bind to P. aeruginosa via its cationic charge, and served as a positive control in these experiments. *P<0.001 vs. bacteria only. Data are the representatives of at least three independent experiments. (D) E. coli and P. aeruginosa bind to plasma PXDN. 4 x 108 of E.coli K12 or P. aeruginosa strain K in 50 μL PBS were added to 50 μL human plasma. The mixtures were incubated at 37°C for 1 h. Bacterial suspensions were spun down at 3099 x g for 5 min, and then washed twice with 500 μL PBS. Samples were subjected to immunoblotting using anti-PXDN antibody. “C” represents positive control of 1 μL of plasma containing 200 ng of PXDN, which was directly loaded onto the gel. Data are the representatives of three independent experiments. (E) Truncated PXDN binds to live P. aeruginosa and E. coli. Recombinant truncated peptides of PXDN 29-250aa or PXDN 251-609aa were added to live bacterial suspensions of P. aeruginosa strain K and E. coli K12, respectively. The mixtures were incubated at 37°C for 1 h. P. aeruginosa and E. coli suspension were centrifuged at 3099 x g for 5 min, and washed twice with PBS. The bacterial lysates were subjected to immunoblotting using anti-His antibody and visualized by chemiluminescence. A mixture containing only 1.3 μM of recombinant PXDN peptide without bacteria served as negative control (to verify no binding of PXDN peptide to the test tube). Data are representatives of three independent experiments.
Results
PXDN binds to gram-negative (GN) bacteria
Our data reveal that PXDN is able to generate hypohalous acids and kills bacterial in vitro [16, 17]. However, the precise mechanism of bactericidal activity of PXDN is unknown. PXDN contains specific N-terminus with five LRRs and four Ig C2 domains, and is predicted in protein-protein interaction and/or protein-pathogen interaction. Its C-terminus, which mainly consists of the peroxidase domain, is responsible for the generation of hypohalous acids [14]. We hypothesize that the N-terminus of PXDN binds to GN bacteria and facilitates bacterial killing by the peroxidase activity at the C-terminus. We sub-cloned full-length PXDN (FL-PXDN) as well as specific constructs containing LRRs and/or Ig C2 domains of PXDN into expression plasmids harboring His-tag (Fig 1A). These recombinant proteins were expressed either in E. coli or human HEK293 cells, and purified using HisPur resin. We first mixed recombinant FL-PXDN with Pseudomonas aeruginosa (P. aeruginosa); bacterial suspensions were spun down by centrifugation. PXDN was detected in the bacterial pellets (Fig 1B), and a dose-dependent increase of peroxidase activity was verified in these fractions (Fig 1C). Additionally, we assessed whether endogenous PXDN in circulating plasma from healthy human volunteers can bind to P. aeruginosa and E. coli. Live P. aeruginosa and E. coli bacterial suspensions were mixed with human plasma. After centrifugation, PXDN was co-precipitated with bacteria, suggesting direct interaction between bacteria and plasma PXDN (Fig 1D). Unlike MPO, the pI value of PXDN is near neutral (pI = ~7.0); thus, the interaction of PXDN with bacteria is unlikely to be due to its surface charge. Ceruloplasmin is a ferroxidase enzyme present in circulating plasma at a concentration of 20 to 60 mg/dL (1.5–4.5 μM) [21]. We utilized this enzyme as an internal control to verify the specificity of the interaction of bacteria with plasma PXDN. As expected, ceruloplasmin did not bind to P. aeruginosa while minimal binding to E. coli was detected (S1 Fig).
We further evaluated whether specific regions in PXDN mediates binding to P. aeruginosa and E. coli. Similarly, recombinant truncated peptides of PXDN 29-250aa and PXDN 251-609aa were incubated with bacteria, respectively. These recombinant peptides were able to bind to P. aeruginosa and E. coli in a dose-dependent manner (Fig 1E). The lower molecular weight bands (approximate 35 kDa) observed in the PXDN 251-609aa group likely represent degradation products of the PXDN 251-609aa peptide (Fig 1E). Together, these data demonstrate that PXDN interacts with GN bacteria via its N-terminus.
PXDN binds to LPS
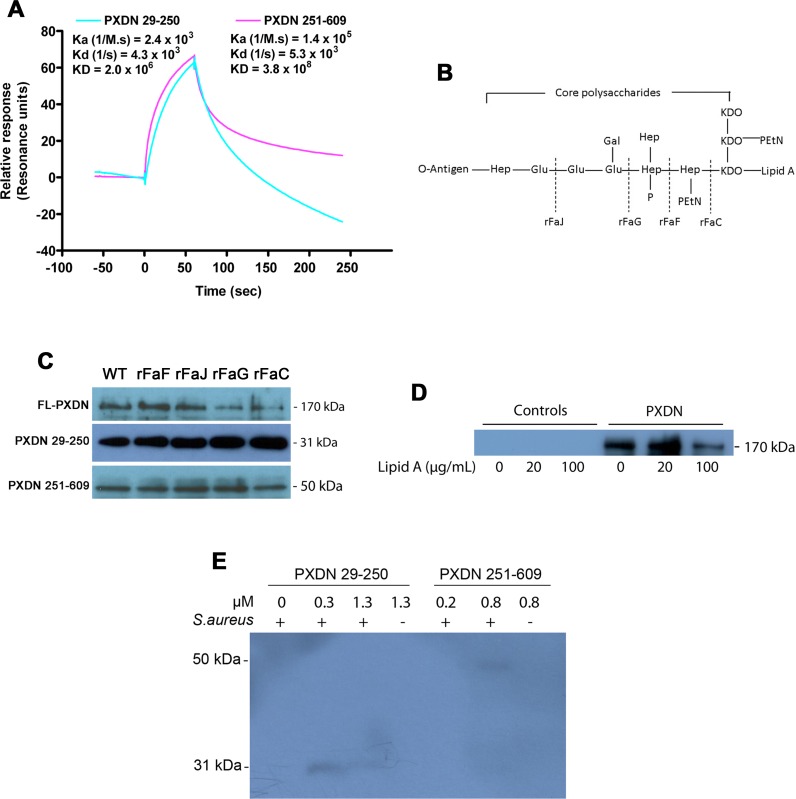
Next, we determined the mechanisms of PXDN binding to P. aeruginosa and E. coli. LPS, cell-surface polysaccharide, maintains outer membrane integrity and mediates host-pathogen interactions. LPS comprises O-antigen, core polysaccharides and lipid A. LPS from E. coli and P. aeruginosa has the same general structure. Lipid A and the proximal region of oligosaccharides are relatively conserved, while O-antigen is highly variable in composition and structure [22]. We hypothesized that LPS serves as a PAMP that directly binds the LRRs and/or Ig domains of PXDN. To determine whether LPS binds to PXDN, we performed surface plasmon resonance (SPR) assays. As shown in Fig 2A, both the LRRs and Ig domains of PXDN were able to bind LPS. The association constants of LRRs and Ig domains of PXDN to LPS are 2.4 x 103 and 1.4 x 105, respectively.
Fig 2. PXDN interacts with LPS.
(A) PXDN binding to LPS determined by SPR. Recombinant peptides of PXDN 29-250aa or PXDN 251-609aa were immobilized on a NTA chip, and LPS (2 μM) was flowed over the chip. The binding data were collected and analyzed with Biacore T200 software. Data are the representatives of three independent experiments. (B) The typical structure of E. coli LPS and genes determining the biosynthesis of LPS. Dash indicates the stage point of LPS synthesis by the corresponding gene. KDO, 3-deoxy-D-manno-oct-2-ulosonic acid; Hep, L-glycero-D-manno-heptose; EtN, ethanolamine; Gal, D-galactose; Glu, D-glucose; P, phosphate. (C) Interaction of PXDN with LPS-deficient strains of E. coli. The binding experiments of truncated PXDN to LPS-deficient E. coli strains were carried out as in Fig 1B and 1E. 100 nM of FL-PXDN or truncated PXDN were utilized. rFaF, rFaJ, rFaG and rFaC are the LPS-deficient E. coli strains with mutation of related gene. “WT” is wild-type of E. coli K12 BW25113, which is the parent strain of LPS-deficient E. coli strains. Data are representatives of three independent experiments. (D) Lipid A inhibits PXDN binding to GN bacteria. Lipid A was mixed with E. coli K12 and PXDN similar to Fig 1B. The pellets were washed twice and subjected to immunoblotting using anti-His antibody. The data are representatives of two independent experiments. (E) The binding capacity of PXDN to GP bacteria is limit. Recombinant peptides of PXDN 29-250aa or PXDN 251-609aa were added to live bacterial suspensions of S. aureus, similar to Fig 1E. The bacterial lysates were subjected to immunoblotting using anti-His antibody and visualized by chemiluminescence. Data are representatives of three independent experiments.
We further queried which component of LPS is responsible for the interaction with PXDN. E. coli LPS structure is determined by a set of genes that encode biosynthesis of LPS components at various stages (Fig 2B) [23]. For example, a mutation in the rFaC gene causes formation of LPS consisting of only lipid A and 3-deoxy-D-manno-oct-2-ulosonic acid (KDO), with an absence of the O-antigen and most of the core polysaccharides. We carried out bacterial binding experiments of FL-PXDN and truncated PXDN with the various mutants of E. coli LPS. FL-PXDN and the truncations of PXDN 29-250aa and PXDN 251-609aa bound to several LPS variants (Fig 2C). Importantly, both full-length and truncated forms of VPO-1 were able to bind to the LPS variant that only contains lipid A (rFaC mutant). Lipid A was able to inhibit PXDN binding to E. coli (Fig 2D). Since gram-positive (GP) bacteria lack of LPS, we determined whether the N-terminus of PXDN binds specifically to GN bacteria, but not to GP bacteria. Binding assays with the GP bacteria, Staphylococcus aureus (S. aureus), were carried using truncated PXDN 29-250aa and 251-609aa (the experiment was performed concurrently with the studies of GN bacteria, as shown in Fig 1E). In contrast to P. aeruginosa and E. coli, the N-terminal domains of both LRRs and Ig domains did not bind to S. aureus (Fig 2E). Taken together, these data strongly support that the N-terminal domains of PXDN bind to LPS of GN bacteria.
LPS activates PXDN
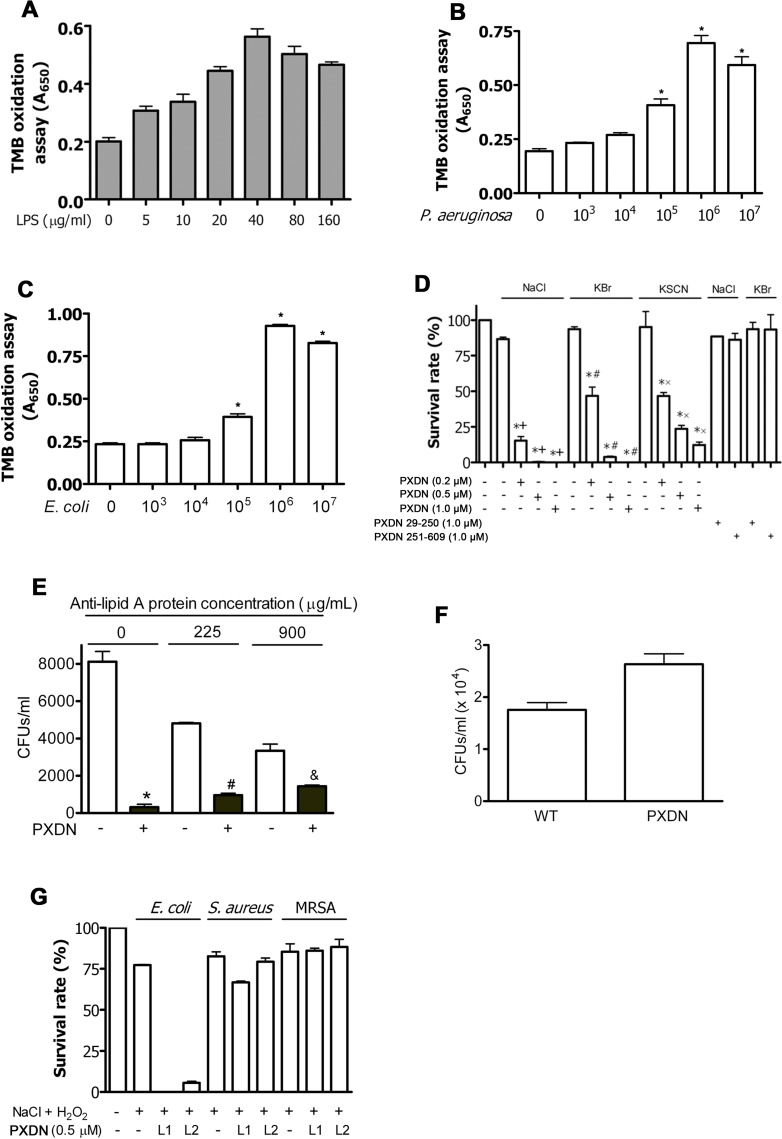
We explored whether bacterial binding induces PXDN activation. LPS was added to reaction mixtures containing PXDN, H2O2 and 3,3’,5,5’-tetramethylbenzidine (TMB), and oxidation products were measured as an index of PXDN-dependent peroxidase activity. PXDN activity increased in a dose-dependent manner with the addition of LPS (Fig 3A). Maximal PXDN activity occurred at an LPS concentration of 40 μg/mL; the induced activation was ~2.8 fold higher than control without LPS (Fig 3A). Higher concentrations of LPS inhibited PXDN activity (Fig 3A). Further, live P. aeruginosa and E. coli at varying colony formation units (CFUs) were added to the reactions. Stimulation of PXDN activation was found to be dependent on the number of CFUs, with PXDN activity increasing approximately 3.7 and 4.2 folds by P. aeruginosa and E. coli, respectively, at 106 CFUs (Fig 3B and 3C). Live bacteria in the absence of PXDN are incapable of oxidizing TMB (S2 Fig). Interestingly, live E. coli did not activate MPO and LPO, which are not known to have specific interacting domains with LPS or bacteria (S3A and S3B Fig). Thus, both LPS and live bacteria are capable of activating PXDN, but not MPO or LPO.
Fig 3. PXDN selectively kills GN bacteria in vitro.
(A) LPS stimulates PXDN activity. Reaction mixtures (100 μL each) containing TMB solution, H2O2, recombinant FL-PXDN (400 nM/heme) and LPS as indicated were carried out at room temperature for 30 min. LPS and recombinant FL-PXDN were pre-incubated at 4°C for 30 min prior to mix with TMB solution. TMB oxidation was recorded at absorbance 650 nm. One-way analysis of variance, P < 0.001 for all comparisons. (B) Live P. aeruginosa stimulates PXDN activity. P. aeruginosa was pre-incubated with recombinant FL-PXDN at RT for 30 min in 20 mM of phosphate buffer without NaCl. The mixture was added into TMB solution (100 μL) and incubated at RT for 30 min. Absorbance at 650 nm was measured. *P<0.05 vs. non-bacteria control. (C) E. coli stimulates PXDN activity. The same experiment was carried out as in “B” where P. aeruginosa was replaced by E. coli. *P<0.05 vs. non-bacteria control. (D) PXDN kills GN bacteria in vitro. P. aeruginosa strain K suspensions were incubated in 50 mM phosphate buffer (pH 6.2) containing indicated amounts of recombinant FL-PXDN or 1 μM of truncated PXDN (PXDN 29-250aa and PXDN 251-609aa), 10 μM H2O2, and halide (140 mM NaCl, 100 μM KBr or 100 μM KSCN) at 37°C for 1 h. Cell mixtures were plated on LB agar plates and incubated at 37°C overnight. The control group contained P. aeruginosa only (lane 1). The CFUs were counted, and relative survival rates were calculated as CFUs in the experimental group divided by those in the control group. *P < 0.001 vs. Control; +P < 0.001 vs. H2O2 + NaCl; #P < 0.001 vs. H2O2 + KBr; xP < 0.001 vs. H2O2 + KSCN. Data are representatives of three independent experiments. (E) Anti-lipid A antibody inhibits bacterial killing by PXDN. E. coli K12 was incubated in 50 mM phosphate buffer (pH 6.2) containing 2 μM recombinant PXDN, 10 μM H2O2, 140 mM NaCl and indicated amounts of anti-lipid A antibody, at 37°C for 1 h. Cell mixtures were plated on LB agar plates and incubated at 37°C overnight. The colonies were counted. Paired Student’s t-test: *P = 0.0031; #P = 0.0016; &P = 0.0464. (F) Sera from PXDN-deficient mice are impaired to kill bacteria. Sera from C57BL/6 and PXDN-deficient mice were used in the experiments. 100 μL reaction mixtures contained 50 μL serum and 50 μL PBS containing P. aeruginosa strain K and 50 μM H2O2. After incubation at 37°C for 1h, the mixture was plated on LB agar plates and incubated at 37°C for overnight. n = 3. P = 0.03. (G) PXDN cannot kill GP bacteria. The bacterial killing experiment was carried out similar to “D” in the presence of H2O2 and NaCl. Two lots of recombinant PXDN were utilized (L1 and L2). E. coli K12 was used as positive control. One-way analysis of variance, P < 0.001 for E. coli; P = 0.1308 for S. aureus; P = 0.6460 for MRSA. All data are representatives of at least three independent experiments.
PXDN kills GN, but not GP bacteria
Since the hPx family plays an important role in host defense [8, 9], we further investigated whether PXDN in the presence of H2O2 is able to kill bacteria, similar to the MPO/H2O2 system in phagocytes [8, 24] and the LPO/H2O2 system at mucosal surfaces [25]. Bacterial suspensions of P. aeruginosa were incubated with H2O2 and halide anion (chloride, bromide or thiocyanate, a pseudohalide), in the absence or presence of PXDN (0.2 to 1 μM). Bactericidal activities were assessed by survival rates of P. aeruginosa relative to control (without PXDN). Survival of P. aeruginosa was dose-dependent, with 0.2 μM PXDN inducing ≥ 50% killing effect under these conditions (Fig 3D). Physiological concentrations of PXDN (~1.1 μM) [20] completely killed the bacteria in the presence of physiological concentrations of Cl- or Br- and 10 μM H2O2 (Fig 3D). PXDN was less efficient in killing P. aeruginosa in the presence of 10 μM H2O2 and 100 μM of SCN-. This is likely explained by the lower oxidizing potency of the product, HOSCN. However, PXDN (0.5 μM), in the presence of 10 μM H2O2 and 500 μM SCN-, completely killed P. aeruginosa (S4 Fig). Neither truncated PXDN 29-250aa nor PXDN 251-609aa had the capacity to kill P. aeruginosa (Fig 3D). Similar capacity for E. coli killing was observed with FL-PXDN (Table 1). Interestingly, anti-lipid A antibody was able to significantly inhibit bacterial killing by PXDN (Fig 3E), consistent with the observation that PXDN binds to lipid A (Fig 2D). In addition, we evaluated bacterial killing of sera. Our data indicate that sera from PXDN-deficient mice is diminished capacity of bacterial killing, comparing with that from WT mice (Fig 3F), supporting a role of circulating PXDN in host defense.
Table 1. Bactericidal activities of PXDN, MPO and LPO.
E. coli K12 was incubated in 50 mM phosphate buffer (pH 6.2) containing halide anion (Cl-, Br- or I-), 10 μM H2O2, and indicated amount of hPx at 37°C for 1 h. Cell mixtures were plated on LB agar plates and incubated at 37°C overnight. The negative control experiment contained E.coli only. The CFUs were counted. The relative survival rate (%) was calculated as CFUs in the experimental group divided by those in the negative control. Data are representatives of at least three independent experiments.
| hPx (nM/heme) | Cl- (100 mM) | Br- (100 μM) | I- (0.25 μM) | SCN- (100 μM) | |
|---|---|---|---|---|---|
|
MPO |
50 | 0.0% | 0.0% | 0.0% | 56.7 ± 2.8% |
| 200 | N/A | N/A | N/A | 48.9 ± 0.9% | |
| 1000 | N/A | N/A | N/A | 4.7 ± 3.2% | |
|
LPO |
50 | 103.7 ± 8.6% | 0.0% | 98.1 ± 10.9% | 104.7 ± 5.9% |
| 200 | 110.5 ± 15.4% | N/A | 4.0 ± 0.7% | 120.4 ± 6.9% | |
| 1000 | 92.3 ± 8.6% | N/A | 0.0% | 82.6 ± 13.1% | |
|
PXDN |
50 | 103.3 ± 7.6% | 0.0% | 62.5 ± 5.3% | 71.8 ± 1.1% |
| 200 | 12.0 ± 1.8% | 0.0% | 0.0% | 46.4 ± 2.2% | |
| 1000 | 0.0% | 0.0% | 0.0% | 32.7 ± 3.4% | |
| 0 | 118.7± 8.5% | 89.6± 4.2% | 97.7± 9.8% | 102.0± 9.3% | |
We further examined whether PXDN is capable of killing GP bacteria, which lack LPS and do not directly bind PXDN (Fig 2E). We carried out the bacterial killing experiments in the presence of H2O2 and Cl-. Two different lots of recombinant FL-PXDN were utilized; both lots were unable to kill S. aureus and methicillin-resistant Staphylococcus aureus (MRSA), while their capacity to kill E. coli was maintained (Figs 3G and S5). Together, these data provide strong evidence that binding of PXDN to LPS of GN bacteria is essential for bacterial killing.
PXDN secreted by lung epithelial cells kills bacteria
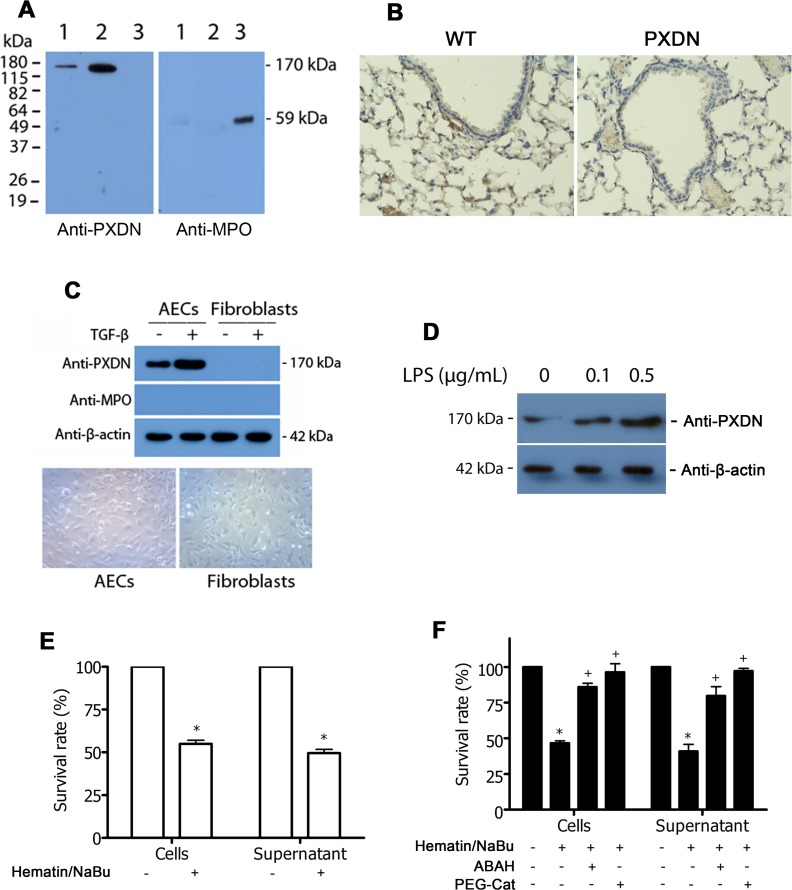
The mammalian lung is endowed with multiple mechanisms of host defense that protect the mucosal epithelial barrier from pathogens, although a role for PXDN has not been established. We investigated the expression and distribution of PXDN in the mammalian lung and its potential role in lung host defense. First, high levels of PXDN were detected in bronchoalveolar lavage fluid (BALF) from normal human volunteers, whereas only trace amounts of MPO were detected (Fig 4A). Second, PXDN expressed in alveolar epithelial cells of WT mice, but not in PXDN-deficient mice (Fig 4B, dark brown). We further determined the expression of PXDN in type II alveolar epithelial cells (AECs). Primary type II AECs and fibroblasts were isolated from lungs of C57BL/6 mice using previously described methods [26]. PXDN was expressed in type II AECs, but not in fibroblasts (Fig 4C). PXDN expression in lung epithelial cells is also supported by the data of proteomic profiling and RNA-seq profiling (https://lungmap.net/). Interestingly, PXDN expression in AECs was induced by LPS in a dose-dependent manner (Fig 4D). We ascertained whether type II AECs are capable of PXDN-mediated P. aeruginosa killing. PXDN was expressed and activated by adding hematin (1 μg/mL) and sodium butyrate (NaBu, 5 mM), since the combination of hematin and NaBu induces PXDN expression and enhances PXDN activity [14]. After stimulation for 24 h, cells and medium were separated for evaluation of PXDN-mediated P. aeruginosa killing. AECs as well as the corresponding supernatants significantly induced P. aeruginosa killing (Fig 4E). 4-aminobenzoic acid hydrazide (ABAH), an inhibitor of hPx enzymes, and catalase, which reduces H2O2 to water, inhibited P. aeruginosa killing (Fig 4F). These studies demonstrate that lung epithelium is a source of PXDN which mediates H2O2-dependent bactericidal activity, a previously unrecognized host defense function of this enzyme.
Fig 4. PXDN is expressed in the lung epithelium and contributes to bacterial killing.
(A) Detection of PXDN in human bronchoalveolar lavage fluid (BALF). Right middle lobe BALF was collected and processed within 2 h. BALF samples were then centrifuged at 400 x g for 10 min at 4°C. BALF were concentrated by Centricon (~10x). Samples of BALF (1), positive control of human plasma containing ~100 ng of PXDN (2), and purified MPO (50 ng) (3) were subjected to immunoblotting using anti-PXDN or anti-MPO antibodies. (B) IHC of the lung. IHC of lung sections from WT and PXDN-deficient mice was performed using anti-PXDN antibody (1:600). Images were taken using BZ-X710 All-in-One Fluorescence Microscope. PXDN: dark brown. Magnification: 400x. (C) PXDN expresses in mouse primary lung type II alveolar epithelial cells (AECs). Upper panel showed immunoblot analysis of AECs and fibroblasts while lower panel showed phase contrast microscopy of AECs (left) and fibroblasts (right); magnification 100x. Cell lysates were subjected to conventional immunoblotting by using anti-PXDN antibody. β-actin was used as loading control. (D) LPS induces PXDN expression. AECs were induced by LPS as indicated. The cell lysates were subject to immunoblot analysis as in (C). (E) Mouse primary type II AECs mediate P. aeruginosa killing. AECs were grown in 12-well plate in DMEM with 10% FBS without antibiotics until 70% confluence. Cells were serum-starved for 16 h prior to bactericidal assays; cells were stimulated with/without TGF-β and hematin/NaBu. Cells and the overlying medium (supernatant) were separated and utilized for evaluation of PXDN-mediated bactericidal killing. 1 mL of fresh DMEM containing 104 P. aeruginosa strain K and 10 μM H2O2 was added to AECs or supernatant. The mixture was incubated at 37°C for 1 h, and then plated on LB agar plates prior to overnight incubation at 37°C. The control group contained untreated AECs and P. aeruginosa. The relative survival rate was calculated as CFUs in the experimental group divided by those in the control group; *P< 0.001 vs. Control. (F) Inhibition of AEC-mediated P. aeruginosa killing by peroxidase inhibitors. The experiments were carried out as in “E” with addition of ABAH (broadly specific inhibitor of heme-containing peroxidases) or PEG-catalase (which reduces H2O2). *P< 0.001 vs. Control; +P < 0.001 vs. hematin/NaBu. Data are representatives of three independent experiments.
Deficiency of PXDN impairs bacterial killing and decreases survival in a murine model of GN bacterial pneumonia
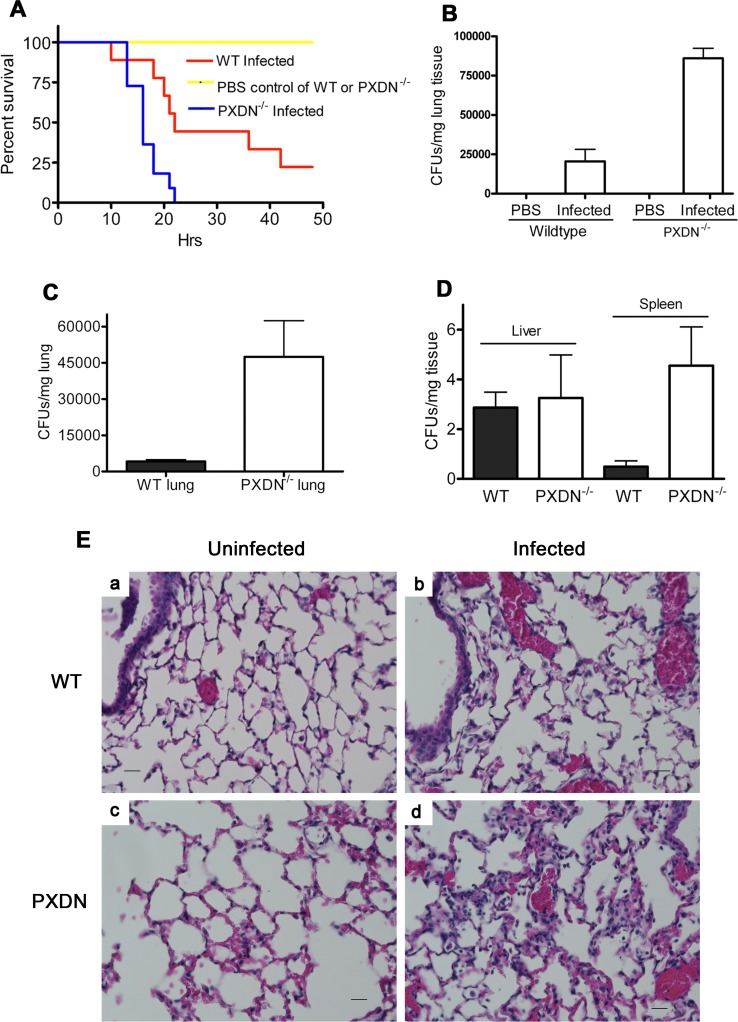
To determine whether PXDN mediates critical host defense functions in vivo, we employed a murine model of GN bacterial pneumonia in wild-type and PXDN mutant mice (PXDNmhdakta048) [19]. P. aeruginosa, a leading cause of GN bacterial pneumonia in humans [27], were intra-tracheally injected into mice to induce acute lung infection. PXDN-deficient mice had markedly diminished survival, with 100% mortality at 24 h following infection while wild-type mice had 44% and 22% survival at 24 and 48 h, respectively (Fig 5A). Wild-type mice that survived acute infection at 48 h appeared to recover completely and remained healthy for several days. The high mortality in PXDN-deficient mice was associated with increased bacterial burden, as evidenced by CFUs of P. aeruginosa in lung tissues (Fig 5B). PBS treatment did not result in death in either strain of mice, and bacteria were absent in the lungs of these mice (Fig 5A and 5B). Intratracheal injection with sublethal dose of P. aeruginosa (3 x 106 CFUs/mouse) showed increased bacterial burden in the lungs of PXDN-deficient mice (Fig 5C). In the liver and spleen, fewer bacterial CFUs were detected from both WT and PXDN-deficient mice; the bacterial number detected from the spleen from PXDN-deficient mice was significantly higher than that from WT mice (Fig 5D). The infected lungs of PXDN-deficient mice revealed more severe tissue injury and neutrophil infiltration, while uninfected lungs showed normal structure (Fig 5E). Together, these studies provide compelling data to support a critical host defense function of PXDN, specifically against GN bacterial pneumonia.
Fig 5. PXDN mutant mice reveal impairment in bacterial clearance during acute lung infection.
(A) Decrease of relative survival rates. PXDN-deficient and C57BL/6 wild-type mice were intratracheally instilled with 7 x 106 of P. aeruginosa strain K or PBS control. Relative survival rates were determined over a period of 48 hours; n = 9–11 per group, P = 0.0068. (B) Lung Bacterial burden. Mice were sacrificed at 20 hours; lungs were aseptically removed, weighed, and homogenized in PBS. Lung tissue suspension was serially diluted and plated on LB agar plates. After incubation at 37°C for 18 h, CFUs were counted and CFUs/mg tissue were calculated; n = 12 from 4 mice per group; one-way analysis of variance, P < 0.0001 for all comparisons. (C and D) Tissue burden on bacterial infection with sublethal dose was carried out by intratracheally infecting the mice with 3 x 106 of P. aeruginosa strain K. After 20 h, lung, liver and spleen were taken as in (B) for detection of bacteria. n = 15–21 from 5–7 mice per group; P (lung) = 0.016; P (liver) = 0.843; P (spleen) = 0.014. (E) H&E staining of the lungs from uninfected and infected mice. Mice were infected as in (C) and the lungs were harvested at 20 h for preparation of staining. a. WT mouse, uninfected; b, WT mouse, infected; c. PXDN-deficient mouse, uninfected; d, PXDN-deficient mouse, infected. Magnification: 400x. Scale: 5μm.
Discussion
The lung is a uniquely vulnerable organ with a very thin, delicate epithelial lining, abundant blood flow, and a vast surface area. The lung resides at the interface of the body and environmental exposures to inhaled or aspirated pathogens. Thus, the lung is an important organ in host defense. Multiple layers of defense in the normal lung are involved in innate immune functions. Loss of one or more of these host defense mechanisms increases the susceptibility of the lung to infections. PXDN is a newly identified hPx, an enzyme family that plays an important role in host defense. Its physiological function is largely unknown, although recent studies implicate this gene in basement membrane synthesis [28, 29]. The present study, for the first time, identifies PXDN as a novel host defense enzyme in the lung with selectively for recognizing and directly killing GN bacteria. The N-terminus of PXDN, which contains five LRRs and four Ig domains, selectively binds to LPS while the C-terminus of PXDN containing the peroxidase domain kills GN bacteria via generation of hypohalous acids. An enzyme containing the molecular structure of both pattern-recognition domain and scavenger domain suggests evolutionary conservation of a dual-function protein capable of both pathogen recognition and killing, expanding our current view of innate immunity.
The original theory of innate immune pattern recognition is based on the interaction between host PRRs and specific PAMPs, and the activation of downstream signaling events and host defense mechanisms [4, 5]. On the other hand, many effectors including complement system, antimicrobial peptides, and lysozymes may bind to important bacterial molecules and directly kill pathogens. For example, neutrophils possess bactericidal permeability-increasing protein (BPI) in their azurophilic granules. BPI is structurally related to LPS binding protein, and avidly binds LPS in GN organisms to directly kill them by compromising membrane integrity [30]. Peptidoglycan recognition proteins (PGRPs) are innate immune molecules present in insects, mollusks, echinoderms, and vertebrates. Mammals have four PGRPs. One mammalian PGRP, PGLYRP-2, is an N-acetylmuramoyl-L-alanine amidase that hydrolyzes bacterial peptidoglycan and reduces its pro-inflammatory activity. The three remaining PGRPs kill bacteria by interacting with cell wall peptidoglycan [31]. Our data support the uniqueness of PXDN, among the known mammalian hPx’es, based on its structural characteristics that allows for its dual-function in pathogen recognition and killing.
PXDN is found in circulating plasma at a concentration of 1.1 ± 0.6 μM in humans and 2.6 ± 0.6 μM in mouse, approximately ~1000-fold higher than MPO [20]. Although its peroxidase activity is ~5–10% of MPO activity [14], the net activity of PXDN may be 50–100 folds higher than MPO in plasma. The high expression of PXDN in alveolar epithelium and in bronchoalveolar lavage fluid suggests that this enzyme may mediate critical host defense functions and mucosal immunity. The ability of PXDN to directly bind GN bacterial pathogens allows for more targeted activation and killing without collateral damage to surrounding tissues.
The concept of targeted killing is further supported by the finding that LPS induces PXDN activation, a phenomenon has not been reported for other members of the hPx family. Our data reveals that LPS is able to activate PXDN by ~4-fold. Thus, the bactericidal activity of PXDN to GN bacteria is great increased in a selective and targeted manner. The mechanism of activation of PXDN by LPS may be due to LPS-mediated conformation changes of PXDN since the appropriate conformation is critical for the catalytic activity of hPx enzymes [8]. The activation of PXDN by LPS has important implications for enhanced bactericidal activity, while limiting damage to host tissues.
P. aeruginosa is a pathogen responsible for a variety of severe infections, including acute lower respiratory tract infections in both immunocompetent and immunocompromised hosts, as well as chronic respiratory infections in select patient populations such as those with cystic fibrosis [32]. High incidence, infection severity and increasing resistance characterize P. aeruginosa infections, highlighting the need for new therapeutic options. Our findings of PXDN killing of GN bacteria, including P. aeruginosa, support therapeutic interventions involving PXDN to augment host defense against such infections.
In summary, PXDN is a novel host defense enzyme in the lung with dual function in pathogen recognition and killing. Further studies are required to determine its role in systemic immunity. The unique structural and functional characteristic of PXDN expands our current understanding of mucosal innate immunity, and has important implications for novel therapeutic strategies.
Materials and methods
Animals
C57BL/6 and PXDNmhdakta048 mutant mice (male and females, 8–12 week-old) were used in the study. PXDNmhdakta048 mice were in C57BL/6 background [19]. Unless otherwise stated, mice were fed with normal chow diet.
Ethics statement
The protocol of animal study was approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham with approval number 20223. The animal care and use are adhered to the regulations and guidelines of International Association of Assessment and Accreditation of Laboratory Animal Care, Office of Laboratory Animal Welfare and the United State Department of Agriculture.
Bacteria and PXDN binding assay
Recombinant FL-PXDN, PXDN 29-250aa or PXDN 251-609aa was added into 100 μL PBS containing 4 x 108 live P. aeruginosa strain K or E. coli K12. The mixture was incubated at RT for 1 h. Bacteria were spun down at 3099 x g for 5 min. The cells were washed twice by 5 x initiating volume of PBS. Bacteria were lysed in 2 x SDS-PAGE loading buffer and the lysates were subject to immunoblot analysis using anti-His antibody. Protein bands were visualized by chemiluminescence. Negative controls contained only PXDN or bacteria. Quantitative analysis was carried out by using ImageJ software (The National Institute of Health). In some experiments, LPS-deficient E. coli stains were utilized. LPS-deficient E. coli strains and their parent strain K12 BW25113 were from The Coli Genetic Stock Center at Yale University. In bacteria and plasma PXDN binding assay, 4 x 108 E. coli or P. aeruginosa in 50 μL PBS were mixed with 50 μl of human plasma. Control groups were cells or plasma alone. In some experiments, lipid A (Sigma-Aldrich Cat. #L5399) was used. In brief, indicated amount of lipid A was mixed with 150 nM of recombinant PXDN in 600 μL PBS. The mixture was shacked at RT for 15 min. Then 2 x 108 live E.coli K12 cells were added into the mixture and incubated at RT for additional 30 min. Bacterial pellets were obtained and subject to immunoblot analysis.
Surface plasmon resonance (SPR)
Analyses were carried out using a Biacore-T200 instrument (GE-Healthcare) at 22°C in PBS. Recombinant PXDN 29-250aa or PXDN 251-609aa (50 μg/mL) were captured onto an NTA Sensor Chip (GE-Healthcare), respectively. LPS (2 μM) was injected over each surface, as well as over a blank surface. Full kinetics was carried out by flowing a serial concentration (range from 0.25 to 10.0 μM) of LPS over the chip. Binding data (Ka, Kd and KD) were collected and analyzed by using the BIAevaluation software (Biacore). All measurements were conducted in triplicate. Rate constants of SPR were calculated as following.
Association rate: d[AB]/dt = ka·[A]·[B]
Dissociation rate: -d[AB]/dt = kd·[AB]
Equilibrium dissociation constant: KD = kd/ka = [A]·[B]/[AB]
3,3’,5,5’-Tetramethylbenzidine (TMB) oxidation assay
hPx as indicated was added into 100 μL of TMB solution (TMB Liquid Substrate System, Sigma-Aldrich), which contains H2O2. Reaction mixture was incubated at RT for 30 min. TMB oxidation was recorded at absorbance 650 nm. In some experiments, LPS (Sigma-Aldrich Cat #L2880, from E. coli 055:B5) or GN bacteria were mixed with 400 nM/heme of recombinant PXDN at RT for 30 min. 20 μL of mixture was added into 100 μL TMB solution. After 30 min, absorbance at 650 nm was recorded.
In vitro killing of bacteria by PXDN
P. aeruginosa strain K and E. coli K12 cells were incubated in 50 mM phosphate buffer (pH 6.2) containing 140 mM NaCl, 10 μM H2O2, and indicated amounts of PXDN at 37°C for 1 h. Cell mixtures were plated on LB agar plates, followed by incubation at 37°C overnight. In control experiments, only H2O2 (10 μM) or Cl- (140 mM) was present. The CFUs were counted and relative survival rates were calculated as CFUs in the experimental group divided by CFUs in the control group. Other halide anions (bromide, iodide and thiocyanate) in addition to Cl- were used as indicated. In some experiments, anti-lipid A antibody was used for inhibition of bacterial killing by PXDN. GP bacteria were also utilized in some bacterial killing experiments.
Bactericidal activity of serum
100 μL reaction mixtures contained 50 μL serum from C57BL/6 or PXDN-deficient mouse and 50 μL PBS containing P. aeruginosa and 50 μM H2O2. After incubation at 37° for 1h, the mixture was plated on LB agar plates and incubated at 37° for overnight. Colonies were counted.
L-012 oxidation assay
Chemiluminescent dye L-012 is a sensitive substrate for measuring heme-containing peroxidase activity. It generates chemiluminescence once oxidation. In present study, 20 μM (final concentration) was added into 100 μL of bacterial suspension containing bound hPx and 20 μM of H2O2. Chemiluminescent light at 450 nm was immediately recorded by a luminometer (Molecular Devices, Sunnyvale, CA).
Lung tissue staining
Mouse lung tissue was harvested and fixed in 10% formalin. The tissue sections (5 μm) were prepared, and Haemotoxylin and Eosin (H&E) staining was performed at the Comparative Pathological Laboratory at the University of Alabama at Birmingham. The Conventional immunohistochemistry (IHC) was carried out by using anti-PXDN antibody (1:600). Images were taken using BZ-X710 All-in-One Fluorescence Microscope (Keyence Corporation of America, Itasca, IL, USA).
Separation of mouse primary lung type II alveolar epithelial cells (AECs)
Mouse primary lung type II AECs were isolated as described in [26] with slight modification. In brief, 4–5 mice were euthanized with CO2. Blood was exsanguinated by clipping abdominal aorta. Trachea and lungs were carefully exposed and lungs were perfused through puncture of right ventricle with 10 mL of sterile PBS until lungs clear of blood. 18G catheter was inserted into trachea. 1 mL of dispase II (5 U/mL) per mouse was instilled into the lung; then 1% warm low melting agarose was instilled. Lungs were carefully removed and incubated in dispase II solution for 45 min. Lung tissue was minced with scissors until the consistency of jelly. Minced lungs were incubated with 5 mL per mouse of DNase I (42 U/mL)/DMEM solution for 10 min; then the suspension was filtered through successive filters (100 μm, 35 μm and 15 μm). Biotin-conjugated anti-CD32/16 (BD Biosciences, Cat. #BD553143, 15.6 μl per mouse) and Biotin-conjugated anti-CD45 (BD Biosciences, Cat. #BD553078, 36 μl per mouse) were added into the filtered suspension and incubated at 37°C for 30 min with gentle shake. The filtered suspension was centrifuged and cells were re-suspended in complete media (5 mL/mouse). 1 mL of streptavidin-magnetic beads (Promega, Cat. #8452) was added into the cells and the mixture was incubated for 30 min. The media containing unbound cells were carefully removed and placed into culture dish. The cells were then incubated at 37°C overnight. Next day, the media containing non-adherent cells were transferred into 50 mL-conical tube. The remaining adherent cells were fibroblast. The cell suspension in 50-mL tube was centrifuged at 344 x g for 10 min at 4°C. The cell pellet was re-suspended in complete media. Cells were placed into fibrinogen coated plates. These cells were type II alveolar cells, generally with ~95% purity, evaluated using anti-surfactant Protein C antibody. All steps were carried out aseptically.
Immunoblot analysis
The conventional immunoblotting assay was carried out using anti-PXDN affinity-purified polyclonal antibody (against residues 49–63 of human PXDN) [20], anti-MPO polyclonal antibody (CALBIOCHEM, Cat #475915) or anti-His antibody (Qiagen). BALF samples were centrifuged at 400 x g for 10 min at 4°C. Resultant supernatants were portioned and stored at -80°C for analysis. BALF was concentrated by Centricon (~10x). Human plasma and purified MPO (Cat. #MY167, Elastin Products Company, Owensville, MO) were as positive controls for PXDN and MPO, respectively. In some experiments, the primary lung type II AECs were stimulated with 2 ng/mL of TGF-β or LPS (0.1 or 0.5 μg/mL) for 24 hrs. Cell lysates were subject to conventional immunoblotting by using anti-PXDN or anti-MPO antibody. β-actin was used as loading control.
PXDN-mediated bacterial killing of primary lung type II AECs
AECs were cultured in 12-well plate in DMEM (Life Technologies, Inc.) supplemented with 10% FBS (Life Technologies, Inc.) without antibiotics. Cells were incubated at 37°C in 5% CO2 until 70% confluence. Cells were serum-starved for 16 hrs. Some cells were induced by addition of TGF-β1 (2 ng/mL) or NaBu (5 mM)/hematin (1 μg/mL) for 24 h to increase the PXDN expression. In some experiments, ABAH (100 μM) or catalase-polyethylene glycol (PEG-cat, 200 U/mL) was added. Cells and medium were separated for evaluation of the bacterial killing, respectively. 100 μL supernatant of medium were incubated with 4 x 104 P. aeruginosa cells containing 10 μM H2O2 at 37°C for 1 h. AECs (~5 x 105) were added 1 mL fresh DMEM plus 4 x 104 P. aeruginosa cells and 10 μM H2O2 and incubated at 37°C for 1 h. The mixture was plated on LB agar plates followed by incubation at 37°C overnight. The CFUs were counted and relative survival rate was calculated as CFUs in the experimental group divided by CFUs in the control group.
Acute lung infection model
P. aeruginosa strain K (a gift of Dr. Jean-Francois Pittet at the Department of Anesthesiology, UAB) was used for acute infections. Inoculums for mouse infections will be prepared as previously described with modification [33]. In Brief, bacteria from LB agar plate were inoculated in 5 mL of LB broth at 37°C with shaking (175 rpm). After 16–18 h of incubation at 37°C, the stationary-phase bacteria were pelleted, washed 3 times with 15 mL of sterile in PBS, and re-suspended in 3 mL sterile PBS. This stock will be diluted in sterile PBS to give an appropriate titer. Female and male C57BL/6 mice or PXDN mutant mice at 8–12 week-old (9-11/group) were used. The mice were briefly anaesthetized with ketamine-xylazine (100 mg/kg ketamine and 6 mg/kg xylazine) via intraperitoneal. The mouse was laid on a board with head elevated at 45°. 30 μL of PBS containing 3.0 x 106 or 7.0 x 106 CFUs of P. aeruginosa strain K was intratracheally instilled with a 29 G gauge needle. Mice were allowed to recover for 30–60 min prior to being returned to the cage.
Bacterial burden
50–100 mg of lung, liver or spleen was aseptically removed from mice with cervical dislocation at 20 h after instillation. The tissue was weighed and homogenized for bacteria clearance analysis. Homogenized tissue was washed with 4 mL sterile PBS. The tissue suspension was filtered through 100 μm Nylon mesh (Fisher Scientific, Cat #22363549) to remove tissue debris. The filtered suspension was centrifuged at 3099 x g at 4°C for 10 min to pellet the bacteria. The pellet was re-suspended in sterile PBS (10 μL/mg tissue). The bacterial resuspension was serial dilution. 50 μL of sample was plated on LB agar plates (triplicate) and the plates were incubated at 37°C for 18 h. Bacterial colonies were counted for analysis.
Statistical analysis
Data were shown as means ± SD, unless otherwise indicated. Quantitative variables were compared by means of Student's t-test for two groups or ANOVA for multiple groups. A value of P < 0.05 was considered significant.
Supporting information
Experiment was carried out as in Fig 1D. 4 x 108 of E. coli K12 or P. aeruginosa strain K in 50 μL PBS was added to 50 μL human plasma. The mixtures were incubated at 37°C for 1 h. Bacteria suspensions were spun down at 3099 x g for 5 min, and then washed twice with 500 μL PBS. Samples were subjected to immunoblotting using anti- PXDN or anti-ceruloplasmin antibodies. “C” represents positive control of 1 μL plasma (containing 200 ng of PXDN and 200–600 ng of ceruloplasmin) directly loaded onto the gel.
(TIF)
Experiments were similar to Fig 3B and 3C. Indicated number of bacteria was added into TMB solution. Absorbance at 650 nm was measured. “V” is recombinant FL-PXDN as positive control. *P < 0.0001 vs. experimental group with 107 bacteria.
(TIF)
TMB oxidation assay was carried out similar to Fig 3B and 3C. Indicated number of E. coli was added into TMB solution containing 50 nM MPO (A) or 50 nM LPO (B). Absorbance at 650 nm was measured. *P > 0.05 vs. control; #P < 0.05 vs. control.
(TIF)
The experiment was similar to that in Fig 3D. P. aeruginosa strain K suspensions were incubated in 50 mM phosphate buffer (pH 6.2) containing 500 nM FL-PXDN, 10 μM H2O2, and 500 μM KSCN) at 37°C for 1 h. Cell mixtures were plated on LB agar plates and incubated at 37°C overnight. The CFUs were counted, and relative survival rates were calculated. n = 3, P = 0.0004.
(TIF)
The bactericidal experiments were carried out as in Fig 3G in the presence of H2O2 and NaCl. Photographs of petri dishes were taken. Data are representatives of three plates. Two lots of recombinant PXDN were used. E. coli was used as positive control.
(TIF)
Acknowledgments
We thank Drs. Joao A. de Andrade and Tracy Luckhardt at the Division of Pulmonary, Allergy and Critical Care Medicine, Department of Medicine, University of Alabama at Birmingham (UAB) for providing BALF and technical support for preparation of mouse primary lung type II AECs, respectively. We thank Dr. Jean-Francois Pittet at the Department of Anesthesiology, UAB for providing P. aeruginosa strain K. We are grateful to Dr. Edlue M. Tabengwa, Multidisciplinary Molecular Interaction Core at UAB, who carried out the analyses with Biacore T200. We thank Dr. Moon Nahm at the Division of Pulmonary, Allergy and Critical Care Medicine, Department of Medicine, UAB for suggestive comments. We thank The Coli Genetic Stock Center at Yale University for providing the LPS-deficient E. coli strains.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the grants from the National Heart, Blood, and Lung Institute, and National Institute of Allergy and Infectious Disease, National Institutes of Health grants, R01HL086836 and R21AI101642, as well as American Heart Association grant 12GRNT12040409 (GC). RS was supported by grant from The International Postdoctoral Exchange Fellowship Program of China (No.2013M542143). VJT was supported by grants from the National Institutes of Health grants, P01HL114470 and R01AG046210. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Franken C, Meijer CJ, Dijkman JH. Tissue distribution of antileukoprotease and lysozyme in humans. J Histochem Cytochem. 1989;37(4):493–8. Epub 1989/04/01. doi: 10.1177/37.4.2926127 . [DOI] [PubMed] [Google Scholar]
- 2.Thompson AB, Bohling T, Payvandi F, Rennard SI. Lower respiratory tract lactoferrin and lysozyme arise primarily in the airways and are elevated in association with chronic bronchitis. J Lab Clin Med. 1990;115(2):148–58. Epub 1990/02/01. . [PubMed] [Google Scholar]
- 3.Martin TR, Frevert CW. Innate immunity in the lungs. Proceedings of the American Thoracic Society. 2005;2(5):403–11. doi: 10.1513/pats.200508-090JS ; PubMed Central PMCID: PMCPMC2713330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54 Pt 1:1–13. Epub 1989/01/01. . [DOI] [PubMed] [Google Scholar]
- 5.Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. Epub 2002/02/28. doi: 10.1146/annurev.immunol.20.083001.084359 . [DOI] [PubMed] [Google Scholar]
- 6.Palsson-McDermott EM, O'Neill. Building an immune system from nine domains. Biochem Soc Trans. 2007;35(Pt 6):1437–44. Epub 2007/11/23. doi: 10.1042/BST0351437 . [DOI] [PubMed] [Google Scholar]
- 7.Lee MS, Kim YJ. Pattern-recognition receptor signaling initiated from extracellular, membrane, and cytoplasmic space. Mol Cells. 2007;23(1):1–10. Epub 2007/04/28. . [PubMed] [Google Scholar]
- 8.Klebanoff SJ. Myeloperoxidase: friend and foe. J Leukoc Biol. 2005;77(5):598–625. doi: 10.1189/jlb.1204697 . [DOI] [PubMed] [Google Scholar]
- 9.Nauseef WM. Myeloperoxidase in human neutrophil host defence. Cell Microbiol. 2014;16(8):1146–55. Epub 2014/05/23. doi: 10.1111/cmi.12312 ; PubMed Central PMCID: PMCPMC4301731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen RC, Stephens JT Jr. Myeloperoxidase selectively binds and selectively kills microbes. Infect Immun. 2011;79(1):474–85. Epub 2010/10/27. doi: 10.1128/IAI.00910-09 ; PubMed Central PMCID: PMCPMC3019908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnhold J, Flemmig J. Human myeloperoxidase in innate and acquired immunity. Arch Biochem Biophys. 2010;500(1):92–106. Epub 2010/04/20. doi: 10.1016/j.abb.2010.04.008 . [DOI] [PubMed] [Google Scholar]
- 12.Wijkstrom-Frei C, El-Chemaly S, Ali-Rachedi R, Gerson C, Cobas MA, Forteza R, et al. Lactoperoxidase and human airway host defense. Am J Respir Cell Mol Biol. 2003;29(2):206–12. Epub 2003/03/11. doi: 10.1165/rcmb.2002-0152OC . [DOI] [PubMed] [Google Scholar]
- 13.Fabian TK, Hermann P, Beck A, Fejerdy P, Fabian G. Salivary defense proteins: their network and role in innate and acquired oral immunity. Int J Mol Sci. 2012;13(4):4295–320. doi: 10.3390/ijms13044295 ; PubMed Central PMCID: PMCPMC3344215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng G, Salerno JC, Cao Z, Pagano PJ, Lambeth JD. Identification and characterization of VPO1, a new animal heme-containing peroxidase. Free Radic Biol Med. 2008;45(12):1682–94. doi: 10.1016/j.freeradbiomed.2008.09.009 ; PubMed Central PMCID: PMCPMC2659527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson RE, Fessler LI, Takagi Y, Blumberg B, Keene DR, Olson PF, et al. Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 1994;13(15):3438–47. ; PubMed Central PMCID: PMCPMC395246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Cao Z, Moore DR, Jackson PL, Barnes S, Lambeth JD, et al. Microbicidal activity of vascular peroxidase 1 in human plasma via generation of hypochlorous acid. Infect Immun. 2012;80(7):2528–37. Epub 2012/04/25. doi: 10.1128/IAI.06337-11 ; PubMed Central PMCID: PMCPMC3416459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Cao Z, Zhang G, Thannickal VJ, Cheng G. Vascular peroxidase 1 catalyzes the formation of hypohalous acids: characterization of its substrate specificity and enzymatic properties. Free Radic Biol Med. 2012;53(10):1954–9. Epub 2012/09/18. doi: 10.1016/j.freeradbiomed.2012.08.597 ; PubMed Central PMCID: PMCPMC3506185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan K, Rudkin A, Parry DA, Burdon KP, McKibbin M, Logan CV, et al. Homozygous mutations in PXDN cause congenital cataract, corneal opacity, and developmental glaucoma. American journal of human genetics. 2011;89(3):464–73. doi: 10.1016/j.ajhg.2011.08.005 ; PubMed Central PMCID: PMCPMC3169830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan X, Sabrautzki S, Horsch M, Fuchs H, Gailus-Durner V, Beckers J, et al. Peroxidasin is essential for eye development in the mouse. Human molecular genetics. 2014;23(21):5597–614. doi: 10.1093/hmg/ddu274 ; PubMed Central PMCID: PMCPMC4189897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng G, Li H, Cao Z, Qiu X, McCormick S, Thannickal VJ, et al. Vascular peroxidase-1 is rapidly secreted, circulates in plasma, and supports dityrosine cross-linking reactions. Free Radic Biol Med. 2011;51(7):1445–53. Epub 2011/07/30. doi: 10.1016/j.freeradbiomed.2011.07.002 ; PubMed Central PMCID: PMCPMC3439998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang WH, Wu Y, Hartiala J, Fan Y, Stewart AF, Roberts R, et al. Clinical and genetic association of serum ceruloplasmin with cardiovascular risk. Arterioscler Thromb Vasc Biol. 2012;32(2):516–22. doi: 10.1161/ATVBAHA.111.237040 ; PubMed Central PMCID: PMCPMC3262121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson SG. Composition and structure of lipopolysaccharides from Pseudomonas aeruginosa. Rev Infect Dis. 1983;5 Suppl 5:S941–9. Epub 1983/11/01. . [DOI] [PubMed] [Google Scholar]
- 23.Klena JD, Ashford RS 2nd, Schnaitman CA. Role of Escherichia coli K-12 rfa genes and the rfp gene of Shigella dysenteriae 1 in generation of lipopolysaccharide core heterogeneity and attachment of O antigen. J Bacteriol. 1992;174(22):7297–307. ; PubMed Central PMCID: PMCPMC207424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klebanoff SJ. Myeloperoxidase-halide-hydrogen peroxide antibacterial system. J Bacteriol. 1968;95(6):2131–8. ; PubMed Central PMCID: PMCPMC315145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerson C, Sabater J, Scuri M, Torbati A, Coffey R, Abraham JW, et al. The lactoperoxidase system functions in bacterial clearance of airways. Am J Respir Cell Mol Biol. 2000;22(6):665–71. doi: 10.1165/ajrcmb.22.6.3980 . [DOI] [PubMed] [Google Scholar]
- 26.Corti M, Brody AR, Harrison JH. Isolation and primary culture of murine alveolar type II cells. Am J Respir Cell Mol Biol. 1996;14(4):309–15. doi: 10.1165/ajrcmb.14.4.8600933 . [DOI] [PubMed] [Google Scholar]
- 27.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22(4):582–610. doi: 10.1128/CMR.00040-09 ; PubMed Central PMCID: PMCPMC2772362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhave G, Cummings CF, Vanacore RM, Kumagai-Cresse C, Ero-Tolliver IA, Rafi M, et al. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat Chem Biol. 2012;8(9):784–90. doi: 10.1038/nchembio.1038 ; PubMed Central PMCID: PMC4128002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCall AS, Cummings CF, Bhave G, Vanacore R, Page-McCaw A, Hudson BG. Bromine is an essential trace element for assembly of collagen IV scaffolds in tissue development and architecture. Cell. 2014;157(6):1380–92. doi: 10.1016/j.cell.2014.05.009 ; PubMed Central PMCID: PMC4144415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elsbach P. The bactericidal/permeability-increasing protein (BPI) in antibacterial host defense. J Leukoc Biol. 1998;64(1):14–8. . [DOI] [PubMed] [Google Scholar]
- 31.Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 2006;7(8):232 doi: 10.1186/gb-2006-7-8-232 ; PubMed Central PMCID: PMCPMC1779587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies JC. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr Respir Rev. 2002;3(2):128–34. . [DOI] [PubMed] [Google Scholar]
- 33.Carles M, Lafargue M, Goolaerts A, Roux J, Song Y, Howard M, et al. Critical role of the small GTPase RhoA in the development of pulmonary edema induced by Pseudomonas aeruginosa in mice. Anesthesiology. 2010;113(5):1134–43. Epub 2010/10/13. doi: 10.1097/ALN.0b013e3181f4171b . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experiment was carried out as in Fig 1D. 4 x 108 of E. coli K12 or P. aeruginosa strain K in 50 μL PBS was added to 50 μL human plasma. The mixtures were incubated at 37°C for 1 h. Bacteria suspensions were spun down at 3099 x g for 5 min, and then washed twice with 500 μL PBS. Samples were subjected to immunoblotting using anti- PXDN or anti-ceruloplasmin antibodies. “C” represents positive control of 1 μL plasma (containing 200 ng of PXDN and 200–600 ng of ceruloplasmin) directly loaded onto the gel.
(TIF)
Experiments were similar to Fig 3B and 3C. Indicated number of bacteria was added into TMB solution. Absorbance at 650 nm was measured. “V” is recombinant FL-PXDN as positive control. *P < 0.0001 vs. experimental group with 107 bacteria.
(TIF)
TMB oxidation assay was carried out similar to Fig 3B and 3C. Indicated number of E. coli was added into TMB solution containing 50 nM MPO (A) or 50 nM LPO (B). Absorbance at 650 nm was measured. *P > 0.05 vs. control; #P < 0.05 vs. control.
(TIF)
The experiment was similar to that in Fig 3D. P. aeruginosa strain K suspensions were incubated in 50 mM phosphate buffer (pH 6.2) containing 500 nM FL-PXDN, 10 μM H2O2, and 500 μM KSCN) at 37°C for 1 h. Cell mixtures were plated on LB agar plates and incubated at 37°C overnight. The CFUs were counted, and relative survival rates were calculated. n = 3, P = 0.0004.
(TIF)
The bactericidal experiments were carried out as in Fig 3G in the presence of H2O2 and NaCl. Photographs of petri dishes were taken. Data are representatives of three plates. Two lots of recombinant PXDN were used. E. coli was used as positive control.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.