Abstract
We recently proposed a dynamic copy-choice model for retroviral recombination in which a steady state between the rates of polymerization and RNA degradation determines the frequency of reverse transcriptase (RT) template switching. The relative contributions of polymerase-dependent and polymerase-independent RNase H activities during reverse transcription and template switching in vivo have not been determined. We developed an in vivo trans-complementation assay in which direct repeat deletion through template switching reconstitutes a functional green fluorescent protein gene in a retroviral vector. Complementation in trans between murine leukemia virus Gag-Pol proteins lacking polymerase and RNase H activities restored viral replication. Because only polymerase-independent RNase H activity is present in this cell line, the relative roles of polymerase-dependent and -independent RNase H activities in template switching could be determined. We also analyzed double mutants possessing polymerase and RNase H mutations that increased and decreased template switching, respectively. The double mutants exhibited low template switching frequency, indicating that the RNase H mutations were dominant. Trans-complementation of the double mutants with polymerase-independent RNase H did not restore the high template switching frequency, indicating that polymerase-dependent RNase H activity was essential for the increased frequency of template switching. Additionally, trans-complementation of RNase H mutants in the presence and absence of hydroxyurea, which slows the rate of reverse transcription, showed that hydroxyurea increased template switching only when polymerase-dependent RNase H activity was present. This is, to our knowledge, the first demonstration of polymerase-dependent RNase H activity in vivo. These results provide strong evidence for a dynamic association between the rates of DNA polymerization and polymerase-dependent RNase H activity, which determines the frequency of in vivo template switching.
Retroviral populations exhibit high levels of variation, allowing the virus to escape host immune systems (1–4) and acquire resistance to antiretroviral drugs. The rate of genetic variation depends on the mutation and recombination rates per replication cycle (5, 6). Frequent homologous recombination has been observed in many different retroviral species and occurs during reverse transcription between two copackaged RNAs (7–10). The reassortment of mutations generated by reverse transcriptases (RTs) through recombination results in greater genetic diversity of retroviral populations.
It has been proposed that RT is evolutionarily selected for low template affinity and processivity, because RT must undergo two template-switching events to complete synthesis of viral DNA; consequently, other internal template-switching events can occur during reverse transcription (11). Because retroviral genomes consist of two copackaged RNAs, template-switching events can be inter- or intramolecular. Both types of template-switching events can result in deletions, deletions with insertions, insertions, and duplications (5, 12, 13), whereas intermolecular template-switching events can result in homologous or nonhomologous recombination (14–17).
Directly repeated sequences in retroviral vectors are often deleted during reverse transcription, which provides a powerful in vivo system for studying RT template switching (12, 18–23). Studies of direct repeat deletion rates in both spleen necrosis virus and murine leukemia virus (MLV) systems have shown that the size of the direct repeat, distance between direct repeats, and secondary structures in the template influence the rate of template switching (18–21).
The polymerase and RNase H activities of MLV RT have previously been shown to be genetically distinct (24). Most polymerase mutations retain normal levels of RNase H activity, and several RNase H mutations retain their polymerase activity (24–27). Two different modes of RNase H activity have been identified from in vitro studies and have been implicated in the degradation of viral RNA during replication (28, 29). The polymerase-dependent RNase H activity occurs on the same RT that is polymerizing DNA (28–35). The DNA polymerization rate is 7–10 times faster than the polymerase-dependent RNase H rate (36), and polymerase-independent RNase H activity degrades RNA into fragments that remain associated with the nascent DNA (28, 33, 37–42).
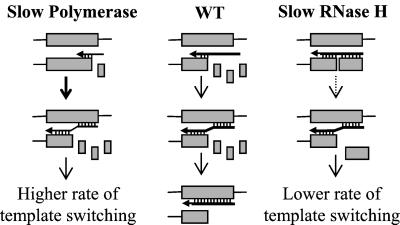
We recently proposed a dynamic copy-choice model for RT template switching in vivo (Fig. 1) (18). The model proposes that base pairing between newly synthesized DNA sequences behind the RT with complementary sequences of the template increases the probability of RT template switching. A proposed steady state between the rate of DNA polymerization and the rate of RNA degradation behind the RT determines the amount of nascent DNA that is able to base pair with the acceptor template. We previously observed that mutations in the RNase H domain reduce the frequency of template switching (18). The reduction in template switching may have been caused by a reduction in the rate of RNA degradation because of the mutations, resulting in the impairment of base pairing between the nascent DNA and the acceptor template. These results clearly showed that RNase H plays a vital role in template switching in vivo. However, it is not known whether the polymerase-dependent and/or -independent RNase H activities are involved in template switching. In this study, we have used an in vivo trans-complementation assay to show that a steady state exists between the rate of DNA polymerization and polymerase-dependent RNase H activity, which determines the frequency of RT template switching. We also provide the first evidence, to our knowledge, for polymerase-dependent RNase H activity in vivo.
Figure 1.
Dynamic copy-choice model for RT template switching. Shaded boxes represent direct repeats in an RNA template. Horizontal arrows represent nascent DNA. The thickness of these arrows indicates relative polymerization rate: the thicker the arrow, the faster the polymerization rate. Small boxes represent degraded RNA by the RNase H domain. In the case of slow RNase H activity, the degraded RNA fragments are shown as larger boxes. Hydrogen bonds between the RNA template and nascent DNA are designated by vertical marks. Vertical arrows of various thicknesses indicate the relative efficiency of template switching.
Materials and Methods
Plasmids.
pRMBNB contains the MLV gag and pol genes (43). pSV-hygro contains the hygromycin phosphotransferase B gene (hygro) expressed from the simian virus (SV) 40 promoter (44). pSV-A-MLV-env contains the amphotropic MLV envelope gene and was obtained from the AIDS Research and Reference Reagents Program (45). pES-GFFP, an MLV-based retroviral vector, contains two directly repeated fragments of green fluorescent protein (GFP) (GIBCO/BRL) (18).
Construction of RT Mutants.
The dNTP-binding site, YXDD motif, and RNase H mutants of MLV RT were derived from pRMBNB by using standard cloning procedures (46) and were previously described (43). Construction of double mutants is available on request.
Cells, Transfections, and Infections.
Transfection, infection, and drug selection of D17 and D17-based cells (dog osteosarcoma cell lines) were performed as previously described (18). A3–4 is a D17-based cell line that contains pES-GFFP and amphotropic MLV envelope expressed from pSV-A-MLV-env. The cell line was constructed as previously described (18).
Determining RT Template Switching in Vivo.
The protocol for determining RT template switching in vivo and treatment with hydroxyurea (HU) was described previously (18). At least three independent experiments were performed, and a minimum of 1,000 colonies were analyzed for each mutant.
GFP Detection by Flow Cytometry.
Cells expressing GFP were quantified by flow cytometry (FACScan, Becton Dickinson), and the results were analyzed by using cellquest software (Becton Dickinson). Briefly, drug-resistant cell colonies were pooled and plated on 100-mm tissue culture dishes; after 24 h, the cells were harvested and resuspended in 1 ml of PBS supplemented with 1% bovine calf serum before flow cytometry.
Results
Trans-Complementation of Polymerase and RNase H Mutants.
An in vivo trans-complementation assay was developed to determine the role of polymerase-dependent and -independent RNase H activity in RT template switching. An MLV-based retroviral vector, pES-GFFP, containing overlapping fragments of GFP designated GF and FP, was constructed (Fig. 2A) (18). pES-GFFP contains an internal ribosomal entry site from encephalomyocarditis virus (47) that drives the expression of the neomycin-resistance gene (neo). When pES-GFFP undergoes reverse transcription, one copy of the directly repeated F portion (250 bp) is deleted at a high frequency, which results in reconstitution of the functional GFP gene.
Figure 2.
Structure of MLV-based direct repeat vector and trans-complementation assay for determining which RNase H mode is important for template switching. (A) The pES-GFFP vector contains MLV-derived cis-acting elements including LTRs. The GFFP and neo genes are transcribed from the LTR promoter, and neo is expressed by the internal ribosomal entry site. During reverse transcription, deletion of one of the “F” repeats (shaded) results in the reconstitution of functional GFP. (B) Experimental protocol. A D150E A3–4 cell line stably expressing the pol− RNase H+ mutant D150E, pES-GFFP, and pSV-A-MLV-env was constructed. Wild-type and mutant pRMBNB constructs were separately cotransfected (Tf) with pSV-hygro into the A3–4 cell line. Virus harvested from the A3–4 cell line was used to infect D17 target cells and placed under G418 selection. After selection, G418-resistant clones were pooled and analyzed by flow cytometry to determine the frequency of direct repeat deletion.
We constructed the D150E A3–4 cell line (Fig. 2B) by first infecting the ES-GFFP vector into a D17 cell line expressing the amphotropic MLV envelope (named A3–4) (18). The MLV mutant D150E, which expresses a polymerase-defective Gag-Pol, was transfected into the A3–4 cell line, and several cell clones were selected. The D150 residue is located in the dNTP-binding site and forms a part of the catalytic triad of the polymerase domain (48). This mutant was shown to display a >10,000-fold reduction in viral titer and possesses low RT activity in vitro (43). Clone C4 D150E A3–4 was used for all subsequent trans-complementation experiments.
For trans-complementation assays (Fig. 2B), the C4 D150E A3–4 cell line was cotransfected with pRMBNB, which expresses wild-type MLV Gag-Pol, or mutants derived from pRMBNB, and pSV-hygro, which confers resistance to hygromycin. After selection with hygromycin, virus was harvested and used to infect D17 target cells. After selection with G418, colonies were pooled and analyzed by flow cytometry to determine the frequency of direct repeat deletion.
The ratio of the D150E mutant Gag-Pol and the transfected wild-type or mutant Gag-Pol present in the virion produced from the C4 D150E A3–4 cell line is unknown. Because the D150E mutant is stably expressed in the cell line, and a pool of transfected cell clones express the other Gag-Pol mutants, we expect a similar level of expression of the D150E mutant in all cells, and a binomial distribution of expression of the other Gag-Pol mutants, depending on the number of copies present in each cell clone and the efficiency of expression. In most experiments, both Gag-Pol mutants must be present in the virion in sufficient quantities to complement each other and complete viral replication.
Transfection of wild-type MLV gag-pol into C4 D150E A3–4 cells resulted in the production of ES-GFFP virus. Infection of D17 target cells with the virus resulted in average viral titers of 4,500 colony-forming units (cfu)/ml. Flow cytometric analysis revealed that 12% of the infected cells expressed GFP, indicating the proportion of the viruses that underwent a template switch that reconstituted the GFP gene (Fig. 3). When the A3–4 cell line was transfected with the D150E polymerase mutant and the D524N RNase H mutant individually, no viral titers were observed. However, when the D524N RNase H mutant was transfected into C4 D150E A3–4 cells expressing the polymerase mutant, the viral titer was rescued to near wild-type levels, and the frequency of template switching was 9%. This result indicated that the polymerase and RNase H mutants can be complemented in trans as previously demonstrated (49) and that polymerase-dependent RNase H activity is not necessary for completion of reverse transcription.
Figure 3.
Trans-complementation and direct repeat deletion assay. The frequency of GFP reconstitution is shown on the left axis and represented by gray bars, whereas the titers are shown as a log scale on the right axis and represented by white bars. The error bars represent the SEM of at least two independent experiments. Squares represent the polymerase domain, and circles represent the RNase H domain of RT. White squares and circles represent wild type; black squares and circles represent replication-defective mutants. Mutants separated by a “/” represent trans-complementation; mutants with a “+” represent mutations contained within the same RT.
The observation that trans-complementation of the D150E polymerase mutant and the D524N RNase H mutant resulted in a template-switching rate that was 75% of the wild-type frequency suggested three possible interpretations. First, polymerase-independent RNase H activity might play a significant role in template switching in the absence of polymerase-dependent RNase H activity. Second, during trans-complementation, a large proportion of the template-switching events could have occurred during plus-strand DNA synthesis and did not involve RNase H activity. This interpretation is consistent with the previous observation that the rates of RT template switching during minus- and plus-strand DNA synthesis are similar (50). Third, the D150E polymerase mutant might have interfered with the processivity of the RT expressed from the D524N RNase H mutant, perhaps by binding to the template or the growing point. The D150E polymerase mutant's effect on the processivity of DNA synthesis could potentially increase RT template switching and compensate for any reduction in template switching by the D524N RNase H mutant.
To determine whether the D150E polymerase mutant interfered with reverse transcription, we constructed a D150E + D524N double mutant lacking both polymerase and RNase H activities, transfected it into the A3–4 cell line, and selected a stably expressing cell clone. As expected, the D150E + D524N A3–4 cell line did not produce a detectable titer (>10,000-fold lower than wild type). When the D150E + D524N double mutant was coexpressed with wild-type RT, a wild-type viral titer was observed, and the frequency of template switching was not significantly different from wild type (11% vs. 12%, P = 0.3; all statistical analyses were performed by using the two-sample t test). Thus, coexpression of the D150E + D524N double mutant did not increase template switching, suggesting that the D150E mutation does not interfere with the processivity of wild-type RT. However, because the D150E + D524N double mutant is not required for viral replication of the wild type, we cannot rule out other reasons for the lack of interference, such as failure to coassemble with wild-type Gag-Pol.
At first glance, the result that trans-complementation of D150E and D524N mutants exhibited 75% of the wild-type template switching frequency suggested that polymerase-independent RNase H activity can account for the majority of RT template switching, and the RT template switching can occur in the absence of polymerase-dependent RNase H activity. However, two additional lines of experimentation described below provide strong evidence that polymerase-dependent RNase H activity is primarily responsible for the majority of RT template switching.
Analysis of Polymerase and Polymerase + RNase H Double Mutants.
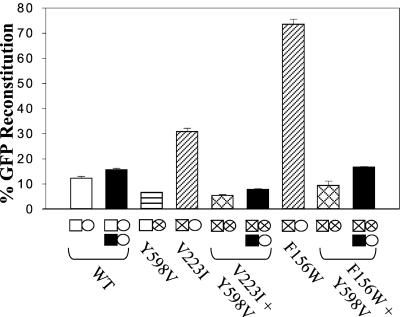
We previously observed that several polymerase mutants increased the rate of template switching, whereas RNase H mutants reduced template switching (18). To determine which activity, DNA polymerization or RNA degradation, plays a more significant role in RT template switching, we constructed RTs containing mutations in both the polymerase and RNase H domains. Two polymerase mutants, V223I and F156W, were selected for construction of the double mutants (viral titers 200 cfu/ml and 2,700 cfu/ml, respectively). In agreement with previous results (18), the template-switching frequencies of V223I and F156W were ≈2.5- (31%) and 6-fold (73%) higher than the wild-type RT, respectively (Fig. 4, hatch bars). The RNase H mutant Y598V (viral titer 1,000 cfu/ml) was also selected for the construction of the double mutants. The template-switching frequency of Y598V was 6.4%, approximately half that of the wild-type RT (Fig. 4, horizontal stripe bar).
Figure 4.
Trans-complementation of polymerase, RNase H, and polymerase + RNase H double mutants. Error bars represent the SEM of a least three independent experiments. Symbols are the same as in Fig. 3. Squares and circles containing an “X” represent mutants that are able to undergo viral replication.
Two double mutants, V223I + Y598V and F156W + Y598V, were constructed (viral titers 10 and 40 cfu/ml, respectively). The frequencies of template switching were determined to be 5 and 9%, respectively (Fig. 4, cross bars). Thus, the Y598V mutation in the RNase H domain drastically reduced the frequency of template switching by the V223I mutant almost 6-fold and the F156W mutant nearly 8-fold. The results indicated that the RNase H mutation is dominant over the polymerase mutation and that RNase H activity is essential for the increased frequencies of RT template switching observed with the V223I and F156W mutants.
To determine whether the high frequency of template switching achieved by the V223I and F156W mutants required polymerase-dependent or -independent RNase H activity, the V223I + Y598V and F156W + Y598V double mutants were trans-complemented with the D150E polymerase mutant (viral titers 2,400 and 360 cfu/ml, respectively). If the Y598V mutation eliminates the polymerase-independent RNase H activity, then providing the polymerase-independent RNase H activity in trans should rescue this defect, and the double mutants should exhibit a high frequency of template switching, similar to the levels observed with the V223I and F156W polymerase mutants. However, if polymerase-dependent RNase H activity was primarily responsible for the high frequency of template switching achieved by V223I and F156W, then providing polymerase-independent RNase H activity in trans should not significantly increase the template-switching frequency of the double mutants.
Both V223I + Y598V and F156W + Y598V double mutants were analyzed, and the rate of RT template switching after trans-complementation was determined to be 8 and 17%, respectively (Fig. 4, black bars). These template-switching frequencies were 1.5- and 1.8-fold higher than the frequencies of the double mutants without trans-complementation; therefore, providing polymerase-independent RNase H activity in trans resulted in modest stimulation of the template-switching frequency (P < 0.05). However, the template-switching frequencies after trans-complementation were still 3.9- and 4.4-fold lower than the frequencies of the V223I and F156W polymerase mutants, respectively. Therefore, providing polymerase-independent RNase H activity in trans was not able to rescue template switching to the levels observed with the polymerase mutants alone, which provides strong evidence that polymerase-dependent RNase H activity is crucial for the majority of RT template switching by the polymerase mutants.
Effect of HU on Template Switching.
To further determine the role of polymerase-dependent RNase H activity in template switching, and to test the idea that a steady state exists between the rate of DNA polymerization and polymerase-dependent RNase H activity, we performed in vivo trans-complementation assays in the presence of HU (Fig. 5). HU has been shown to deplete all four cellular nucleotide pools (51), significantly reduce the rate of polymerization by RT during viral replication (19), and increase the frequency of template switching of wild-type RT and polymerase mutants, but not RNase H mutants (18, 19).
Figure 5.
Effect of HU on the frequency of direct repeat deletion during trans-complementation. Gray bars represent experiments performed without HU, whereas black bars represent experiments performed with HU. Error bars represent the SEM of at least three independent experiments. Symbols are the same as in Fig. 3.
If there is a dynamic association between the rates of DNA polymerization and polymerase-dependent RNase H activity, and if this association determines the frequency of RT template switching, then infection of target cells in the presence of HU should increase the frequency of template switching for RTs that possess polymerase-dependent RNase H activity. However, in the absence of polymerase-dependent RNase H activity, as in the case of the D150E mutant complemented with the D524N mutant, HU treatment should not influence the frequency of RT template switching.
The wild-type virus titers were on average 4,500 cfu/ml, and HU treatment of the target cells resulted in ≈19- to 30-fold reduction in viral titers. HU treatment increased template switching of wild-type RT from 12 to 31% (P < 0.05) (Fig. 5). In addition, HU treatment increased template switching of virus containing wild-type and D150E polymerase mutant RTs from 15 to 34% (P < 0.05). However, when the D150E polymerase mutant was complemented in trans with the D524N RNase H mutant, HU treatment did not increase template switching (9 vs. 10%, P = 0.42). This result indicated that slowing down polymerization with HU treatment increased the template-switching frequency only when polymerase-dependent RNase H activity was present.
Analysis of the polymerase and RNase H double mutants (Fig. 4) and the experiments in which HU was used to slow down the rate of DNA polymerization (Fig. 5) provided strong evidence that there is a dynamic association between the rate of DNA polymerization and polymerase-dependent RNase H activity in vivo, which determines the frequency of RT template switching. However, we also observed that RT template switching could occur in the absence of polymerase-dependent RNase H activity at a rate that was 75% of wild type (Fig. 3). To further delineate the role of polymerase-independent RNase H activity, we performed the trans-complementation experiment with RNase H mutants.
Effect of Trans-Complementation of Wild-Type RT and RNase H Mutants on Template Switching.
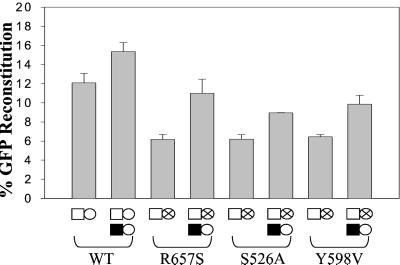
To determine whether the polymerase-independent RNase H activity contributed to RT template switching, we expressed the wild-type RT and the D150E polymerase mutant in the same virus producer cell line. The virus produced from these cells contained a mixture of the wild-type Gag-Pol and the D150E mutant, which could provide polymerase-independent RNase H activity in trans. These viruses contain both polymerase-dependent and -independent RNase H activities. However, all RTs in the virion have polymerase-independent activity, but only the RTs expressed from wild-type gag-pol have polymerase-dependent RNase H activity. Thus, in comparison to the wild type, the ratio of polymerase-independent and -dependent RNase H activities should be higher.
We observed that the RT template-switching frequency increased from 12 to 15% (P < 0.05) (Fig. 6), suggesting that the presence of additional polymerase-independent activity resulted in a slight (1.25-fold) increase in the frequency of RT template switching. Next, the RNase H mutants R657S, S526A, and Y598V were complemented in trans by the D150E mutant. These mutants were able to undergo viral replication and previously displayed a reduced frequency of RT template switching that was 50% of the wild-type frequency (18). Trans-complementation increased the RT template switching of R657S from 6 to 11% (P < 0.05), S526A from 6 to 9% (P < 0.05), and Y598V from 6 to 10% (P < 0.05), indicating that the additional polymerase-independent RNase H activity increased RT template switching by 1.4- to 1.8-fold. These results suggest that polymerase-independent RNase H activity could contribute to RT template switching. However, polymerase-independent RNase H activity was quantitatively less important than the dynamic association between DNA polymerase and polymerase-dependent RNase H activity, because the increased RT template-switching frequencies did not go above the wild-type RT level of 15% and were significantly less in comparison to the effects of polymerase mutants and HU treatment (2.3- to 6-fold).
Figure 6.
Trans-complementation of RNase H mutants. Wild-type and RNase H mutants were trans-complemented with D150E to determine the effects of providing polymerase-independent RNase H in trans. Error bars represent the SEM of at least three independent experiments. Circles containing an “X” represent RNase H mutants that are able to undergo viral replication. Symbols are the same as in Fig. 3.
Discussion
Model for in Vivo RT Template Switching.
We propose a revised dynamic copy-choice model for retroviral recombination (Fig. 7). Wild-type RT copies the RNA template as polymerase-dependent RNase H activity cleaves the RNA/DNA hybrid (Fig. 7A). The polymerase-dependent RNase H cleavages leave fragments of viral RNA associated with nascent DNA. Polymerase-independent RNase H activity then cleaves the remaining fragments of RNA further, removing the RNA from the nascent DNA so that it is available for base-pairing with the acceptor RNA template. Interactions of the nascent DNA with the acceptor RNA lead to the template-switching frequency observed with the wild-type RT.
Figure 7.
Model for in vivo template switching. (A) Wild type, (B) wild type + HU, and (C) D150E trans-complemented with D524N in the presence and absence of HU. Shaded boxes represent direct repeats within the RNA template, with the top box signifying the acceptor template and the bottom box the donor template. Horizontal lines between the shaded boxes represent nascent DNA. Small shaded boxes represent RNA that has been degraded by the RNase H domain. Vertical lines represent hydrogen bonds between the nascent DNA and RNA. RT symbols are the same as in Fig. 3. RTs annealed to the bottom of the shaded box represent polymerase-independent RNase H activity.
In the presence of HU, the cellular dNTP pools are depleted, which slows down DNA polymerization; the reduction in the rate of DNA synthesis with HU treatment (or other mechanisms such as RNA secondary structure or mutations in the polymerase domain) leads to more efficient RNA cleavage by polymerase-dependent RNase H activity (Fig. 7B). The polymerase-independent RNase H activity further degrades the template RNA. It is possible that the additional cleavages by the polymerase-dependent RNase H activity provide more 5′ RNA ends for RT to bind, leading to more efficient RNA degradation by polymerase-independent RNase H activity, as indicated by previous studies (52). Therefore, increased polymerase-dependent RNase H activity directly leads to more efficient degradation of the RNA, increasing the probability of base pairing between nascent DNA and acceptor RNA and a higher frequency of RT template switching.
During trans-complementation of the D524N and D150E mutants, polymerase-dependent cleavages of the RNA template are drastically reduced or eliminated (Fig. 7C). Although polymerase-independent RNase H activity presumably degrades the RNA template eventually, it is kinetically a much slower event, relative to the rate of DNA polymerization. If RT copies past the directly repeated region before the nascent DNA base pairs with the acceptor RNA, a template switch cannot take place. The results indicate that reducing the rate of DNA polymerization with HU did not provide sufficient time for polymerase-independent RNase H activity to effectively degrade the template RNA and increase template switching.
The frequency of template switching after trans-complementation of the D524N and D150E mutants was 9%, which was somewhat higher than the approximate 6% frequencies observed for the nonlethal RNase H mutants (P < 0.05). We hypothesize that the nonlethal RNase H mutants have lower levels of polymerase-dependent as well as -independent RNase H activities. In contrast, during trans-complementation of the D524N RNase H mutant, the D150E polymerase mutant can provide wild-type levels of polymerase-independent RNase H activity. Thus, the presence of wild-type levels of polymerase-independent RNase H activity during trans-complementation can explain the slightly higher levels of template switching observed relative to the nonlethal RNase H mutants.
It was previously demonstrated in vitro that HIV-1 polymerase-independent RNase H activity proceeds in a stepwise manner in which primary cuts occur first (18 nt from the 5′ end of RNA), followed by additional cuts including internal cuts (41). It was shown that the primary cuts require a 5′ end of the RNA fragment that is recessed on a DNA template (39). In the absence of polymerase-dependent RNase H activity, only one 5′ end of RNA would be present, and the number of primary cuts would be limited. Thus, in the absence of polymerase-dependent RNase H activity, the rate-limiting step in the degradation of RNA would be internal cuts made by the polymerase-independent RNase H activity to generate new 5′ ends, which would then be used to initiate additional cuts and further degrade the viral RNA.
As described previously, the dynamic copy-choice model is consistent with the forced-copy-choice model (53) and several previous observations indicating that RT pause sites and RNA secondary structures promote template switching (18).
Dynamic Association Between DNA Polymerization and Polymerase-Dependent RNase H activities.
The results of these studies indicate that a steady state exists between the rate of DNA polymerization and polymerase-dependent RNase H activity in vivo, and this steady state is the primary determinant of the frequency of RT template switching. When polymerase-dependent RNase H activity was present, reducing the rate of DNA polymerization with polymerase mutations or with HU treatment significantly increased the frequency of template switching. Furthermore, in the absence of polymerase-dependent RNase H activity, slowing down the rate of DNA polymerization had little effect on the frequency of template switching. Previous in vitro studies have shown that the rate of DNA polymerization is 7–10 times faster than the rate of polymerase-dependent RNase H cleavages by HIV-1 RT (36). Our results suggest that slowing down the rate of DNA polymerization leads to more frequent polymerase-dependent cleavages by RNase H, perhaps by allowing the RNase H enzyme more time to carry out the cleavage reaction. Whether the additional cleavages that occur by slowing down DNA polymerization are evenly distributed along the template is unknown.
The results also show that polymerase-independent RNase H activity can contribute to template switching, because trans-complementation of nonlethal RNase H mutants as well as polymerase + RNase H double mutants resulted in slightly higher frequencies of template switching. In these experiments, the polymerase-independent RNase H activity was sufficient to rescue viral titers to wild-type levels, indicating that minus- and plus-strand transfers were carried out efficiently. Unlike the template-switching events during the elongation phase of reverse transcription, minus- and plus-strand transfer events occur when RT reaches the end of a template. Therefore, these transfer events do not depend on the rate of DNA polymerization and could be efficiently catalyzed by the kinetically slower polymerase-independent RNase H activity.
Studies analyzing other factors that may be involved in RT template switching, such as the role of nucleocapsid protein and RNA secondary structure, are currently in progress. Understanding the mechanisms of retroviral recombination through RT template switching could provide new targets for antiviral drug therapy. Our studies suggest that RNase H inhibitors may reduce the rate of recombination in retroviral populations and decrease viral variation, which could suppress the generation of multidrug-resistant viruses.
Acknowledgments
We especially thank Wei-Shau Hu and John Coffin for intellectual input and valuable discussion of this manuscript, and Anne Arthur for editorial expertise and revisions. This work was supported by the HIV Drug Resistance Program, National Cancer Institute.
Abbreviations
- RT
reverse transcriptase
- MLV
murine leukemia virus
- HU
hydroxyurea
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schinazi R F, Larder B A, Mellors J W. Int Antiviral News. 2000;8:65–91. [Google Scholar]
- 2.McKeating J A, Gow J, Goudsmit J, Pearl L H, Mulder C, Weiss R A. AIDS. 1989;3:777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Phillips R E, Rowland-Jones S, Nixon D F, Gotch F M, Edwards J P, Ogunlesi A O, Elvin J G, Rothbard J A, Bangham C R, Rizza C R, et al. Nature (London) 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 4.Reitz M S, Wilson C, Naugle C, Gallo R C, Robert-Guroff M. Cell. 1988;54:57–63. doi: 10.1016/0092-8674(88)90179-1. [DOI] [PubMed] [Google Scholar]
- 5.Pathak V K, Hu W S. Semin Virol. 1997;8:141–150. [Google Scholar]
- 6.Mansky L M. J Gen Virol. 1998;79:1337–1345. doi: 10.1099/0022-1317-79-6-1337. [DOI] [PubMed] [Google Scholar]
- 7.Anderson J A, Bowman E H, Hu W S. J Virol. 1998;72:1195–1202. doi: 10.1128/jvi.72.2.1195-1202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clavel F, Hoggan M D, Willey R L, Strebel K, Martin M A, Repaske R. J Virol. 1989;63:1455–1459. doi: 10.1128/jvi.63.3.1455-1459.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt P K. Virology. 1971;46:947–952. doi: 10.1016/0042-6822(71)90093-6. [DOI] [PubMed] [Google Scholar]
- 10.Hu W S, Temin H M. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temin H M. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathak V K, Temin H M. Proc Natl Acad Sci USA. 1990;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parthasarathi S, Varela-Echavarria A, Ron Y, Preston B D, Dougherty J P. J Virol. 1995;69:7991–8000. doi: 10.1128/jvi.69.12.7991-8000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu W S, Bowman E H, Delviks K A, Pathak V K. J Virol. 1997;71:6028–6036. doi: 10.1128/jvi.71.8.6028-6036.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz R A, Skalka A M. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 16.Temin H M. Trends Genet. 1991;7:71–74. doi: 10.1016/0168-9525(91)90272-R. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Temin H M. Science. 1993;259:234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]
- 18.Svarovskaia E S, Delviks K A, Hwang C K, Pathak V K. J Virol. 2000;74:7171–7178. doi: 10.1128/jvi.74.15.7171-7178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeiffer J K, Topping R S, Shin N H, Telesnitsky A. J Virol. 1999;73:8441–8447. doi: 10.1128/jvi.73.10.8441-8447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeiffer J K, Georgiadis M M, Telesnitsky A. J Virol. 2000;74:9629–9636. doi: 10.1128/jvi.74.20.9629-9636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delviks K A, Pathak V K. J Virol. 1999;73:7923–7932. doi: 10.1128/jvi.73.10.7923-7932.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Sapp C M. J Virol. 1999;73:5912–5917. doi: 10.1128/jvi.73.7.5912-5917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Boeke J D. Proc Natl Acad Sci USA. 1987;84:8553–8557. doi: 10.1073/pnas.84.23.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanese N, Goff S P. Proc Natl Acad Sci USA. 1988;85:1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotewicz M L, Sampson C M, D'Alessio J M, Gerard G F. Nucleic Acids Res. 1988;16:265–277. doi: 10.1093/nar/16.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin J G, Crouch R J, Post K, Hu S C, McKelvin D, Zweig M, Court D L, Gerwin B I. J Virol. 1988;62:4376–4380. doi: 10.1128/jvi.62.11.4376-4380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hizi A, Hughes S H, Shaharabany M. Virology. 1990;175:575–580. doi: 10.1016/0042-6822(90)90444-v. [DOI] [PubMed] [Google Scholar]
- 28.Furfine E S, Reardon J E. J Biol Chem. 1991;266:406–412. [PubMed] [Google Scholar]
- 29.Gopalakrishnan V, Peliska J A, Benkovic S J. Proc Natl Acad Sci USA. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeStefano J J, Buiser R G, Mallaber L M, Bambara R A, Fay P J. J Biol Chem. 1991;266:24295–24301. [PubMed] [Google Scholar]
- 31.Oyama F, Kikuchi R, Crouch R J, Uchida T. J Biol Chem. 1989;264:18808–18817. [PubMed] [Google Scholar]
- 32.Wohrl B M, Volkmann S, Moelling K. J Mol Biol. 1991;220:801–818. doi: 10.1016/0022-2836(91)90119-q. [DOI] [PubMed] [Google Scholar]
- 33.Peliska J A, Benkovic S J. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 34.Post K, Guo J, Kalman E, Uchida T, Crouch R J, Levin J G. Biochemistry. 1993;32:5508–5517. doi: 10.1021/bi00072a004. [DOI] [PubMed] [Google Scholar]
- 35.DeStefano J J, Buiser R G, Mallaber L M, Myers T W, Bambara R A, Fay P J. J Biol Chem. 1991;266:7423–7431. [PubMed] [Google Scholar]
- 36.Kati W M, Johnson K A, Jerva L F, Anderson K S. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 37.Kelleher C D, Champoux J J. J Biol Chem. 2000;275:13061–13070. doi: 10.1074/jbc.275.17.13061. [DOI] [PubMed] [Google Scholar]
- 38.Schultz S J, Zhang M, Kelleher C D, Champoux J J. J Biol Chem. 1999;274:34547–34555. doi: 10.1074/jbc.274.49.34547. [DOI] [PubMed] [Google Scholar]
- 39.Palaniappan C, Fuentes G M, Rodriguez-Rodriguez L, Fay P J, Bambara R A. J Biol Chem. 1996;271:2063–2070. [PubMed] [Google Scholar]
- 40.DeStefano J J, Bambara R A, Fay P J. Biochemistry. 1993;32:6908–6915. doi: 10.1021/bi00078a014. [DOI] [PubMed] [Google Scholar]
- 41.Wisniewski M, Balakrishnan M, Palaniappan C, Fay P J, Bambara R A. J Biol Chem. 2000;275:37664–37671. doi: 10.1074/jbc.M007381200. [DOI] [PubMed] [Google Scholar]
- 42.Wisniewski M, Balakrishnan M, Palaniappan C, Fay P J, Bambara R A. Proc Natl Acad Sci USA. 2000;97:11978–11983. doi: 10.1073/pnas.210392297. . (First Published October 17, 2000; 10.1073/pnas.210392297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halvas E K, Svarovskaia E S, Pathak V K. J Virol. 2000;74:10349–10358. doi: 10.1128/jvi.74.22.10349-10358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gritz L, Davies J. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 45.Landau N R, Page K A, Littman D R. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 47.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 49.Telesnitsky A, Goff S P. EMBO J. 1993;12:4433–4438. doi: 10.1002/j.1460-2075.1993.tb06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowman R R, Hu W S, Pathak V K. J Virol. 1998;72:5198–5206. doi: 10.1128/jvi.72.6.5198-5206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Julias J G, Pathak V K. J Virol. 1998;72:7941–7949. doi: 10.1128/jvi.72.10.7941-7949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuentes G M, Fay P J, Bambara R A. Nucleic Acids Res. 1996;24:1719–1726. doi: 10.1093/nar/24.9.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coffin J M. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]