Abstract
The NAM, ATAF1/2, and CUC2 (NAC) transcription factors form a large plant-specific gene family, which is involved in the regulation of tissue development in response to biotic and abiotic stress. To date, there have been no comprehensive studies investigating chromosomal location, gene structure, gene phylogeny, conserved motifs, or gene expression of NAC in pepper (Capsicum annuum L.). The recent release of the complete genome sequence of pepper allowed us to perform a genome-wide investigation of Capsicum annuum L. NAC (CaNAC) proteins. In the present study, a comprehensive analysis of the CaNAC gene family in pepper was performed, and a total of 104 CaNAC genes were identified. Genome mapping analysis revealed that CaNAC genes were enriched on four chromosomes (chromosomes 1, 2, 3, and 6). In addition, phylogenetic analysis of the NAC domains from pepper, potato, Arabidopsis, and rice showed that CaNAC genes could be clustered into three groups (I, II, and III). Group III, which contained 24 CaNAC genes, was exclusive to the Solanaceae plant family. Gene structure and protein motif analyses showed that these genes were relatively conserved within each subgroup. The number of introns in CaNAC genes varied from 0 to 8, with 83 (78.9%) of CaNAC genes containing two or less introns. Promoter analysis confirmed that CaNAC genes are involved in pepper growth, development, and biotic or abiotic stress responses. Further, the expression of 22 selected CaNAC genes in response to seven different biotic and abiotic stresses [salt, heat shock, drought, Phytophthora capsici, abscisic acid, salicylic acid (SA), and methyl jasmonate (MeJA)] was evaluated by quantitative RT-PCR to determine their stress-related expression patterns. Several putative stress-responsive CaNAC genes, including CaNAC72 and CaNAC27, which are orthologs of the known stress-responsive Arabidopsis gene ANAC055 and potato gene StNAC30, respectively, were highly regulated by treatment with different types of stress. Our results also showed that CaNAC36 plays an important role in the interaction network, interacting with 48 genes. Most of these genes are in the mitogen-activated protein kinase (MAPK) family. Taken together, our results provide a platform for further studies to identify the biological functions of CaNAC genes.
Keywords: pepper, transcription factor, NAC family, phylogenetic, gene expression, interaction network
1. Introduction
Transcriptional regulation of gene expression controls many important cellular processes in plants, such as environmental stress responses, signal transduction, and cellular morphogenesis. Transcription factors (TFs) regulate gene expression by binding to specific cis-acting promoter elements, resulting in the activation or repression of the transcriptional rates of target genes, thereby facilitating the evolution and adaption of complex developmental systems in plant responses to environmental stress. Many plant-specific transcription factors, such as NAC, WRKY, AP2/EREBP, and MYB proteins have been identified and shown to play an important role in defense responses to stress [1,2].
NAC proteins are encoded by one of the largest plant-specific transcription factor gene families and contain both highly conserved N-terminal DNA binding domains and variable C-terminal domains. The conserved N-terminal DNA binding domain consists of five subdomains, A through E. On the basis of the structure of the domains and subdomains, NACs can be classified into two main groups (I and II), which can be further divided into 14 and 4 subgroups according to their tree topology in Arabidopsis and rice [3].
Since NAC transcription factors were originally identified as petunia NAM [4] and Arabidopsis CUC2 [5], many NAC proteins have been identified from virtually all classes of plants. As an increasing number of whole plant genomes are sequenced, genome-wide identification and functional analysis of NAC transcription factors have been accomplished in species such as Arabidopsis thaliana [3], rice [6], soybean [7], apple [8], foxtail miller [9], potato [10], Chinese cabbage [11], cassava [12], maize [13], melon [14], and wheat [15].
Many studies of NAC identification and functional analysis have shown that NAC proteins play significant roles in signaling and regulation of gene expression in response to biotic and abiotic stresses. Studies have shown that in Arabidopsis, the NAC transcription factor ANAC096 cooperates with the bZIP-type TF abscisic acid responsive element (ABRE) binding factor and the ABRE binding protein (ABF/AREB) to help plants survive under dehydration and osmotic stress conditions [16]. The overexpression of ANAC055 significantly improved drought resistance in transgenic plants [17]. In addition, the transcription of SNAC3 is induced by drought, high temperature, salinity stress, and ABA treatment in rice. Overexpression of SNAC3 resulted in enhanced tolerance to high temperatures, drought, and oxidative stress, whereas suppression of SNAC3 by RNAi resulted in increased sensitivity to these stresses [18]. In apple, a total of 180 NAC genes were identified, and 17 of 29 selected MdNAC genes were differentially expressed in response to at least one abiotic stress (low temperature, drought, high salinity, or exogenous ABA) [8]. In cassava, the expression profiles of MeNAC genes under various conditions (osmotic, salt, cold, ABA, and H2O2) suggest that different MeNAC genes may participate in different signaling or stress responses, and most of the cassava NAC genes can be significantly induced by multiple stressors, such as ABA and H2O2 treatment [12]. In maize, three genes, ZmNAC18, ZmNAC51, and ZmNAC145 were upregulated in the drought-tolerant genotype and downregulated in the susceptible genotype. These three genes are closely linked to SNAC1 and OsNAC6 in rice, which have been reported as drought responsive, and act as transcriptional activators/regulators of genes that encode proteins involved in the regulation of stress responses and the production of osmolytes [13]. In potato, 48 StNAC genes were expressed under different abiotic stress treatments (salt, mannitol, and heat), and 44 StNAC genes were expressed under biotic stress conditions with P. infestans inoculum (Pi isolate US8:Pi02-007), acibenzolar-s-methyl (BTH) and DL-β-amino-n-butyric acid (BABA) [10]. Meanwhile, the NAC transcription factor SISRN1 positively regulates the defense response against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000, but negatively regulates the oxidative and drought stress response in tomato [19].
Compared to potato and tomato, the few NAC genes identified were shown to display tissue-specific, inducible expression patterns in Capsicum. The first NAC gene, i.e., CaNAC1, was cloned by reverse RNA gel blot analysis. It was shown that the expression of the CaNAC1 gene is rapidly and specifically induced during incompatible interactions between pepper and bacterial or viral pathogens. Additionally, CaNAC1 was strongly induced by exogenously applied SA and ethephon (ET), whereas MeJA only had a transient effect [20]. The full-length cDNA of CaNAC72, a gene encoded by NAC transcription factors, was isolated from the normalized cDNA library. It was shown that this gene shares 70% amino acid sequence identity with the AtNAC072 gene of Arabidopsis. The expression of CaNAC72 was induced by hormones (SA, MeJA, ET, and ABA), dehydration, high salinity, cold, heat, and Ralstonia solanacearum inoculation [21]. The transcription expression of CaNAC2 was strongly induced by abiotic stress treatments, such as cold, salt, and ABA, but was inhibited by osmotic stress and SA treatment. Viral silencing of CaNAC2 in pepper seedlings resulted in increased susceptibility to cold stress and delayed salt-induced leaf chlorophyll degradation [22]. Although 73 and 45 putative NAC sequences were identified from a wild Mexican pepper accession “Chiltepin” and a Chinese inbred derivative “Zunla-1”, respectively [23], detailed analyses including gene classification, chromosome distribution, gene duplication, gene phylogeny, gene structure, conserved motif composition, and gene expression under various abiotic and biotic stress conditions was lacking.
Pepper (Capsicum annuum L.) is the second most widely cultivated Solanaceous vegetable worldwide, after tomato. However, when in the reproductive stage, pepper is sensitive to biotic and abiotic stresses such as pathogens, drought, cold, and high temperatures. The NAC transcription factors are one of the largest families of transcriptional regulators in plants, and members of the NAC gene family are thought to play important roles in the regulation of transcriptional reprogramming associated with plant abiotic and biotic stress responses [24]. Thus, it is necessary to identify NAC transcription factors and determine the molecular mechanisms involved in the multiple regulatory biological processes in pepper. Drafts of the C. annuum L. genome sequence were reported recently [25,26]. In the present study, we searched two pepper genome sequences to identify the NAC genes (CaNAC). Detailed analyses were then conducted, including gene classification, chromosome distribution, gene structure, gene duplication, gene phylogeny, conserved motif composition, and cis-acting elements in the promoter. Further, we analyzed the expression of the identified 22 CaNAC genes under abiotic or biotic stress conditions (salt, heat shock, drought, Phytophthora capsici, ABA, SA, and MeJA), and predicted the protein–protein interaction network of CaNACs. These results will provide a platform for the future identification of the biological functions of NAC genes in pepper.
2. Results
2.1. Identification of NAC Family Members in the Capsicum Annuum Genome
A comprehensive analysis was performed to identify NAC genes in the pepper genome sequences downloaded from PGP (Available online: http://peppergenome.snu.ac.kr) and PGD (Available online: http://peppersequence.genomics.cn). A total of 104 non-redundant putative NAC genes were identified using two pepper genome databases and named CaNAC1 to CaNAC104. The NAC gene family in the pepper is comprised of >100 genes, like those reported for Arabidopsis, rice, potato, apple, and soybean. The annotation IDs of each CaNAC in two pepper genome databases (PGP and PGD) are given in Table 1. Ten genes (CaNAC8, CaNAC9, CaNAC42, CaNAC44, CaNAC47, CaNAC48, CaNAC58, CaNAC64, CaNAC68, CaNAC89) have only one annotation ID because of the specificity of their gene sequences. It is interesting to note that these ten genes only exist in the PGD database. The nucleotide and protein sequences of CaNAC gene family were also (File S1).
Table 1.
NAC transcript factor family in pepper.
| Gene | Annotation ID | Chr | Group | Introns | PI | AA | WT | At Orthologous Gene | e-Value |
|---|---|---|---|---|---|---|---|---|---|
| CaNAC1 | Capana00g000175/Ca10g20640 | chr 10 | II(26) | 1 | 4.21 | 569 | 64.52 | AT2G43000 | 0.35 |
| CaNAC2 | Capana00g001681/Ca10g20530 | chr 10 | II(26) | 0 | 8.47 | 100 | 11.80 | AT2G43000 | 0.097 |
| CaNAC3 | Capana00g001918/Ca04g14740 | chr 04 | I(8) | 1 | 4.56 | 586 | 65.85 | AT1G34190 | 1 × 10−13 |
| CaNAC4 | Capana00g003238/Ca07g03900 | chr 07 | - | 0 | 7.45 | 137 | 16.09 | - | - |
| CaNAC5 | Capana00g004670/Ca06g17870 | chr 06 | I(14) | 2 | 6.42 | 317 | 36.12 | AT3G15170 | 4 × 10−21 |
| CaNAC6 | Capana00g004843/Ca06g14960 | chr 06 | II(25) | 0 | 4.88 | 326 | 37.03 | AT5G04400 | 3.3 |
| CaNAC7 | Capana00g004888/Ca06g15350 | chr 06 | II(23) | 0 | 4.36 | 534 | 60.80 | - | - |
| CaNAC8 | Capana01g000033 | chr 01 | I(12) | 2 | 6.79 | 420 | 48.06 | AT1G32770 | 1 × 10−18 |
| CaNAC9 | Capana01g000034 | chr 01 | I(12) | 2 | 6.79 | 420 | 48.06 | AT1G32770 | 1 × 10−18 |
| CaNAC10 | Capana01g000515/Ca01g04740 | chr 01 | I(15) | 2 | 7.34 | 336 | 38.87 | AT1G62700 | 2 × 10−47 |
| CaNAC11 | Capana01g000650/Ca06g18770 | chr 01 | I(14) | 3 | 7.57 | 350 | 38.97 | AT5G39610 | 5 × 10−30 |
| CaNAC12 | Capana01g001228/Ca01g09650 | chr 01 | I(14) | 2 | 7.70 | 352 | 39.20 | AT5G18270 | 7 × 10−15 |
| CaNAC13 | Capana01g001521/Ca01g11960 | chr 01 | II(25) | 0 | 5.36 | 276 | 31.72 | AT1G60340 | 2.7 |
| CaNAC14 | Capana01g002000/Ca00g54690 | chr 01 | I(3) | 2 | 10.12 | 248 | 28.34 | AT5G13180 | 1 × 10−8 |
| CaNAC15 | Capana01g002406/Ca01g17160 | chr 01 | I(12) | 1 | 8.07 | 303 | 34.94 | AT4G10350 | 6 × 10−57 |
| CaNAC16 | Capana01g003266/Ca01g26660 | chr 01 | II(26) | 0 | 4.08 | 461 | 52.28 | AT2G43000 | 0.29 |
| CaNAC17 | Capana01g004120/Ca01g30550 | chr 01 | I(14) | 2 | 9.15 | 353 | 39.92 | AT2G24430 | 1 × 10−21 |
| CaNAC18 | Capana01g004489/Ca01g34750 | chr 01 | I(12) | 1 | 6.57 | 313 | 35.95 | AT1G32770 | 3 × 10−25 |
| CaNAC19 | Capana02g000057/Ca02g06450 | chr 02 | I(9) | 5 | 4.92 | 577 | 66.03 | AT3G03200 | 2 × 10−9 |
| CaNAC20 | Capana02g000302/Ca02g03060 | chr 02 | I(2) | 1 | 6.52 | 252 | 29.08 | AT1G62700 | 4 × 10−5 |
| CaNAC21 | Capana02g001393/Ca02g04820 | chr 02 | I(16) | 2 | 6.26 | 322 | 36.74 | AT1G34190 | 6 × 10−11 |
| CaNAC22 | Capana02g002153/Ca02g17910 | chr 02 | II(25) | 0 | 8.56 | 246 | 28.72 | AT5G39690 | 8.5 |
| CaNAC23 | Capana02g002277/Ca02g20290 | chr 02 | I(9) | 5 | 6.27 | 632 | 72.58 | AT3G03200 | 1 × 10−16 |
| CaNAC24 | Capana02g002611/Ca00g89790 | chr 02 | I(15) | 0 | 5.17 | 348 | 39.97 | AT2G18060 | 6 × 10−85 |
| CaNAC25 | Capana02g002612/Ca00g89790 | chr 02 | I(15) | 1 | 5.17 | 348 | 39.97 | AT2G18060 | 1 × 10−75 |
| CaNAC26 | Capana02g002682/Ca02g22730 | chr 02 | II(23) | 0 | 7.09 | 296 | 33.97 | - | - |
| CaNAC27 | Capana02g003352/Ca02g28070 | chr 02 | I(14) | 2 | 7.74 | 326 | 37.31 | AT5G07680 | 3 × 10−37 |
| CaNAC28 | Capana02g003374/Ca02g28290 | chr 02 | I(16) | 1 | 6.52 | 397 | 44.85 | AT5G39820 | 2 × 10−11 |
| CaNAC29 | Capana02g003557/Ca02g30090 | chr 02 | II(19) | 5 | 4.81 | 387 | 43.11 | AT5G18270 | 2 × 10−4 |
| CaNAC30 | Capana03g000802/Ca12g13470 | chr 03 | I(14) | 2 | 7.53 | 332 | 38.10 | AT3G29035 | 3 × 10−34 |
| CaNAC31 | Capana03g000991/Ca03g28100 | chr 03 | - | 0 | 4.38 | 236 | 26.39 | - | - |
| CaNAC32 | Capana03g001014/Ca03g27130 | chr 03 | II(23) | 0 | 4.94 | 245 | 27.96 | - | - |
| CaNAC33 | Capana03g001027/Ca03g27300 | chr 03 | II(23) | 0 | 4.67 | 284 | 32.05 | - | - |
| CaNAC34 | Capana03g001525/Ca03g23090 | chr 03 | I(9) | 1 | 4.77 | 248 | 28.75 | AT3G17730 | 9 × 10−34 |
| CaNAC35 | Capana03g001657/Ca03g22350 | chr 03 | I(3) | 2 | 6.64 | 237 | 27.06 | AT4G17980 | 4 × 10-5 |
| CaNAC36 | Capana03g001780/Ca03g21000 | chr 03 | I(10) | 5 | 4.65 | 632 | 69.87 | AT4G35580 | 0.002 |
| CaNAC37 | Capang03g002541/Ca03g15410 | chr 03 | I(15) | 1 | 9.14 | 170 | 19.75 | AT1G12260 | 5 × 10−19 |
| CaNAC38 | Capana03g002678/Ca02g15520 | chr 03 | II(20) | 2 | 8.46 | 290 | 33.07 | AT1G28470 | 8 × 10−7 |
| CaNAC39 | Capana03g003003/Ca03g14470 | chr 03 | I(12) | 2 | 7.24 | 326 | 37.00 | AT1G79580 | 3 × 10−19 |
| CaNAC40 | Capang03g003315/Ca00g46270 | chr 03 | I(13) | 2 | 6.24 | 290 | 33.35 | AT4G28530 | 3 × 10−6 |
| CaNAC41 | Capana04g000051/Ca04g23340 | chr 04 | - | 2 | 6.71 | 390 | 44.12 | AT5G18270 | 5 × 10−21 |
| CaNAC42 | Capana04g000414 | chr 04 | II(18) | 4 | 4.67 | 350 | 38.80 | - | - |
| CaNAC43 | Capana04g000417/Ca04g20070 | chr 04 | II(18) | 4 | 4.42 | 371 | 40.96 | - | - |
| CaNAC44 | Capana04g000587 | chr 04 | I(13) | 2 | 6.04 | 279 | 31.13 | - | - |
| CaNAC45 | Capana04g001537/Ca00g03050 | chr 04 | I(6) | 2 | 8.33 | 290 | 33.39 | AT1G01720 | 9 × 10−16 |
| CaNAC46 | Capana05g000569/Ca05g04410 | chr 05 | I(6) | 2 | 7.45 | 306 | 34.95 | AT5G08790 | 1 × 10−39 |
| CaNAC47 | Capana05g001291 | chr 05 | - | 0 | 4.26 | 140 | 16.32 | - | - |
| CaNAC48 | Capana05g001292 | chr 05 | - | 2 | 6.16 | 459 | 51.28 | - | - |
| CaNAC49 | Capana05g001365/Ca05g09500 | chr 05 | II(20) | 2 | 6.94 | 486 | 53.97 | AT4G29230 | 3 × 10−47 |
| CaNAC50 | Capana05g001593/Ca11g04580 | chr 05 | I(16) | 2 | 7.13 | 410 | 46.22 | AT1G26870 | 3 × 10−25 |
| CaNAC51 | Capana05g002476/Ca00g86410 | chr 05 | I(8) | 5 | 4.44 | 551 | 61.62 | AT3G10500 | 3 × 10−8 |
| CaNAC52 | Capana05g002477/Ca00g86400 | chr 05 | I(8) | 3 | 5.26 | 396 | 44.64 | AT5G07680 | 4 × 10-12 |
| CaNAC53 | Capana06g000136/Ca06g27940 | chr 06 | I(13) | 2 | 6.14 | 287 | 33.46 | AT4G28530 | 2 × 10−10 |
| CaNAC54 | Capana06g000341/Ca06g25420 | chr 06 | II(26) | 0 | 4.40 | 467 | 53.68 | AT1G34180 | 0.073 |
| CaNAC55 | Capana06g000625/Ca06g22780 | chr 06 | I(11) | 3 | 9.14 | 408 | 45.91 | AT3G44290 | 3 × 10−16 |
| CaNAC56 | Capana06g000752/Ca06g21590 | chr 06 | I(10) | 5 | 4.56 | 637 | 70.12 | AT3G10490 | 0.006 |
| CaNAC57 | Capana06g001075/Ca06g17290 | chr 06 | II(17) | 2 | 5.08 | 379 | 42.94 | AT3G15170 | 0.93 |
| CaNAC58 | Capana06g001093 | chr 06 | II(17) | 1 | 4.54 | 133 | 15.37 | AT3G15170 | 0.08 |
| CaNAC59 | Capana06g001387/Ca06g14050 | chr 06 | I(15) | 2 | 6.24 | 121 | 37.60 | AT4G36160 | 1 × 10−18 |
| CaNAC60 | Capang06g001560/Ca06g13110 | chr 06 | I(9) | 3 | 5.45 | 342 | 38.48 | AT1G32510 | 2 × 10−10 |
| CaNAC61 | Capana06g001739/Ca06g11310 | chr 06 | I(6) | 2 | 6.72 | 294 | 33.67 | AT5G08790 | 3 × 10−12 |
| CaNAC62 | Capana06g002411/Ca06g06410 | chr 06 | I(15) | 2 | 6.63 | 347 | 40.20 | AT1G12260 | 8 × 10−29 |
| CaNAC63 | Capana06g002441/Ca00g77790 | chr 06 | I(9) | 3 | 5.24 | 334 | 37.91 | AT5G63790 | 1 × 10−5 |
| CaNAC64 | Capana06g002936 | chr 06 | - | 1 | 9.83 | 136 | 15.88 | AT3G10490 | 0.005 |
| CaNAC65 | Capana06g003012/Ca06g01680 | chr 06 | I(13) | 2 | 6.24 | 290 | 33.39 | AT4G28530 | 3 × 10−12 |
| CaNAC66 | Capana07g000058/Ca07g00500 | chr 07 | II(17) | 2 | 5.42 | 415 | 47.22 | AT3G15500 | 4 × 10−9 |
| CaNAC67 | Capana07g000215/Ca07g02120 | chr 07 | II(22) | 0 | 4.42 | 196 | 22.85 | AT1G34180 | 5 × 10−4 |
| CaNAC68 | Capana07g001015 | chr 07 | - | 1 | 10.03 | 238 | 26.98 | AT5G39820 | 1 × 10−8 |
| CaNAC69 | Capana07g001769/Ca00g84200 | chr 07 | II(20) | 3 | 7.65 | 325 | 36.58 | AT4G28500 | 1 × 10−21 |
| CaNAC70 | Capana07g002072/Ca07g16790 | chr 07 | II(25) | 0 | 9.70 | 235 | 27.48 | AT1G34190 | 1.5 |
| CaNAC71 | Capana07g002159/Ca07g17460 | chr 07 | I(14) | 1 | 8.85 | 323 | 36.23 | AT5G53950 | 5 × 10−70 |
| CaNAC72 | Capana07g002219/Ca07g18020 | chr 07 | I(5) | 2 | 7.44 | 338 | 37.67 | AT3G15500 | 5 × 10−27 |
| CaNAC73 | Capana07g002220/Ca07g02180 | chr 07 | I(7) | 8 | 8.70 | 340 | 38.00 | - | - |
| CaNAC74 | Capana07g002471/Ca07g21320 | chr 07 | I(16) | 2 | 7.54 | 323 | 37.20 | AT2G43000 | 3 × 10−9 |
| CaNAC75 | Capana07g002485/Ca07g21470 | chr 07 | I(13) | 2 | 8.61 | 136 | 15.53 | AT1G56010 | 3 × 10−18 |
| CaNAC76 | Capana08g000884/Ca08g06780 | chr 08 | II(24) | 0 | 7.30 | 232 | 26.86 | - | - |
| CaNAC77 | Capana08g000885/Ca08g06790 | chr 08 | II(24) | 0 | 6.79 | 178 | 20.64 | - | - |
| CaNAC78 | Capana08g001727/Ca08g13660 | chr 08 | I(2) | 1 | 7.16 | 408 | 46.32 | AT2G02450 | 3 × 10−75 |
| CaNAC79 | Capana08g001999/Ca08g15460 | chr 08 | II(21) | 2 | 4.48 | 195 | 22.63 | AT5G64530 | 5 × 10−10 |
| CaNAC80 | Capana09g000105/Ca09g18600 | chr 09 | II(21) | 1 | 4.92 | 314 | 36.51 | - | - |
| CaNAC81 | Capana09g000936/Ca09g12970 | chr 09 | I(5) | 2 | 9.45 | 350 | 39.62 | AT4G27410 | 9 × 10−44 |
| CaNAC82 | Capana09g002022/Ca03g21000 | chr 09 | - | 0 | 10.32 | 103 | 12.00 | AT4G35580 | 3 × 10−4 |
| CaNAC83 | Capana09g002444/Ca09g00160 | chr 09 | I(1) | 1 | 9.83 | 161 | 18.66 | AT5G39610 | 0.002 |
| CaNAC84 | Capana10g000063/Ca10g00460 | chr 10 | I(7) | 4 | 8.41 | 342 | 39.05 | AT1G61110 | 2 × 10−1 |
| CaNAC85 | Capana10g000683/Ca11g08290 | chr 10 | I(12) | 2 | 6.46 | 341 | 39.09 | AT2G46770 | 2 × 10−51 |
| CaNAC86 | Capana10g001138/Ca10g09910 | chr 10 | I(2) | 1 | 6.68 | 299 | 34.40 | AT3G12977 | 0.047 |
| CaNAC87 | Capana10g002356/Ca10g20640 | chr 10 | II(26) | 0 | 4.17 | 578 | 65.18 | AT2G43000 | 0.023 |
| CaNAC88 | Capana10g002363/Ca10g20640 | chr 10 | II(26) | 1 | 5.72 | 421 | 48.33 | AT2G43000 | 0.26 |
| CaNAC89 | Capana11g000328 | chr 11 | I(1) | 2 | 8.56 | 256 | 29.67 | AT3G44350 | 7 × 10−13 |
| CaNAC90 | Capana11g000346/Ca11g16490 | chr 11 | I(11) | 3 | 8.94 | 414 | 46.60 | AT3G44290 | 8 × 10−14 |
| CaNAC91 | Capana11g000795/Ca11g12260 | chr 11 | II(17) | 2 | 6.51 | 344 | 40.34 | AT1G34180 | 2× 10−5 |
| CaNAC92 | Capana11g001813/Ca11g04440 | chr 11 | I(4) | 2 | 7.57 | 294 | 33.85 | AT1G69490 | 2 × 10−26 |
| CaNAC93 | Capana11g001932/Ca10g13700 | chr 11 | II(24) | 0 | 10.07 | 167 | 19.90 | - | - |
| CaNAC94 | Capana11g001935/Ca10g13720 | chr 11 | II(24) | 0 | 6.68 | 218 | 25.63 | - | - |
| CaNAC95 | Capana11g002231/Ca11g01550 | chr 11 | II(19) | 3 | 5.12 | 481 | 54.73 | AT1G25580 | 1 × 10−15 |
| CaNAC96 | Capana12g002058/Ca12g07920 | chr 12 | - | 3 | 4.44 | 719 | 79.51 | AT1G33280 | 0.007 |
| CaNAC97 | Capana12g002348/Ca12g05890 | chr 12 | II(22) | 0 | 10.15 | 154 | 18.04 | - | - |
| CaNAC98 | Capana12g002352/Ca12g05910 | chr 12 | II(22) | 0 | 4.75 | 269 | 31.41 | AT5G62380 | 0.011 |
| CaNAC99 | Capana12g002357/Ca12g06210 | chr 12 | II(22) | 0 | 9.35 | 224 | 26.75 | AT1G34180 | 0.14 |
| CaNAC100 | Capana12g002360/Ca12g05910 | chr 12 | II(22) | 0 | 4.63 | 268 | 31.17 | AT5G62380 | 0.011 |
| CaNAC101 | Capana12g002456/Ca12g04970 | chr 12 | I(8) | 5 | 4.35 | 476 | 52.88 | AT3G10500 | 1 × 10−9 |
| CaNAC102 | Capana12g002457/Ca12g04950 | chr 12 | I(8) | 3 | 5.17 | 426 | 47.63 | AT1G34180 | 1 × 10−9 |
| CaNAC103 | Capana12g002917/Ca12g00080 | chr 12 | II(21) | 2 | 4.27 | 195 | 22.09 | - | - |
| CaNAC104 | Capana12g002918/Ca12g00080 | chr 12 | II(21) | 2 | 4.27 | 195 | 22.96 | - | - |
Chr: chromosome; WT: molecular weight (kDa); AA: amino acid; PI: isoelectric point.
Typically, the NAC proteins have an N-terminal NAC domain and a C-terminal activation domain. The NAC domain can be further divided into five subdomains, A to E [3]. All of the putative 104 NAC genes were analyzed to confirm the presence of the NAC domain, and 97 CaNAC genes, containing a complete N-terminal NAC domain, were identified; another seven genes did not have complete domains. Thereinto, CaNAC48 has only a D subdomain, CaNAC4 and CaNAC47 have D and E subdomains, CaNAC64 and CaNAC82 have C and D subdomains, and CaNAC31 and CaNAC68 have C, D, and E subdomains. Similar phenomena were observed in the potato, where 13 StNACs lack conserved A and/or B subdomains, and four StNACs do not contain conserved C and/or D subdomains [10].
The 104 identified CaNAC proteins varied greatly in length, from 100 amino acids (aa, AA) (CaNAC2) to 719 aa (CaNAC96); the average NAC protein length was 331.9 aa. The relative WT (molecular weight) varied from 11.80 kDa (CaNAC2) to 79.51 kDa (CaNAC96), and the PI (isoelectric point) ranged from 4.08 (CaNAC16) to 10.32 (CaNAC82). The detailed information about CaNAC proteins, including gene loci accession number in PGP or PGD, chromosome location, gene classification, introns, PI, AA, WT, and Arabidopsis thaliana orthologous genes or orthologs, are listed in Table 1.
2.2. Chromosomal Distribution and Duplication of CaNAC Genes
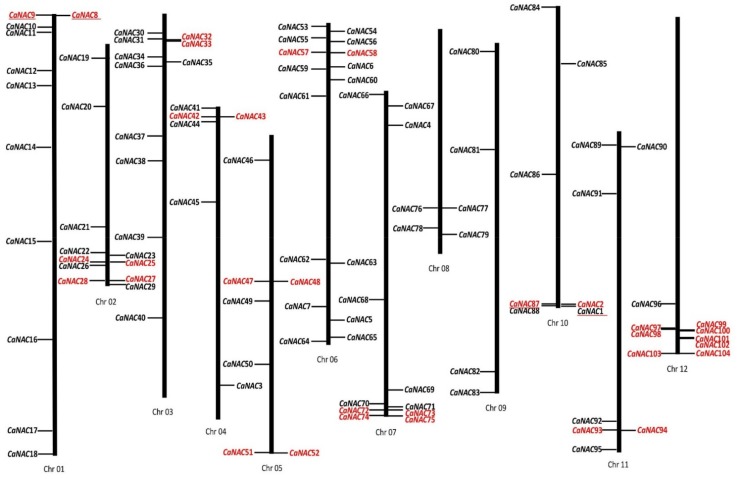
CaNACs were widely distributed within the Capsicum annuum reference genome, with all 104 CaNAC genes mapped to 12 chromosomes (Figure 1). Chromosomes 1, 2, 3, 6, and 7 contained relatively more CaNAC genes, with 11, 11, 11, 16, and 11 genes, respectively, compared to chromosomes 8 and 9 which each possessed only four of the genes. Interestingly, CaNAC genes were enriched on chromosomes 2 and 6. The sequenced size of chromosome 2 is 156.37 M, and it contains 11 (10.6%) of the 104 CaNAC genes; the sequenced size of chromosome 6 is 209.35 M, and it contains 16 (15.4%) CaNAC genes. Further, chromosomes 2 and 6 account for only 4.87 and 6.53% of the assembled pepper genome (3.13 G). Similar to pepper, chromosome 2 in potato also contains the largest number of StNAC genes, comprising 14 members (~13%), whereas chromosome 9 in potato contains only three members (~3%) [10]. These data suggest that in Solanaceae, NAC genes are enriched on chromosome 2, but not chromosome 9; however, the distribution of NAC genes in other Solanaceae family plants, such as tomato and eggplant, has yet to be determined.
Figure 1.
Mapping of the NAC gene family on Capsicum annuum L. chromosomes. The size of each chromosome is indicated by its relative length. Tandemly duplicated genes are indicated in red. Segmental duplicated genes are underlined.
Further, we determined the tandem and segmental duplications of CaNAC genes along the 12 pepper chromosomes. As shown in Figure 1, 16 CaNAC gene clusters (genes labeled in red) containing 32 tandemly duplicated genes were identified on 10 chromosomes, except for chromosomes 8 and 9. Interestingly, we identified eight tandemly duplicated genes within four gene clusters on chromosome 12. The sequences of the clustered members are highly alike, suggesting the use of gene duplication in the expansion of the CaNAC family. However, the number of segmental duplicated genes was lower compared to the number of tandem duplicated genes, containing only two pairs and four genes (CaNAC1 to CaNAC87 and CaNAC8 to CaNAC9) (Figure 1).
2.3. Classification of CaNACs
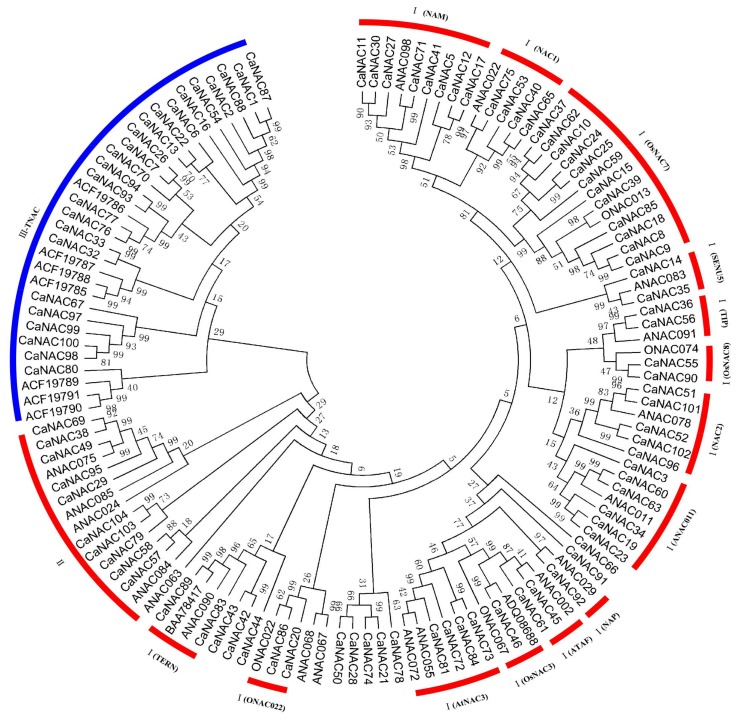
The most prominent structural feature of NAC proteins is the conserved NAC domain, which is divided into five subdomains at its N-terminal region. The phylogenetic relationship of the 97 CaNAC proteins and 31 known representative NAC proteins belonging to different subgroups from Arabidopsis, rice, and potato was examined by multiple sequence alignment of their NAC domain sequences. Because of short sequences or the lack of complete subdomains, the other seven NAC genes cannot be used for phylogenetic tree construction. Based on the ANAC, ONAC, and TNAC classification [10] and NAC domain alignments of CaNACs, 97 CaNACs were classified into three main groups (Groups I, II, and III) which contained 63, 10, and 24 pepper NAC members, respectively (Figure 2). Further, the CaNAC domains in each main group could be divided into several subgroups based on similarities in NAC domain structures.
Figure 2.
Unrooted phylogenetic tree representing the relationships among the NAC domains of pepper, potato, rice, and Arabidopsis. The amino acid sequences of all NAC domains were aligned with Clustal W (University College Dublin, Belfield, Dublin, Ireland), and the phylogenetics tree was constructed using the neighbor-joining method in MEGA 6.06 (Tokyo Metropolitan University, Hachioji, Tokyo, Japan). The red arcs indicate different groups or subgroups of NAC domains.
Fourteen subgroups in Group I, and four subgroups in Group II have been identified in Arabidopsis and rice [3]. In this study, 14 subgroups were also identified in the Group I NAC gene family in pepper. The number of CaNAC genes in each subgroup varied greatly, as shown in Figure 2. Subgroup I (OsNAC3) and I (NAP) possessed only one gene, respectively. Subgroup I (OsNAC7) and I (NAM) contained relatively more CaNAC genes, with 12 and 8 genes, respectively. Further, we identified four subgroups in the pepper Group II NAC gene family, which is similar to Arabidopsis and rice, where the Group II NAC gene family contains the four subgroups (ANAC001, ONAC003, ONAC001, and ANAC063). Interestingly, we found that Group III contains 24 CaNAC genes, but no Arabidopsis and rice NAC. The 24 CaNAC genes are included into the TNAC subfamily, which is thought to be exclusive to the Solanaceae family [10,27]. Similar phenomena were observed in tobacco and potato. Here, we confirmed the existence of a Solanaceae-specific NAC subfamily.
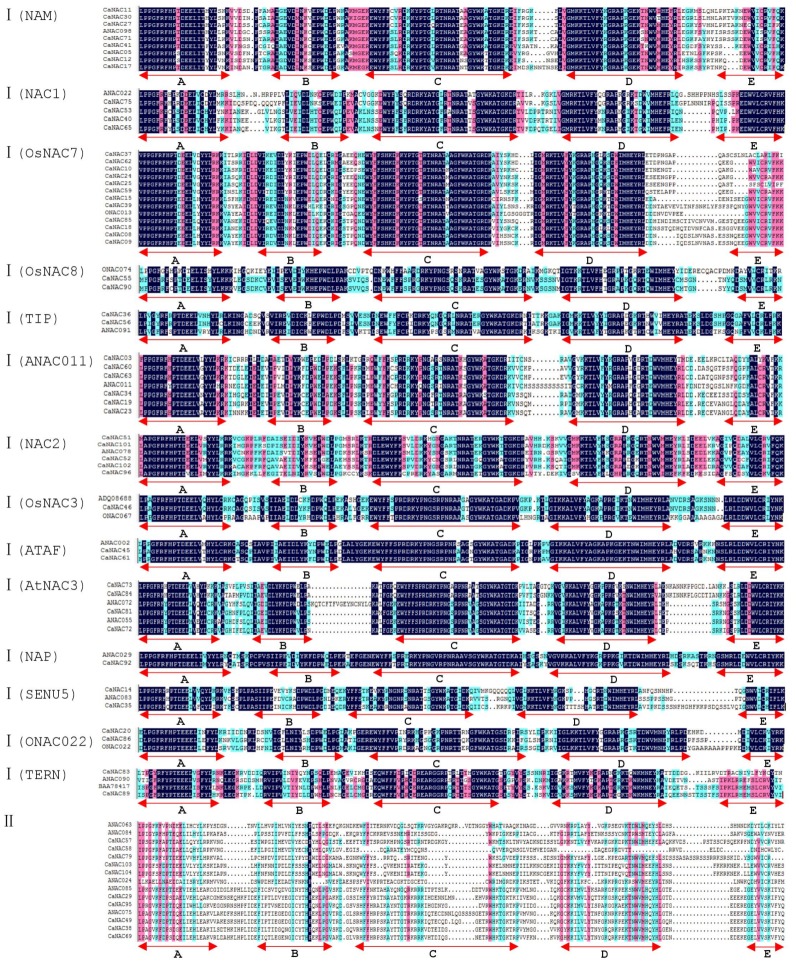
Further, we determined the characteristic features of each group and subgroup using the sequence alignments of the quantified sequences of NAC domains. As shown in Figure 3, subdomains A, C, and D were tightly conserved, whereas subdomains B and E were relatively divergent, except in subgroups 4, 5, and 6 of Group I. The sequences of subdomains B and E in subgroups 4, 5, and 6 were almost identical, akin to observations in Arabidopsis [3]. In this regard, pepper is similar to Arabidopsis and rice. Compared with Group I, the sequences of NAC in Group II from Arabidopsis or rice have low similarity to CaNAC in pepper, except for subgroup ANAC075 (Figure 3).
Figure 3.
Sequence alignment of two groups of CaNAC domains amino acid sequences. Subdomains A to E are shown by arrows above the sequences. Amino acids in the consensus sequences that are common to all groups are shown in black (=100%). Amino acids in the consensus sequences that are common to 20–25 subgroups (≥75%) are shown in blue, and those common to 13–19 (≥50%) are shown in green.
2.4. Gene Structure and Motif Composition Analysis of CaNAC Proteins
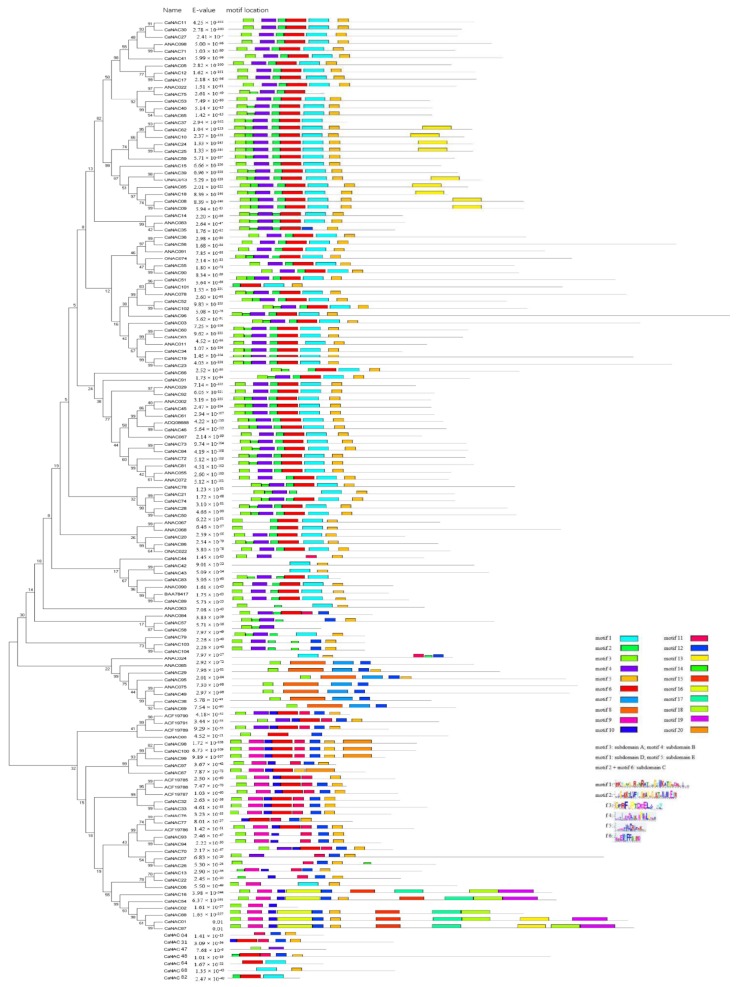
To better understand the similarity and diversity of CaNAC proteins, the intron/exon distribution of CaNAC was examined, and the conserved motifs of NAC family proteins in pepper, potato, Arabidopsis, and rice were also investigated using MEME version 4.12.0 online software (Available online: http://meme-suite.org/tools/meme). Gene structure analysis indicated that the number of introns of CaNAC ranged from 0 to 8 (Table 1). In particular, 26 and 39 CaNAC genes contained zero and two introns, respectively. Eighty-three (78.9%) CaNAC genes contained two or less introns, which is similar to cassava and banana, indicating low structural diversity of CaNAC genes. In addition, the CaNAC73 gene possessed eight introns. As shown in Figure 4 and File S2, a total of 20 distinct motifs were identified, and a schematic overview of the identified motifs is provided. Interestingly, most of the conserved motifs located in the N-terminal of NAC proteins are highly conserved for DNA-binding, and a similar motif composition is shared by the same subgroups, indicating functional similarities among members of the same subgroups.
Figure 4.
Alignment of multiple CaNAC and selected ONAC, ANAC domain amino acid sequences, schematic diagram of amino acid motifs of CaNAC and ONAC, ANAC protein groups or subgroups. Motif analysis was performed using Meme 4.12.0 online software (Available online: http://meme-suite.org/tools/meme) as described in the methods. The NAC proteins are listed on the left. The different-colored boxes represent different motifs and their position in each NAC sequence. The sequences of key motifs (motif 1, motif 2, motif 3, motif 4, motif 5, and motif 6) are shown on the bottom right of the figure. A detailed motif introduction for all CaNAC protein is shown in File S2.
CaNAC1, CaNAC54, and CaNAC87 contained relatively more motifs, (11, 11, and 10 motifs, respectively), compared to CaNAC42, CaNAC43, and CaNAC47, which contained only 2 motifs each. Among the 20 motifs in Group I, motif 3, motif 4, motif 1, and motif 5 comprised the subdomains A, B, D, and E, respectively, and motifs 2 and 6 together comprised subdomain C. The number and kind of motifs were almost identical in Group I. However, motif 13 was unique in subgroup I (OsNAC7). Compared to Group I, the number and kind of motifs were diverse in Group II and III. Motif 3 existed in all CaNAC genes, where motifs 2 and 4 were only represented in subgroup II (ANAC084). Further, motifs 1 and 6 were not found in Group II. Two unique motifs (motifs 7 and 8) were identified in subgroup II (ONAC003), and motifs 8, 7, and 12 comprised the subdomain B, C, and D, respectively. Motif 12 was identified in most CaNAC genes in Group II and III. In Group III, motifs 3, 5, 9, 10, and 11 were found in most CaNAC genes. Motif 9 comprised subdomain B, motifs 6 and 10 together comprised subdomain C, and motifs 11 and 12 together comprised subdomain D. Interestingly, we found that five motifs (15, 16, 17, 18, and 19) were always represented in five genes and that subdomain C in these five CaNAC genes was comprised of motif 16.
Seven NAC genes, CaNAC4, CaNAC31, CaNAC47, CaNAC48, CaNAC64, CaNAC68, and CaNAC82 cannot be divided into any subgroups because they cannot be used for phylogenetic tree construction as a consequence of their shortened sequences or lack of complete subdomains. However, as displayed schematically in Figure 4, CaNAC64, CaNAC68, and CaNAC82 can be divided into Group I because they contain motif 2. Overall, the number and kind of motifs in Group I, Group II, and Group III are quite different, indicating that the functions of these three main groups have been diversified.
2.5. Expression Profiling of CaNAC Genes under Biotic and Abiotic Stresses
We analyzed the expression patterns of 22 CaNACs belonging to the different subgroups under normal growth conditions and various biotic and abiotic stress conditions using real-time quantitative RT-PCR.
2.5.1. Expression of Patterns of CaNAC Genes under Salt, Heat, and Drought Treatment
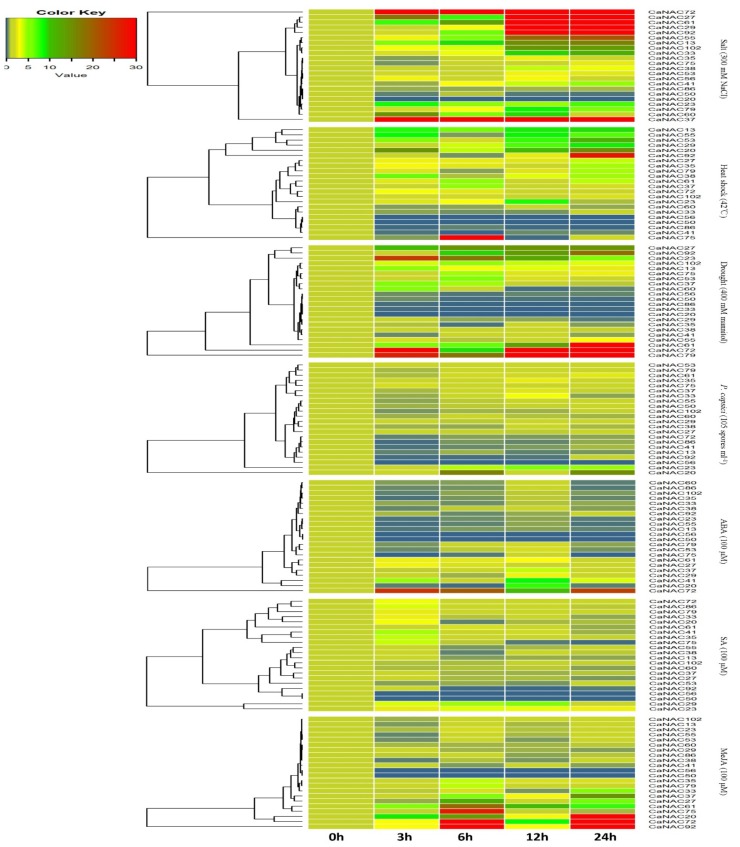
Under salt treatment, only the expression of one gene, CaNAC20, was not detectable in the leaves of the treatment groups. However, the expression of 10 genes was significantly upregulated in response to stress treatment, increasing on average by more than 10-fold after treatment (Figure 5, genes labeled in red or dark green). Of note, CaNAC72 was induced by more than 600-fold after 12 h of salt treatment. As shown in Figure 5, the expression of CaNAC50 and CaNAC86 were downregulated, while the expression of CaNAC50 significantly decreased by 0.2-fold after 24 h of treatment.
Figure 5.
Heat map showing CaNAC genes expression pattern in pepper under seven different types of stress. The leaves or roots of the seedlings (six true leaves) were used to test the changes of CaNAC gene expression levels using real-time PCR at different timepoints (0, 3, 6, 12 and 24 h) with salt, heat shock, abscisic acid (ABA), salicylic acid (SA), methyl jasmonate (MeJA), drought, and Phytophthora capsici treatment, respectively. Actin1 was used as an internal control. qRT-PCR data are shown relative to 0 h. The relative expression levels were calculated using the 2(−∆Ct) method. The heat map was created using R language.
Under heat shock treatment, as shown in Figure 5, CaNAC50 and CaNAC56 were not expressed in the leaves of the treated plants. The expression of four genes (CaNAC13, CaNAC20, CaNAC29, and CaNAC53) was significantly upregulated following stress treatment. The expression of CaNAC27, CaNAC35, CaNAC37, CaNAC61, CaNAC72, and CaNAC102 was also upregulated. Meanwhile, the expression value of seven genes (CaNAC13, CaNAC20, CaNAC27, CaNAC29, CaNAC35, CaNAC53, and CaNA102) peaked after 24 h of treatment. In addition, the expression of CaNAC41 and CaNAC86 was significantly decreased by 0.02- and 0.06-fold after 12 and 24 h of heat shock treatment, respectively.
As shown in Figure 5, the expression of CaNAC20 was not detectable in the roots after drought treatment. Six genes (CaNAC23, CaNAC27, CaNAC61, CaNAC72, CaNAC79, and CaNAC92) were expressed with relatively higher intensities and showed significant upregulation; CaNAC72 was induced by more than 77-fold after 3 h, and CaNAC79 was induced by more than 110-fold after 12 h of drought treatment. Conversely, the expression of six genes (CaNAC29, CaNAC33, CaNAC35, CaNAC50, CaNAC56, and CaNAC86) was downregulated, significantly decreasing by 0.65-, 0.04-, 0.17-, 0.13-, 0.2-, and 0.06-fold after 3 h of drought treatment, respectively.
2.5.2. Expression Patterns of CaNAC Genes under P. capsici Inoculation
As shown in Figure 5, CaNAC56 expression was not detectable in the roots of plants in the P. capsici inoculation treatment groups. The expression of five genes (CaNAC20, CaNAC23, CaNAC35, CaNAC53, and CaNA75) was upregulated, and six genes (CaNAC13, CaNAC41, CaNAC72, CaNAC86, and CaNA102) were downregulated. The expression value of seven genes (CaNAC23, CaNAC29, CaNAC33, CaNAC35, CaNAC37, CaNAC38, and CaNA53) peaked after 12 h of treatment. Among these genes, CaNAC23 was induced by more than 5.0-fold after 12 h of P. capsici inoculation treatment. Further, the expression value of nine genes (CaNAC13, CaNAC41, CaNAC50, CaNAC55, CaNAC60, CaNAC61, CaNAC72, CaNAC79, CaNAC86, and CaNA102) peaked after 3 h of treatment.
2.5.3. Expression Patterns of CaNAC Genes under Hormone Treatment
Under ABA treatment, the expression of five genes (CaNAC27, CaNAC37, CaNAC41, CaNAC61, and CaNAC72) was upregulated, and seven genes (CaNAC13, CaNAC23, CaNAC33, CaNAC35, CaNAC38, CaNAC55, and CaNAC102) were downregulated. The expression value of two genes (CaNAC27 and CaNAC72) and three genes (CaNAC37, CaNAC41, and CaNAC61) peaked after 6 h and 12 h of treatment, respectively. In addition, the expression of five other genes (CaNAC20, CaNAC53, CaNAC60, CaNAC75, and CaNAC86) was downregulated, except after 12 h of treatment. As shown in Figure 5, CaNAC50 and CaNAC56 were not expressed in the leaves in the treatment groups.
Under SA treatment, the expression of five genes (CaNAC23, CaNAC29, CaNAC35, CaNAC72, and CaNAC79) was upregulated and two genes (CaNAC60 and CaNAC102) were downregulated. The expression of six other genes (CaNAC13, CaNAC20, CaNAC27, CaNAC37, CaNAC75, and CaNAC92) were upregulated at first, and then downregulated. The expression value of twelve genes (CaNAC13, CaNAC20, CaNAC27, CaNAC33, CaNAC35, CaNAC37, CaNAC41, CaNAC72, CaNAC75, CaNAC79, CaNAC86, and CaNAC92) peaked after 3 h of treatment. In addition, CaNAC20, CaNAC35, CaNAC41, CaNAC72, CaNAC75, and CaNAC86 were induced by more than 3-fold after 3 h of SA treatment. As shown in Figure 5, CaNAC50 and CaNAC56 were also not expressed in the leaves in the treatment groups.
Under MeJA treatment, the expression of seven genes (CaNAC20, CaNAC35, CaNAC37, CaNAC61, CaNAC72, CaNAC79, and CaNAC92) was upregulated and two genes (CaNAC29 and CaNAC60) were downregulated. The expression value of four genes (CaNAC35, CaNAC61, CaNAC72, and CaNAC79) and three genes (CaNAC20, CaNAC37, and CaNAC92) peaked after 6 and 12 h of treatment, respectively. Following 12 h of MeJA treatment, we observed significant upregulation of CaNAC20 (65-fold) and CaNAC92 (180-fold) expression. In addition, the expression of five other genes (CaNAC13, CaNAC23, CaNAC33, CaNAC53, and CaNAC55) were down–up–down–up-regulated after treatment. As shown in Figure 5, CaNAC50 and CaNAC56 were also not expressed in the leaves after treatment.
2.6. Analysis of Stress-Related Cis-Elements in the CaNAC Promoters
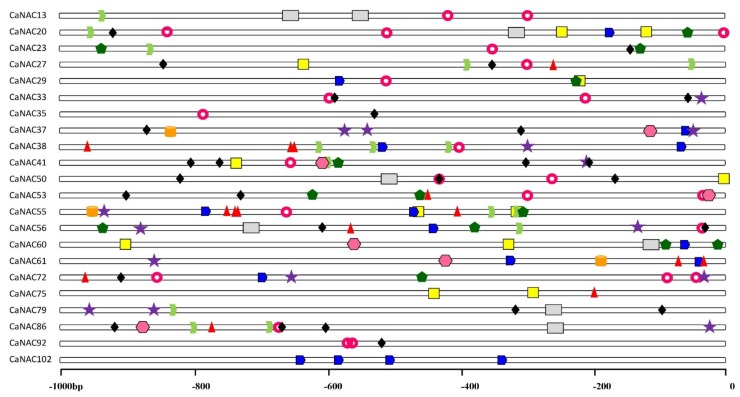
We scanned the cis-elements involved in the activation of defense-related genes in the promoter regions of CaNAC to better understand potential regulatory mechanisms of CaNAC genes in response to abiotic or biotic stress responses. The promoter regions (−1000 bp upstream of the translation start site) of all the CaNAC genes to which gene expression analysis was applied were examined. The predicted cis-elements in the promoter regions of twenty-two CaNAC genes are shown in Figure 6.
Figure 6.
Predicted cis-elements in the promoter regions of CaNAC genes. The promoter sequences (−1000 bp) of 22 CaNAC genes were analyzed. The names of the CaNAC genes are shown on the left side of the figure. The number at the bottom indicates the number of the nucleotides to the translation initiation codon, ATG. The elements are identified as follows: green dovetail for the CGTCA element, red triangle for the ABRE element, yellow square for the W-box element, purple five-pointed star for the TCA element, black diamond for the TC-rich repeats element, dark green pentagon for the MBS element, orange oval for the WUN element, blue pentagon for the GARE element, pink circular crown for the HSEs element, the grey rectangle for the ERE element, the rose red hexagon for the LTR element.
The ABRE element was found in nine selected promoter regions of CaNAC27, CaNAC38, CaNAC53, CaNAC55, CaNAC56, CaNAC61, CaNAC72, CaNAC75, and CaNAC86. In addition, three ABRE elements were identified in CaNAC38, and four were found in CaNAC55. The CGTCA element was found in 10 of the selected promoter regions of CaNAC13, CaNAC20, CaNAC23, CaNAC27, CaNAC38, CaNAC41, CaNAC55, CaNAC56, CaNAC79, and CaNAC86, and at least two CGTAC elements were found in the CaNAC27, CaNAC38, CaNAC55, and CaNAC86 genes. The TCA element was found in 10 selected promoter regions of CaNAC33, CaNAC37, CaNAC38, CaNAC41, CaNAC55, CaNAC56, CaNAC61, CaNAC72, CaNAC79, and CaNAC86. In addition, the GARE element was found in 10 selected regions of CaNAC20, CaNAC29, CaNAC37, CaNAC38, CaNAC55, CaNAC56, CaNAC60, CaNAC61, CaNAC72, and CaNAC102. It is well known that ABRE, CGTCA, TCA, and GARE cis-acting elements are involved in hormone responsiveness. We consistently found these four elements in CaNAC38, CaNAC55, and CaNAC56, thus we suspect that these three NAC genes may play a key role in regulating responsiveness to hormones.
As shown in Figure 6, HSE, TC-rich, MBS, and LTR elements were found in sixteen, fourteen, nine, and five selected promoter regions of CaNAC genes, respectively. The promoter region of CaNAC20 contains three HSE elements, and that of CaNAC41 contains four TC-rich elements. These results suggest that CaNAC20 and CaNAC41 may play a key role in regulating responsiveness to biotic stress. In addition, other stress-related cis-elements were detected, including a W-box element in eight genes (CaNAC20, CaNAC27, CaNAC29, CaNAC41, CaNAC50, CaNAC55, CaNAC60, and CaNAC75), a WUN element in three genes (CaNAC37, CaNAC55, and CaNAC61), and an ERE element in seven genes (CaNAC13, CaNAC20, CaNAC50, CaNAC56, CaNAC60, CaNAC79, and CaNAC86).
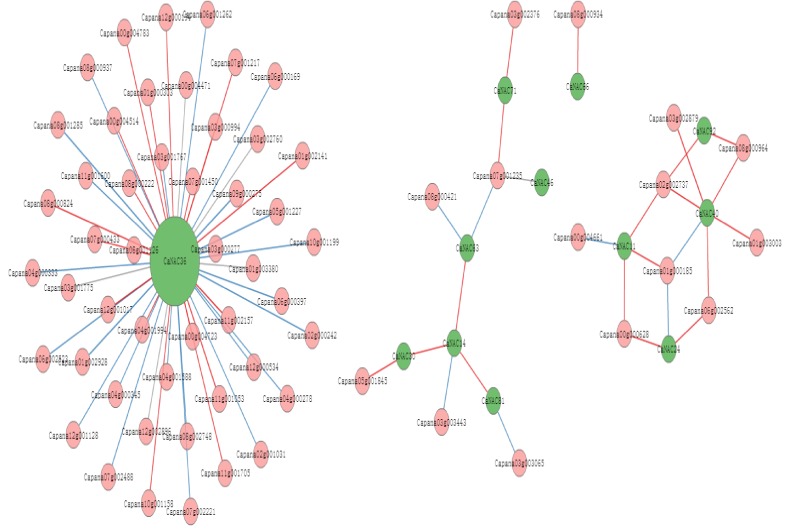
2.7. The Interaction Network of CaNAC Genes
An interaction network of CaNAC genes was constructed by making a comparison to orthologs of Arabidopsis, to better understand the relationships among the CaNAC gene family members. As shown in Table 1, 80.8% (84/104) of CaNAC genes were orthologous to 41.9% (49/117) of Arabidopsis NAC genes, indicating diversity in the sequences of NAC genes in Arabidopsis and pepper and that the sequences of CaNAC genes were tightly conserved. Positive, negative, and unknown correlations were marked by red, blue, and gray lines in Figure 7, which represent Pearson correlation coefficients of >0, <0, and no correlation, respectively. As shown in Figure 7, a complex functional relationship was observed in the interaction network of CaNAC genes; 35 pairs of interacting genes showed positive correlation, 30 showed negative correlation, and 6 were unknown. This suggests that CaNAC36 plays a key role in the interaction network, as it fully interacted with 48 genes.
Figure 7.
The interaction network of CaNAC genes in pepper according to the orthologs in Arabidopsis. The green circles represent the CaNAC genes, and the pink circles represent the pepper genes interacting with CaNAC genes. The red lines correspond to PCC > 0 (Pearson Correlation Coefficient), the blue lines mean PCC < 0, and the gray lines indicate that the PCC is unknown.
CaNAC36 interacted with the Ras-related protein RAB1A gene (Capana07g000433), OsRAN2, a gene orthologous to Capana07g000433 in rice, which enhances cold tolerance by promoting the export of intranuclear tubulin and maintaining cell division under cold stress [28]. CaNAC36 also interacted with the SNARE protein gene (Capana12g000194), GsVAMP72, a gene orthologous to Capana12g000194 in wild soybeans, which is involved in regulating plant salt tolerance and ABA sensitivity [29]. CaNAC14 directly interacted with three CaNAC genes (CaNAC81, CaNAC30, and CaNAC63), and CaNAC40 interacted with mitogen-activated protein kinase genes (Capana02g002737, Capana06g002562, and Capana08g000964), which play important roles in plant growth, development, and in response to a variety of biotic and abiotic stresses [30]. No other direct interactions among CaNAC genes were observed.
3. Discussion
3.1. NAC Gene Expansion and Evolution in Pepper
It has been demonstrated that NAC transcription factors play in important role in regulating stress tolerance against various abiotic and biotic challenges [9]. As a result of the availability of whole genome sequences in recent years, more and more NAC gene families have been identified from many different species of plants, such as Arabidopsis, rice, apple, and potato. However, very little is known about the NAC family in pepper. In a recently published report, a total of 45 putative NAC sequences were identified from the Pepper Genome Database (PGD, http://peppersequence. genomics.cn) [23]. In the present study, we further revealed an expanded NAC gene family in pepper, with a total of 104 members in the pepper genome. However, the NAC gene family in pepper is by far the smallest compared to those estimated for other plant species, which include 117 members in Arabidopsis [3], 151 in rice [6], and 110 in potato [10]. In particular, the genome size of pepper (3.13 Gb) is much larger than that of Arabidopsis (genome size 125 Mb), rice (480 Mb), and potato (840 Mb). By studying subgroups of NAC genes in Arabidopsis and rice, we found that the number of common subgroup CaNAC genes in pepper was much lower than expected as compared to Arabidopsis and rice, especially in subgroups 2, 8, 12, and 14. The number of the identified CaNAC genes in subgroups 2, 8, 12, and 14 were 3, 5, 6, and 8 in pepper, respectively. However, they are 6, 10, 13, and 13 in Arabidopsis, and 7, 8, 6, and 11 in rice, respectively. In our studies, we also identified two new subgroups, subgroup15 and subgroup16, which could not be further divided into subgroups in Group I. A similar phenomenon has been observed in other plants such as potato and is possibly related to the specificity of each species [10]. Further, we also found that 23 NAC genes belong to the Solanaceae specific subfamily (Figure 2). The number of NAC genes is lower in pepper as compared to other Solanaceae specific subfamilies such as tobacco (50 NAC genes) and potato (36 NAC genes). Our results provide further evidence that there is one NAC subfamily which is exclusive to the Solanaceae family.
It is well known that gene duplication events are important in the rapid expansion and evolution of gene families [25]. Tandem gene duplication of NAC transcription factors has been observed in many plant species, such as Arabidopsis, rice, Populus, potato, etc. In potato, one of the most important Solanaceae family plants, 24.5% of StNAC (27/110) were found to be tandemly duplicated [10]. In our investigation, 30.8% (32/104) of CaNAC genes were found to evolve from tandem gene duplication (Figure 1). Therefore, tandem gene duplication plays an important role in NAC gene family expansion in pepper. However, in comparing the number of NAC genes with the size of the genome, the reason for the low number of NAC genes might be due to whole-genome duplication (WGD) events in pepper [25,26].
3.2. CaNAC Proteins Play Important Roles in Various Biological Processes
Most of the NAC proteins that have been studied to date are involved in the response to abiotic and biotic stress and in the regulation of developmental processes. However, very few NAC transcription factors involved in the biotic stress response have been identified in pepper. Therefore, the goal of this study was to obtain more insight into the expression patterns and putative functions of CaNAC genes in response to biotic or abiotic stresses. The expression levels of 22 CaNAC genes under seven stress treatments (salt, heat shock, drought, Phytophthora capsici, ABA, SA, and MeJA) were calculated (Figure 5). Almost all 22 CaNAC genes were induced by these treatments, except CaNAC20 under salt and drought treatment, CaNAC50 and CaNAC56 under heat shock and hormone treatment, and CaNAC56 under Phytophthora capsici infection. These results indicate that the accumulation of CaNAC effectively reduces the damage from biotic or abiotic stress.
In the present study, the expression of the CaNAC72 gene was upregulated under stress treatment, except for Phytophthora capsici infection. Under salt and drought treatments (Figure 5), CaNAC72 gene showed relatively higher intensities, increasing more than 600- and 70-fold, respectively. We also found that CaNAC92 was significantly upregulated by salt (60-fold increase) and drought treatment (18-fold increase). As shown in Table 1, two Arabidopsis NAC genes, AT3G15500 (ANAC055) and AT1G69490 (ANAC029), were orthologous genes to CaNAC72 and CaNAC92, respectively. According to the phylogenetic tree analysis, CaNAC72 and CaNAC92 were attributed to subgroups 1(5) and 1(4), which shared high identity with subgroups AtNAC3 and AtNAP of Arabidopsis, respectively. In a previous study, Kim (2014) reported that the expression of AtNAP and ANAC055 increased in a leaf during aging, and that these two genes were senescence-associated NAC transcription factors, as candidate downstream components of ETHYLENE-INSENSITIVE2 (EIN2) [31]. We suspect that NAC genes from subgroups AtNAC3 and AtNAP may play important roles not only in response to abiotic stress, but also in plant development. However, this theory requires further investigation to be confirmed.
In tomato, the expression of SISRN1 was significantly induced by infection with B. cinerea or P. syringae pv. tomato DC3000, leading to a 6- to 8-fold higher expression than in the mock-inoculated plants [19]. SISRN1 has high identity with CaNAC23, and CaNAC23 also exhibited an increased expression pattern under P. capsici infection stress in this study. These results suggest that CaNAC23 can be induced by pathogens. Further, we also found that the expression of CaNAC23 was significantly induced by salt, drought, and heat shock treatment. Thus, CaNAC23 genes may play important roles in response to abiotic or biotic stress. Similarly, the expression of StNAC030 was significantly induced by salt, heat, and ABA [10] in potato, and CaNAC27, the gene orthologous to StNAC030, was also significantly induced by salt, heat, and ABA. CaNAC27 was also induced by drought and MeJA in the present study, suggesting that CaNAC27 genes may play important roles in response to abiotic or biotic stress. Thus, we suspect that orthologous NAC genes in the Solanaceae family may have similar gene functions; however, their functional characterization would be required to ascertain if they play similar roles in plant processes.
It was reported that two maize NAC transcription factors, namely, ZmNAC41 and ZmNAC100, are induced in leaves infected with Colletotrichum graminicola [32]. However, sequence information about these ZmNAC transcription factors is unavailable, therefore we could not identify orthologous genes in the CaNAC family. These results imply that these NAC members might be regulators of responses to various biotic stresses. To date, there are only three CaNAC genes with expression models that have been characterized after biotic or abiotic inoculation treatment, while the biological and cellular functions of most CaNAC genes remain largely unknown [20,21,22]. The current investigation identified several CaNAC genes that might be involved in stress defense and provides clues for the selection of candidate genes for further studies.
4. Materials and Methods
4.1. Sequence Database Searches
Arabidopsis NAC protein sequences were downloaded from TAIR (Available online: ftp://ftp.arabidopsis.org) [33]. Rice NAC protein sequences were obtained from rice full-length cDNA data (KOME) [34]. The pepper annotated genome sequences were obtained from the Pepper Genome Database (PGP, available online: http://peppergenome.snu.ac.kr and PGD, available online: http://peppersequence. genomics.cn) [25,26]. In a previous study, we obtained 44.6 Gb databases from RNA-Seq using resistant and sensitive gene pools against P. capsici in pepper and identified 47 NAC transcription factors after gene annotations [35]. In the current study, 47 pepper NAC proteins were used as query sequences for searching candidate sequences (e value was ≤e−10) against the two pepper genome databases using the BLAST tool (https://solgenomics.net/tools/blast/. The candidate sequences were confirmed for the presence of NAC domains using the Hmmsearch program (HMMER 3.1, available online: http://hmmer.janelia.org/). The putative NAC sequences, confirmed by Hmmsearch in the two pepper genome databases, were in turn used to search the pepper predicted proteins until no new sequences were found. The NAC sequences in two different pepper genome databases were then blasted using DNAMAN software (8.0, Lynnon Biosoft, San Ramon, CA, USA).
4.2. Multiple Sequence Alignment, Gene Chromosomal Location, and Phylogenetics Analysis
Approximately 160 amino acid sequences spanning the NAC core domain of all CaNAC proteins, 20 selected Arabidopsis NAC proteins and rice NAC proteins belonging to different group/subgroups [3], and 9 tobacco NAC proteins belonging to the Solanaceae specific NAC subfamily, were used to create multiple protein sequence alignments using Clustal X 2.1 (University College Dublin, Belfield, Dublin, Ireland) with default settings [36]. The gene chromosomal locations were obtained by the pepper gene annotation giff3 file, downloaded from the Pepper Genome Databases (Available online: http://peppersequence.genomics.cn). The NAC domain boundary was defined following a previously described method [3]. The neighbor-joining method was used to construct the phylogenetic tree, based on the amino acid sequence of NAC domains using MEGA 6.06 [37]. The parameters used in tree construction were the Jones-Thornton-Taylor (JTT) model plus gamma-distributed rates, as determined by ProTest 3.0 and 1000 bootstraps [38]. The tandemly and segmentally duplicated genes were defined as an array of two or more homologous genes within a 100-kb range distance, or low-copy repeats of DNA segments (blocks of sequence ≥1 kb in length and showing ≥90% sequence identity), respectively [39].
4.3. Motif Composition Analysis of CaNAC Proteins
The MEME 4.12.0 online program (Available online: http://meme-suite.org/) was used for the identification of motifs in the C. annuum NAC protein sequences. The optimized parameters of MEME were employed as follows: number of repetitions, any; maximum number of motifs, 20; optimum width of each motif, between 6 and 50 residues [40].
4.4. Treatments of Pepper Plants with Various Biotic and Abiotic Stresses
Accession PI201234, which is highly resistant to P. capsici, was used throughout this study [41]. The seedlings were grown in a growth chamber for 26 °C (16 h/day) and 20 °C (8 h/night) till reaching the age of six true leaves and were then treated with heat shock (42 °C), salt (300 Mm NaCl), drought (400 Mm mannitol), P. capsici (105 spores mL−1), salicylic acid (SA, 100 μM), methyl jasmonate (MeJA, 100 μM), and abscisic acid (ABA, 100 μM) for 24 h. The samples were prepared and collected using the method reported in our previous study [41]. The roots treated with drought or P. capsici spore suspension inoculation, along with the leaves treated with salt, heat shock, SA, MeJA, or ABA were also collected separately at 0, 3, 6, 12, and 24 h for RNA isolation [42].
4.5. Real-Time Quantitative RT-PCR
Total RNA extraction, cDNA synthesis and real-time quantitative PCR (qRT-PCR) analysis were carried out using the methods reported in our previous study [43]. A total of 22 CaNACs belonging to the different subgroups were randomly selected for gene expression analysis under abiotic or biotic stresses, and the data of three biological replicates were analyzed. Actin1 (NCBI accession number GQ339766) was used as the internal reference gene.
4.6. Search for Cis-Acting Elements in the Promoters of CaNAC Genes
Upstream regions (−1000 bp) of the CaNAC genes which were selected for gene expression analysis, were derived from Sol Genomics Network (Available online: https://solgenomics.net/) and were searched for regulatory elements, including CGTCA (cis-acting regulatory element involved in the MeJA-responsiveness), ABRE (cis-acting element involved in the abscisic acid responsiveness), W-box (binding site for the WRKY transcription factor in the defense response), TCA (cis-acting element involved in salicylic acid responsiveness), TC-rich repeats (cis-acting element involved in the defense and stress responsiveness), MBS (MYB binding site involved in drought induction), WUN (wound-responsive element), GARE (gibberellins-responsive element), HSE (cis-acting element involved in heat stress responsiveness), ERE (ethylene-responsive element), and LTR (low temperature-responsive element), in the promoters using the PlantCARE (Available online: http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE databases [44,45].
4.7. Prediction of CaNACs Protein–Protein Interaction Network
An interaction network of CaNAC proteins was constructed to understand the relationships among CaNAC gene family members and other genes. As described in detail previously [46], the interolog from Arabidopsis was used for predicting protein–protein interaction networks of CaNAC genes. An interaction network of Arabidopsis NAC proteins was constructed, the Arabidopsis NAC proteins were mapped to CaNAC proteins on the basis of their homologous relationships, and an interaction network of CaNAC proteins was drawn by Cytoscape.
5. Conclusions
A genome-wide analysis of the CaNAC gene family in pepper was performed to reveal gene location, gene structure, gene phylogeny, conserved motifs, stress-related cis-element, gene expression response to seven different abiotic or biotic stresses, and interaction network of CaNACs proteins. The vast majority of the 22 CaNAC genes showed a response to different stress types, displaying distinct expression patterns. In addition, one NAC subfamily that is exclusive to the Solanaceae family was identified.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (31601787), Natural Science Foundation of Jiangsu Province (BK20141380), and the Modern Agro-industry Technology Research System (No. CARS-25).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/4/1028/s1.
Author Contributions
Shubin Wang and John C. Snyder designed the project and did literature research; Weiping Diao performed the main part of data acquisition, statistical analysis, and manuscript editing; Jinbing Liu and Baogui Pan performed the main part of the experimental studies; Guangjun Guo, Wei Ge, and Mohammad Hasan Salman Ali Dawood participated in the research and analyzed the data. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Puranik S., Sahu P.P., Srivastava P.S., Prasad M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A., Roychoudhury A. WRKY Proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015;2015:807560. doi: 10.1155/2015/807560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hisako O., Kouji S., Koji D., Toshifumi N., Yasuhiro O., Kazuo M., Kenichi M., Naoki O., Jun K., Piero C., et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 4.Souer E., Houwelingen A., Kloos D., Mol J., Koes R. The No Apical Meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/S0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 5.Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohammed N., Ramaswamy M., Akhter M.S., Kouji S., Hiroaki K., Hisako O., Shoshi K. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Le D.T., Nishiyama R., Watanabe Y., Mochida K., Yamaguchi S.K., Shinozaki K., Tran L.S.P. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18:263–276. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su H.Y., Zhang S.Z., Yuan X.W., Chen C.T., Wang X.F., Hao Y.J. Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1, 2-CUC2 transcription factor family in apple. Plant Physiol. Biochem. 2013;71:11–21. doi: 10.1016/j.plaphy.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Puranik S., Sahu P.P., Mandal S.N., Venkata S.B., Parida S.K., Prasad M. Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.) PLoS ONE. 2013;8:e64594. doi: 10.1371/journal.pone.0064594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh A.K., Sharma V., Pal A.K., Acharya V., Ahuja P.S. Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.) DNA Res. 2013;20:403–423. doi: 10.1093/dnares/dst019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J., Wang F., Li M.Y., Jiang Q., Tan G.F., Xiong A.S. Genome wide analysis of the NAC transcription factor family in Chinese cabbage of elucidate responses to temperature stress. Sci. Hortic. 2014;165:82–90. doi: 10.1016/j.scienta.2013.11.005. [DOI] [Google Scholar]
- 12.Hu W., Wei Y.X., Xia Z.Q., Yan Y., Hou Z.W., Zou M.L., Lu C., Wang W.Q., Peng M. Genome-wide identification and expression analysis of the NAC transcription factor family in cassava. PLoS ONE. 2015;10:e0136993. doi: 10.1371/journal.pone.0136993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiriga K., Sharma R., Kumar K., Yadav S.K., Hossain F., Thirunavukkarasu N. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene. 2014;2:407–417. doi: 10.1016/j.mgene.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei S.W., Gao L.W., Zhang Y.D., Zhang F.R., Yang X., Huang D.F. Genome-wide investigation of the NAC transcription factor family in melon (Cucumis melon) and their expression analysis under salt stress. Plant Cell Rep. 2016;35:1827–1839. doi: 10.1007/s00299-016-1997-8. [DOI] [PubMed] [Google Scholar]
- 15.Borrill P., Harrington S.A., Uauy C. Genome-wide sequence and expression analysis of the NAC transcription factor family in polyploidy wheat. G3 Genes Genomes Genet. 2017;7:3019–3029. doi: 10.1534/g3.117.043679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Z.Y., Kim S.Y., Hyeon D.Y., Kim D.H., Dong T., Park Y., Jin J.B., Joo S., Kim S., Hong J.C., et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell. 2013;25:4708–4724. doi: 10.1105/tpc.113.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Y.J., Liao K.F., Du H., Xu Y., Song H.Z., Li X.H., Xiong L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2013;66:6803–6817. doi: 10.1093/jxb/erv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B., Ouyang Z.G., Zhang Y.F., Li X.H., Hong Y.B., Huang L., Liu S.X., Zhang H.J., Li D.Y., Song F.M. Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS ONE. 2014;9:e102067. doi: 10.1371/journal.pone.0102067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh S.K., Lee S., Yu S.H., Choi D. Expression of a novel NAC motif-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta. 2005;222:876–887. doi: 10.1007/s00425-005-0030-1. [DOI] [PubMed] [Google Scholar]
- 21.Lin J. Master’s Thesis. Fujian Agriculture and Forestry University; Fuzhou, China: 2012. The Cloning and Function Characterization of Transcription Factor CaNAC72 of Capsicum annuum. [Google Scholar]
- 22.Guo W.L., Wang S.B., Chen R.G., Chen B.H., Du X.H., Yin Y.X., Gong Z.H., Zhang Y.Y. Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 2015;6:755. doi: 10.3389/fpls.2015.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra S.S., Gadnayak A., Mandal S., Dehury B. Genome wide screening, identification and characterization of NAC transcription factors from Capsicum Species: An in silico approach. Int. J. Curr. Microbiol. Appl. Sci. 2016;5:527–562. doi: 10.20546/ijcmas.2016.506.059. [DOI] [Google Scholar]
- 24.Nuruzzaman M., Sharoni A.M., Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013;4:248. doi: 10.3389/fmicb.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S., Park M. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014;46:270–278. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- 26.Qin C., Yu C., Shen Y. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA. 2014;111:5135–5140. doi: 10.1073/pnas.1400975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rushton P.J., Bokowiec M.T., Han S.C. Tobacco transcription factors: Novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 2008;147:280–295. doi: 10.1104/pp.107.114041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen N., Xu Y.Y., Wang X., Du C., Du J.Z., Yuan M., Xu Z.H., Chong K. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ. 2011;34:52–64. doi: 10.1111/j.1365-3040.2010.02225.x. [DOI] [PubMed] [Google Scholar]
- 29.Sun X.L., Ji W., Ding X.D., Bai X., Cai H., Yang S.S., Qian X., Sun M.Z., Zhu Y.M. GsVAMP72, a novel Glycine soja R-SNARE protein, is involved in regulating plant salt tolerance and ABA sensitivity. Plant Cell Tissue Organ Cult. 2013;113:199–215. doi: 10.1007/s11240-012-0260-4. [DOI] [Google Scholar]
- 30.Chen L.H., Hu W., Tan S.L., Wang M., Ma Z.B., Zhou S.Y., Deng X.M., Zhang Y., Huang C., Yang G.X., et al. Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS ONE. 2012;7:e46744. doi: 10.1371/journal.pone.0046744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H.J., Hong S.H., Kim Y.W., Lee I.H., Jun J.H., Phee B., Rupak T. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE- INSENSITIVE2 mediated leaf senescence signaling in Arabidopsis. J. Exp. Bot. 2014;65:4023–4036. doi: 10.1093/jxb/eru112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voitsik A.M., Muench S., Deising H.B., Voll L.M. Two recently duplicated maize NAC transcription factor paralogs are induced in response to Colletotrichum graminicola infection. BMC Plant Biol. 2013;13:85. doi: 10.1186/1471-2229-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R. The Arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuchi S., Satoh K., Nagata T. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;310:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- 35.Diao W.P., Wang S.B., Liu J.B., Pan B.G., Guo G.J., Ge W., Gao C.Z. Identification and expression analysis of NAC transcription factor induced by Phytophthora blight in pepper. Mol. Plant Breed. 2018 in press. [Google Scholar]
- 36.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. ClustalW and ClustalX version2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darriba D., Taboada G.L., Doallo R., Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Q., Zhu Z.L., Kasahara M., Zhang Z. Segmental duplications in the silkworm genome. BMC Genomics. 2013;14:521. doi: 10.1186/1471-2164-14-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey T.L., Boden M., Buske F.A., Frith M. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barksdale T.H., Papavizas G.C., Johnston S.A. Resistance to foliar blight and crown rot of pepper caused by Phytophthora capsici. Plant Dis. 1984;68:506–509. doi: 10.1094/PD-68-506. [DOI] [Google Scholar]
- 42.Diao W.P., Wang S.B., Liu J.B., Pan B.G., Guo G.J., Ge W. Genome-wide identification and expression analysis of WRKY gene family in Capsicum annuum L. Front. Plant Sci. 2016;7:211. doi: 10.3389/fpls.2016.00211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Wang S.B., Liu K.W., Diao W.P., Zhi L., Ge W., Liu J.B., Pan B.G., Wan H.J., Chen J.F. Evaluation of appropriate reference genes for gene expression studies in pepper by quantitative real-time PCR. Mol. Breed. 2012;30:1393–1400. [Google Scholar]
- 44.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Peer Y.V., Rouze P., Rombauts S. Plant CARE, a data base of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo M., Liu J.H., Lu J.P., Zhai Y.F., Wang H., Gong Z.H., Wang S.B., Lu M.H. Genome-wide analysis of the CaHsp20 gene family in pepper: Comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 2015;6:806. doi: 10.3389/fpls.2015.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.