Abstract
Rheumatoid arthritis (RA) is the second common rheumatic immune disease with chronic, invasive inflammatory characteristics. Non-steroidal anti-inflammatory drugs (NSAIDs), slow-acting anti-rheumatic drugs (SAARDs), or glucocorticoid drugs can improve RA patients’ symptoms, but fail to cure. Broton’s tyrosine kinase (BTK) inhibitors have been proven to be an efficacious target against autoimmune indications and B-cell malignancies. Among the current 11 clinical drugs, only BMS-986142, classified as a carbazole derivative, is used for treating RA. To design novel and highly potent carbazole inhibitors, molecular docking and three dimensional quantitative structure–activity relationship (3D-QSAR) were applied to explore a dataset of 132 new carbazole carboxamide derivatives. The established comparative molecular field analysis (CoMFA) (q2 = 0.761, r2 = 0.933) and comparative molecular similarity indices analysis (CoMSIA) (q2 = 0.891, r2 = 0.988) models obtained high predictive and satisfactory values. CoMFA/CoMSIA contour maps demonstrated that bulky substitutions and hydrogen-bond donors were preferred at R1 and 1-position, respectively, and introducing hydrophilic substitutions at R1 and R4 was important for improving BTK inhibitory activities. These results will contribute to the design of novel and highly potent BTK inhibitors.
Keywords: rheumatoid arthritis (RA), Broton’s tyrosine kinase (BTK), carbazole carboxamide derivatives, 3D-QSAR, comparative molecular field analysis (CoMFA), comparative molecular similarity indices analysis (CoMSIA)
1. Introduction
Rheumatoid arthritis (RA) is an autoimmune destructive disease by affecting the joints, causing progressive, symmetric, erosive destruction of cartilage and bone [1]. RA has affected about 24.5 million people as of 2015, and the condition newly develops in approximately 1% of the population each year [2]. Two main classes of traditional medications were used for treatment of RA: first-line drugs (involved non-steroidal anti-inflammatory drugs (NSAIDs) and corticosteroids) and second-line drugs (also referred to as disease-modifying anti rheumatic drugs or disease-modifying anti rheumatic drugs (DMARDs)) [3]. However, these two classes of medicine possess some serious side effects, such as increased susceptibility to bruising, abdominal pain, and even risk of infections and bleeding [4]. Therefore, it is increasingly crucial to develop novel drugs with improved efficacy and safety in RA treatment.
Broton’s tyrosine kinase (BTK) is a member of the Tyrosine-protein kinase (TEC) kinase family and plays a critical role in the B-cell development and activation through mediating the downstream signaling cascade of B-cell receptors (BCRs) [5,6]. The increase in BTK expression can cause the chronic activation of the BCR signaling pathway, which affects B-cell proliferation and differentiation [7]. As a result, it can cause a lack of antibodies in the body, which finally gives rise to RA and other inflammatory diseases [8]. Therefore, inhibiting BTK activities to keep the normal function of the BCR signaling pathway is an effective way to treat RA. Recently, BTK inhibitors have been of increased interest in the clinical study of B-cell tumors and immune disease. Ibrutinib [8,9], acalabrutinib [10], ONO-4059 [11], spebtutinib [12], HM71224 [13], and BMS-986142 [14] have advanced into clinical trials, and their reported chemical structures are shown in Figure 1. As candidate drugs for treating RA, only BMS-986142 has advanced into Clinical Phase I with improved oral exposure, kinase selectivity, and high BTK potency [15]. Compared with NSAIDs and DMARDs, BMS-986142 has advantages of increased safety and efficacy as well as the less dependence on medication [16]. Therefore, exploring novel and highly potent BTK inhibitors for RA treatment is an important and promising prospect.
Figure 1.
The chemical structures of several Broton’s tyrosine kinase (BTK) inhibitors that have entered into clinical trials.
Here we report on molecular modeling studies performed by comparative molecular field analysis (CoMFA) [17] and comparative molecular similarity indices analysis (CoMSIA) [18] modules, as well as docking results, to investigate the three-dimensional quantitative structure–activity relationship (3D-QSAR) between carbazole inhibitors (BMS-986142 analogues) and BTK.
2. Results and Discussion
2.1. Molecular Docking
The aim of the molecular docking was to predict the binding affinity and interactions of carbazoles known to modulate the activity of BTK. The accuracy of the docking program was confirmed by comparing the predicted compound (76, green) and ligand (red) extracted from the crystal structure of BTK (Protein Data Bank ID: 5JRS). The result, revealing excellent agreement, is shown in Figure 2A and confirms that the selected experimental parameters and procedures used for molecular docking and alignment were reasonable. As depicted in Figure 2A, the common carbazole rings of 76 and 79 as well as experimental ligand were in the same position and mainly interacted with residues Glu475, Tyr476, and Met477.
Figure 2.
(A) The binding pose prediction of 76 (green) compared to ligand (red) found in an X-ray crystal structure; the position of 79 (yellow) in the active site of the protein and the binding pocket of BTK enzyme. (B) Docking-based alignment of dataset molecules. Hydrogen bonds are represented as yellow dotted lines, and main protein residues are labeled with ball and stick forms. Section A: hinge region; Sections B and C: hydrophobic pocket; Section D: floor loop.
To explain the binding mode, 79 (IC50 = 0.22 nM) was selected for more detailed analysis, since it was the most representative inhibitor in the active site of the protein. Based on Figure 2A, the carbazole ring of 79 interacted with −C = O and N–H of Met 477 and −C = O of Gly475 by hydrogen bonds in the hinge region, and interacted with the benzene ring of Tyr 466 by a conjugate effect; among them, Gly475 and Met477 [19] are two significant gatekeeper residues in BTK enzyme. The hydroxyl group at R4 also had a hydrogen-bond interaction with Ala478. Chlorine atom at R6 formed a hydrophobic interaction with Glu407 and Asp539. The benzene ring’s ortho-groups at R1 also interacted with Cys527 and Leu528 through a hydrophobic effect. At the bottom of the pocket, a substituent at the meta-position of the benzene ring was well filled in a floor loop formed by Asn484, Leu483, and Arg525. All these action characteristics proved that 79 was the most active molecule in the dataset.
As shown in Figure 2B, the selected 132 molecules demonstrate similar features after they are aligned on the common substructure and interact with Gly475 and Met477 through hydrogen-bond actions. The activities factors are groups at R4 trending toward different directions and groups at R6 forming hydrophobic interactions with different residues. Substituents at R1 occupied in sites of the floor loop area are also different. These diverse elements resulted in the selected 132 molecules used to perform molecular modeling studies possessing multiple IC50 values.
2.2. 3D-QSAR Analysis Studies
The aligned dataset was subjected to establish 3D-QSAR modeling using partial least squares (PLS) statistics with different field contribution values. In order to select the best field combination model and avoid the over-fitting problem, the stability statistics including cross-validated correlation coefficient (q2), non-cross-validated correlation coefficient (r2), a standard error of estimate (SEE), an optimum number of components (NOC), and F statistical values were taken into consideration. As a rule of thumb, q2 and r2 should have higher values, while SEE should have smaller error values. Therefore, reasonable CoMFA (q2 = 0.761, NOC = 6, r2 = 0.933) and CoMSIA (q2 = 0.891, NOC = 9, r2 = 0.988) models were developed for the selected training set and the test set. The detailed statistical summary of the CoMFA and CoMSIA analysis are shown in Table 1.
Table 1.
Detailed statistical summary of the comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) models.
| CoMFA | NOC | q2 | r2 | SEE | F Value | Field Contributions | ||||
| S | E | H | D | A | ||||||
| S+E | 6 | 0.761 | 0.933 | 0.202 | 291.45 | 0.46 | 0.54 | - | - | - |
| CoMSIA | NOC | q2 | r2 | SEE | F Value | Field Contributions | ||||
| S | E | H | D | A | ||||||
| S+E | 5 | 0.851 | 0.941 | 0.188 | 404.01 | 0.198 | 0.802 | - | - | - |
| S+E+H | 7 | 0.862 | 0.972 | 0.132 | 606.51 | 0.110 | 0.554 | 0.336 | - | - |
| S+E+D | 4 | 0.863 | 0.930 | 0.205 | 420.26 | 0.117 | 0.515 | - | 0.367 | - |
| S+E+A | 7 | 0.863 | 0.974 | 0.127 | 657.51 | 0.122 | 0.535 | - | - | 0.342 |
| S+E+H+D | 9 | 0.875 | 0.985 | 0.095 | 920.97 | 0.069 | 0.424 | 0.235 | 0.272 | - |
| S+E+H+A | 9 | 0.880 | 0.986 | 0.095 | 923.65 | 0.073 | 0.411 | 0.254 | - | 0.262 |
| S+E+D+A | 10 | 0.878 | 0.985 | 0.092 | 1031.44 | 0.078 | 0.400 | - | 0.270 | 0.253 |
| S+E+H+D+A | 9 | 0.891 | 0.988 | 0.088 | 1076.36 | 0.053 | 0.342 | 0.193 | 0.208 | 0.203 |
q2: cross-validated correlation coefficient; NOC: optimum number of components; r2: non cross-validated correlation coefficient; SEE: standard error of estimation; F value: F-test value. S = steric; E = electrostatic; H = hydrophobic; A = acceptor; D = donor. Final chosen model for CoMSIA analysis is indicated in bold font.
A reasonable CoMFA model was established on the basis of satisfactory statistical values including q2, r2, and SEE values (0.761, 0.933, and 0.202, respectively). When steric, electrostatic, hydrophobic, and H-bond acceptor and donor fields were all employed in the CoMSIA model, q2, r2, and SEE values also acquired good results (0.891, 0.988, and 0.088, respectively), which confirmed that the CoMSIA model was reliable and reasonable.
2.3. Contour Map Analysis
Contour maps for CoMFA and CoMSIA were generated to visualize the information in 3D-QSAR models. The maps of the 3D-QSAR models based on PLS analysis provided a comprehensive understanding of the key structural requirements responsible for the biological activity and are depicted in the following.
2.3.1. CoMFA Contour Map Analysis
CoMFA contour maps are vividly displayed in different color areas and illustrate whether the substituted groups are reasonable. Steric contour maps and electrostatic contour maps are shown in Figure 3A,B compared with 79.
Figure 3.
CoMFA StDev*Coeff contour maps. (A) Steric contour map (green: favored; yellow: disfavored). (B) Electrostatic contour map (blue: favored; red: disfavored). Compound 79 is shown as a capped sticks model.
In the CoMFA steric contour map (Figure 3A), green represents favored bulky groups and yellow represents the opposite. Green contour maps appeared at 9H of carbazole and R1, indicating that more bulky groups in these regions could improve activity. This possibly explained that inhibitory activity of 53 (IC50 = 18 nM), 54 (IC50 = 18 nM), and 55 (IC50 = 17 nM) with a methyl at the benzene ring of R1 was twentyfold more potent compared with 127 (IC50 = 390 nM) with a hydrogen atom at this position. Besides, a yellow contour at R3 suggests that adding a bulky substitution in this region can decrease inhibitory activity, which may explain why the activities of 101–104 (IC50: 110–461 nM) with an added morpholinone or piperazinone group at R3 dropped sharply.
In the CoMFA electrostatic contour maps (Figure 3B), blue contours located near 1-position and R3 imply that positive substitutions in these region can increase the activity of the inhibitors. This may explain why 104 (IC50 = 110 nM) with a piperazin substituent at R3 was more potent than 102 (IC50 = 308 nM) with morpholin in the same position. Inversely, the red contour in the ortho- and meta-positions of the benzene ring at R1 suggested that negative atoms can increase the activity. This was in accordance with the fact that 84 (IC50 = 032 nM), 87 (IC50 = 0.25 nM), 129 (IC50 = 0.4 nM), and 130 (IC50 = 0.9 nM) possessing nitrogen (negative) atoms at R1 demonstrated high BTK inhibition activity.
2.3.2. CoMSIA Contour Map Analysis
CoMSIA StDev*Coeff contour map analysis of steric, electrostatic, hydrophobic, and H-bond donor and H-bond acceptor fields are revealed in the following images, with 79 as the template molecule in the active site of BTK.
In the CoMSIA steric contour map (Figure 4A), the carbazole ring of 79, sheathed by a giant green block, indicates that the bulky groups here can increase the activity. Yellow contours near the extensional area of R3 suggest the unfavorable influence of bulky groups. In Figure 4B, the electron-donating group and electron-withdrawing group covered by blue and red contours were represented at 1-position and ortho-position of the benzene ring at R1, respectively. Compared to the steric/electrostatic contour maps of CoMFA and CoMSIA, they are very similar, except that the largest green field also involved an outstretched space in the carbazole scaffold, which means that adding bulky groups to this region improved activity.
Figure 4.
CoMSIA StDev*Coeff contour maps. (A) Steric contour map (green: favored; yellow: disfavored). (B) Electrostatic contour map (blue: favored; red: disfavored). Compound 79 is shown as a capped sticks model.
The hydrophobic contour map from CoMSIA is shown in Figure 5. Orange contours near the benzene ring of R1 and the hydrocarbyl of R4, as well as the extension space of R3, indicate that the hydrophobic groups in those areas are beneficial for inhibitory activities. This is consistent with the fact that 95–100 (IC50: 0.35–2.0 nM), possessing halogen and hydrocarbyl substituents in these areas, have more potent activities than 54 (IC50 = 18 nM) and 55 (IC50 = 17 nM) with the hydroxyl and amino groups. White contours around R1 reveal that the hydrophobic groups here do not help to enhance the activity. Hence, 121 (IC50 = 16 nM), 122 (IC50 = 15 nM), 124 (IC50 = 17 nM), and 125 (IC50 = 16 nM), possessing aromatic halogen substitutions at this position, held lower activity levels than 129–132 (IC50: 0.4–1.0 nM).
Figure 5.
CoMSIA StDev*Coeff contour maps: Hydrophobic contour map (orange: favored; white: disfavored). Compound 79 is shown as a capped sticks model.
The H-bond donor and acceptor of the CoMSIA contour map are shown in Figure 6A,B, respectively. The remarkable cyan contour on the top of the carbazole ring implies that the presence of hydrogen-bond donor groups might enhance bioactivity. This could be validated if it is found that 1–132 possess hydrogen atoms as hydrogen-bond donor groups in the same positions. The magenta contours around 1-position and meta-position of the benzene ring at R1 show that H-bond acceptor groups in these places can increase the activity of inhibitors. This might explain why 74 (IC50 = 0.79 nM) and 75 (IC50 = 1.2 nM) with two carbonyl substituents at R1 displayed better IC50 values than 1 (IC50 = 44 nM).
Figure 6.
CoMSIA StDev*Coeff contour maps. (A) H-bond donor map (cyan: favored; purple: disfavored). (B) H-bond acceptor map (magenta: favored; brown: disfavored). Compound 79 is shown as a capped sticks model.
2.4. Model Validation of CoMFA and CoMSIA Models
The experimental and predicted activity values of CoMFA and CoMSIA models are depicted in Table 2, and their scatter plots are shown in Figure 7.
Table 2.
The experimental and predicted activity values for the developed models.
| Training Set Compounds | pIC50 | CoMFA | CoMSIA | ||
| Predicted | Residuals | Predicted | Residuals | ||
| 1 | 4.3565 | 4.304 | 0.0525 | 4.286 | 0.0705 |
| 3 | 5.6778 | 5.554 | 0.1239 | 5.730 | −0.0526 |
| 4 | 5.8239 | 5.526 | 0.2981 | 5.790 | 0.0344 |
| 5 | 5.6576 | 5.649 | 0.0082 | 5.673 | −0.0157 |
| 6 | 5.7447 | 5.758 | −0.0135 | 5.753 | −0.008 |
| 7 | 5.6576 | 5.524 | 0.1337 | 5.697 | −0.0393 |
| 8 | 5.6576 | 5.751 | −0.0938 | 5.659 | −0.0011 |
| 9 | 5.6576 | 5.735 | −0.0776 | 5.656 | 0.0015 |
| 10 | 5.6383 | 5.493 | 0.1456 | 5.640 | −0.0015 |
| 11 | 5.7959 | 6.058 | −0.2625 | 5.957 | −0.1615 |
| 12 | 6.0969 | 6.060 | 0.0366 | 6.102 | −0.0048 |
| 13 | 5.6990 | 5.631 | 0.0675 | 5.653 | 0.0459 |
| 14 | 6.0000 | 6.042 | −0.0422 | 5.925 | 0.0755 |
| 15 | 6.1551 | 6.234 | −0.0789 | 6.087 | 0.0681 |
| 17 | 5.8861 | 5.641 | 0.2452 | 5.885 | 0.0008 |
| 18 | 5.6990 | 5.583 | 0.1156 | 5.704 | −0.0049 |
| 19 | 5.7447 | 5.821 | −0.0766 | 5.761 | −0.0162 |
| 20 | 5.5376 | 5.698 | −0.16 | 5.638 | −0.1001 |
| 22 | 5.8539 | 5.615 | 0.2385 | 5.855 | −0.0012 |
| 23 | 5.7213 | 5.465 | 0.2565 | 5.751 | −0.0294 |
| 24 | 5.7959 | 5.917 | −0.1211 | 5.731 | 0.0644 |
| 25 | 5.5229 | 5.901 | −0.3785 | 5.516 | 0.0073 |
| 26 | 5.7213 | 5.876 | −0.1547 | 5.908 | −0.1865 |
| 28 | 6.0924 | 6.208 | −0.1156 | 6.099 | 0.0066 |
| 29 | 6.2076 | 6.337 | −0.1291 | 6.36 | −0.1519 |
| 31 | 5.5528 | 5.597 | −0.0445 | 5.654 | −0.1009 |
| 32 | 5.0458 | 5.081 | −0.0354 | 5.010 | 0.0357 |
| 33 | 5.5850 | 5.744 | −0.1586 | 5.518 | 0.0669 |
| 34 | 5.8239 | 6.272 | −0.4485 | 6.017 | −0.1929 |
| 36 | 5.7695 | 5.951 | −0.1814 | 5.642 | 0.1275 |
| 37 | 6.2007 | 6.017 | 0.1837 | 6.080 | 0.1203 |
| 38 | 6.2840 | 6.026 | 0.2581 | 6.119 | 0.165 |
| 39 | 6.0269 | 5.864 | 0.1626 | 6.068 | −0.0413 |
| 40 | 6.0000 | 6.121 | −0.1213 | 5.939 | 0.0609 |
| 42 | 5.7695 | 5.786 | −0.0166 | 5.831 | −0.0613 |
| 43 | 5.7959 | 5.791 | 0.0051 | 5.737 | 0.059 |
| 44 | 5.7959 | 5.513 | 0.2828 | 5.760 | 0.0358 |
| 45 | 6.0315 | 6.176 | −0.1442 | 6.172 | −0.1406 |
| 46 | 5.6383 | 6.035 | −0.397 | 5.728 | −0.0898 |
| 47 | 5.8239 | 5.844 | −0.02 | 5.909 | −0.0852 |
| 48 | 5.5850 | 5.787 | −0.2015 | 5.695 | −0.1097 |
| 49 | 5.3010 | 5.244 | 0.0572 | 5.325 | −0.0241 |
| 50 | 5.3010 | 5.393 | −0.0917 | 5.275 | 0.026 |
| 52 | 4.7959 | 5.112 | −0.3163 | 4.728 | 0.0682 |
| 54 | 4.7447 | 4.654 | 0.0905 | 4.849 | −0.1045 |
| 55 | 4.7695 | 4.684 | 0.0851 | 4.826 | −0.0568 |
| 56 | 4.8239 | 4.779 | 0.045 | 4.703 | 0.1207 |
| 57 | 4.7959 | 4.792 | 0.0037 | 4.902 | −0.1061 |
| 59 | 4.7695 | 4.634 | 0.1357 | 4.657 | 0.1121 |
| 60 | 5.6990 | 5.694 | 0.0045 | 5.628 | 0.0712 |
| 61 | 5.6990 | 5.440 | 0.2595 | 5.636 | 0.0632 |
| 62 | 4.7959 | 4.760 | 0.0359 | 4.871 | −0.0747 |
| 63 | 4.7695 | 4.812 | −0.0421 | 4.753 | 0.0165 |
| 64 | 5.3010 | 5.582 | −0.2807 | 5.326 | −0.0249 |
| 65 | 4.8239 | 4.686 | 0.1384 | 4.849 | −0.0248 |
| 66 | 5.7213 | 5.880 | −0.159 | 5.762 | −0.0407 |
| 67 | 5.6990 | 5.454 | 0.2446 | 5.714 | −0.0154 |
| 68 | 5.6778 | 5.651 | 0.0267 | 5.790 | −0.1123 |
| 69 | 5.8539 | 5.991 | −0.1368 | 5.792 | 0.0624 |
| 70 | 5.7695 | 5.587 | 0.183 | 5.727 | 0.0424 |
| 71 | 5.7959 | 5.955 | −0.1589 | 5.760 | 0.0362 |
| 72 | 6.1805 | 6.234 | −0.0532 | 6.087 | 0.0931 |
| 73 | 6.3872 | 6.487 | −0.1002 | 6.355 | 0.0321 |
| 76 | 5.3979 | 5.391 | 0.007 | 5.528 | −0.1299 |
| 77 | 6.3468 | 6.220 | 0.1271 | 6.272 | 0.0749 |
| 79 | 6.6576 | 6.305 | 0.3527 | 6.596 | 0.0612 |
| 80 | 6.1135 | 6.136 | −0.0222 | 6.213 | −0.0992 |
| 81 | 6.3188 | 6.131 | 0.188 | 6.402 | −0.0837 |
| 83 | 6.2291 | 6.107 | 0.1225 | 6.339 | −0.1095 |
| 85 | 6.0000 | 6.025 | −0.0254 | 6.029 | −0.0285 |
| 86 | 6.3098 | 6.404 | −0.0941 | 6.365 | −0.0557 |
| 89 | 5.8861 | 6.291 | −0.4053 | 6.112 | −0.2261 |
| 90 | 6.0915 | 6.111 | −0.0196 | 6.131 | −0.0396 |
| 92 | 6.3566 | 6.171 | 0.186 | 6.281 | 0.0751 |
| 93 | 6.2602 | 6.274 | −0.0138 | 6.251 | −0.0092 |
| 94 | 6.0706 | 6.208 | −0.1374 | 6.099 | −0.0288 |
| 95 | 6.0410 | 5.983 | 0.0577 | 5.981 | 0.0595 |
| 96 | 6.0000 | 5.468 | 0.5325 | 5.981 | 0.0187 |
| 97 | 6.0458 | 6.086 | −0.0406 | 6.038 | 0.0077 |
| 98 | 5.6990 | 5.934 | −0.2348 | 5.638 | 0.0608 |
| 99 | 6.3468 | 6.242 | 0.1046 | 6.366 | −0.0187 |
| 100 | 6.4559 | 6.074 | 0.3816 | 6.496 | −0.0397 |
| 105 | 4.5528 | 4.125 | 0.4278 | 4.538 | 0.015 |
| 108 | 4.8239 | 4.736 | 0.0878 | 4.826 | −0.0023 |
| 109 | 4.4949 | 4.403 | 0.0923 | 4.379 | 0.1156 |
| 110 | 4.6198 | 4.870 | −0.2505 | 4.523 | 0.0971 |
| 111 | 4.1487 | 3.873 | 0.2756 | 4.082 | 0.0663 |
| 112 | 4.2291 | 4.703 | −0.4742 | 4.182 | 0.0468 |
| 113 | 3.9872 | 4.031 | −0.0437 | 3.984 | 0.0036 |
| 117 | 4.7212 | 4.481 | 0.2403 | 4.758 | −0.0366 |
| 119 | 4.7959 | 4.747 | 0.0485 | 4.809 | −0.0131 |
| 121 | 4.7959 | 4.881 | −0.0849 | 4.889 | −0.0936 |
| 122 | 4.8239 | 4.695 | 0.1286 | 4.871 | −0.0473 |
| 123 | 4.7959 | 4.888 | −0.0918 | 4.820 | −0.0243 |
| 124 | 4.7695 | 5.019 | −0.2491 | 4.821 | −0.0511 |
| 125 | 4.7959 | 5.032 | −0.2358 | 4.751 | 0.0454 |
| 129 | 6.3979 | 6.202 | 0.1955 | 6.281 | 0.1174 |
| 130 | 6.0458 | 6.086 | −0.0406 | 6.038 | 0.0077 |
| 131 | 6.0000 | 5.468 | 0.5325 | 5.981 | 0.0187 |
| 132 | 6.5850 | 6.559 | 0.0255 | 6.608 | −0.0229 |
| Test Set Compounds | pIC50 | CoMFA | CoMSIA | ||
| Predicted | Residuals | Predicted | Residuals | ||
| 2 * | 5.3010 | 5.505 | −0.2035 | 5.296 | 0.0055 |
| 16 * | 5.8861 | 5.526 | 0.3598 | 5.871 | 0.0147 |
| 21 * | 5.4948 | 5.751 | −0.2562 | 5.648 | −0.1528 |
| 27 * | 5.7213 | 5.631 | 0.0898 | 5.618 | 0.1034 |
| 30 * | 5.585 | 5.522 | 0.0629 | 5.554 | 0.0312 |
| 35 * | 5.6383 | 6.035 | −0.397 | 5.728 | −0.0898 |
| 41 * | 5.6778 | 5.696 | −0.0185 | 5.738 | −0.0601 |
| 51 * | 5.6990 | 5.581 | 0.1182 | 5.596 | 0.103 |
| 53 * | 4.7447 | 4.732 | 0.0132 | 4.745 | −0.0001 |
| 58 * | 5.5229 | 5.515 | 0.0076 | 5.364 | 0.1585 |
| 74 * | 6.1024 | 6.085 | 0.0172 | 6.059 | 0.043 |
| 75 * | 5.9208 | 5.971 | −0.0502 | 5.797 | 0.124 |
| 78 * | 6.3372 | 6.220 | 0.1176 | 6.272 | 0.0654 |
| 82 * | 6.4559 | 6.312 | 0.1437 | 6.417 | 0.0393 |
| 84 * | 6.4948 | 6.451 | 0.0443 | 6.432 | 0.0631 |
| 87 * | 6.6021 | 6.472 | 0.1298 | 6.523 | 0.079 |
| 88 * | 6.0000 | 6.316 | −0.3157 | 6.074 | −0.0739 |
| 91 * | 6.2076 | 6.337 | −0.1291 | 6.360 | −0.1519 |
| 101 * | 3.7235 | 3.525 | 0.1989 | 3.684 | 0.0392 |
| 102 * | 3.5114 | 3.734 | −0.2221 | 3.580 | −0.0684 |
| 103 * | 3.3363 | 3.648 | −0.3118 | 3.523 | −0.1868 |
| 104 * | 3.9586 | 3.937 | 0.0218 | 3.940 | 0.0186 |
| 106 * | 4.0555 | 3.886 | 0.1694 | 4.104 | −0.048 |
| 107 * | 3.4214 | 3.776 | −0.3549 | 3.506 | −0.0851 |
| 114 * | 4.3565 | 4.110 | 0.2468 | 4.286 | 0.0708 |
| 115 * | 3.8962 | 3.795 | 0.1014 | 3.799 | 0.0972 |
| 116 * | 3.9957 | 4.494 | −0.4982 | 4.254 | −0.2585 |
| 118 * | 4.6576 | 4.579 | 0.0789 | 4.488 | 0.1701 |
| 120 * | 4.9208 | 4.915 | 0.0063 | 4.831 | 0.0897 |
| 126 * | 4.7959 | 4.816 | −0.0204 | 4.685 | 0.1113 |
| 127 * | 3.4089 | 3.904 | −0.4953 | 3.483 | −0.0741 |
| 128 * | 4.8239 | 4.873 | −0.0489 | 4.758 | 0.0663 |
* Test set.
Figure 7.
Correlation between the predicted and experimental activities of the training and test set compounds. (A) The scatter plot of CoMFA. (B) The scatter plot of CoMSIA. Black squares represent the training set; red triangles represent the test set.
Based on the above data, the correlation coefficient between the predicted and experimental activities generated by the CoMFA models were 0.94073 and its analytical error was 0.32893, which confirmed that the established CoMFA models are reliable and reasonable. Similarly, the correlation coefficient and analytical error of the CoMSIA model were 0.99181 and 0.04793, respectively, and these two values verify that the CoMSIA models are accurate and reliable. Both CoMFA and CoMSIA models can be further used to predict activities of newly designed inhibitors.
3. Materials and Methods
3.1. Collection of the Dataset
A series of carbazole-carboxamide-based BTK inhibitors (BMS-986142 analogues) were used for the study. The 132 selected molecules [14,20,21,22,23] had a basic tricyclic skeleton and a similar binding mode with the BTK enzyme, which could be well superimposed in the alignment module. These BMS-986142 analogues were evenly distributed in an inhibitory activity range from 0.1 to 1000 nM. These compounds were optimized by energy minimization with a tripos force field in Sybyl-X 2.0 [24] and generated three-dimensional conformations after docking into the BTK-enzyme-binding site. The biological data expressed as IC50 values were converted into pIC50 (−log IC50) values, which were used as dependent variables in the following QSAR analyses [25]. The selected 132 BTK inhibitors were divided into a test set consisting of 32 molecules for model validation and a training set including 100 compounds for model generation. Thirty-two compounds in the test set were selected randomly and included compounds with a uniformly distributed range of pIC50 values from 3.336 to 6.658, covering more than 3 log units, which is fit for 3D QSAR studies [26]. The conformation of the most active compound, 79, was selected as a template structure to sketch the rest of the molecules [27]. The complete dataset (1–132) taken for study is shown in Table 3.
Table 3.
Chemical structures of 1–132 with their pIC50.

| |||||||
|---|---|---|---|---|---|---|---|
| Mol. | R1 | R2 | R3 | R4 | R6 | IC50 (nM) | pIC50 |
| 1 |
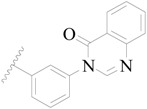
|
H | H |

|
H | 44 | 4.357 |
| 2 * |

|
H | H |

|
H | 5.0 | 5.301 |
| 3 |

|
H | H |

|
H | 2.1 | 5.678 |
| 4 |

|
H | H |

|
H | 15 | 5.824 |
| 5 |

|
H | H |

|
H | 2.2 | 5.658 |
| 6 |

|
H | H |

|
H | 1.8 | 5.745 |
| 7 |

|
H | H |

|
H | 2.2 | 5.658 |
| 8 |

|
H | H |

|
H | 2.2 | 5.658 |
| 9 |

|
H | H |

|
H | 2.2 | 5.658 |
| 10 |

|
H | H |

|
H | 2.3 | 5.638 |
| 11 |

|
H | H |

|
H | 1.6 | 5.796 |
| 12 |

|
H | H |

|
H | 0.8 | 6.097 |
| 13 |

|
H | H |

|
H | 2.0 | 5.699 |
| 14 |

|
H | H |

|
H | 1.0 | 6.000 |
| 15 |

|
H | H |

|
H | 0.7 | 6.155 |
| 16 * |

|
H | H |

|
H | 1.3 | 5.886 |
| 17 |

|
H | H |

|
H | 1.3 | 5.886 |
| 18 |

|
H | H |

|
H | 2.0 | 5.699 |
| 19 |

|
H | H |

|
H | 1.8 | 5.745 |
| 20 |

|
H | H |

|
H | 2.9 | 5.538 |
| 21 * |

|
H | H |

|
H | 3.2 | 5.495 |
| 22 |

|
H | H |

|
H | 1.4 | 5.854 |
| 23 |

|
H | H |

|
H | 1.9 | 5.721 |
| 24 |

|
H | H |

|
H | 1.6 | 5.796 |
| 25 |

|
H | H | CH2OH | H | 3.0 | 5.523 |
| 26 |

|
H | H |

|
H | 1.9 | 5.721 |
| 27 * |

|
H | H |

|
H | 1.9 | 5.721 |
| 28 |

|
H | H |

|
H | 0.81 | 6.092 |
| 29 |

|
H | H |

|
H | 0.62 | 6.208 |
| 30 * |

|
H | H |

|
H | 2.6 | 5.585 |
| 31 |

|
H | H |

|
H | 2.8 | 5.553 |
| 32 |

|
H | H |

|
H | 9.0 | 5.046 |
| 33 |

|
H | H |

|
H | 2.6 | 5.585 |
| 34 |

|
H | H |

|
H | 1.5 | 5.824 |
| 35 * |

|
H | H |

|
H | 2.3 | 5.638 |
| 36 |

|
H | H |

|
H | 1.7 | 5.770 |
| 37 |

|
H | H |

|
H | 0.63 | 6.201 |
| 38 |

|
H | H |

|
H | 0.52 | 6.284 |
| 39 |

|
H | H |

|
H | 0.94 | 6.027 |
| 40 |

|
H | H |

|
H | 1.0 | 6.000 |
| 41 * |

|
H | H |

|
H | 2.1 | 5.678 |
| 42 |

|
H | H |

|
H | 1.7 | 5.770 |
| 43 |

|
H | H |

|
H | 1.6 | 5.796 |
| 44 |

|
H | H |

|
H | 1.6 | 5.796 |
| 45 |

|
H | H |

|
H | 0.93 | 6.032 |
| 46 |

|
H | H |

|
H | 2.3 | 5.638 |
| 47 |

|
H | H |

|
H | 1.5 | 5.824 |
| 48 |

|
H | H |

|
H | 2.6 | 5.585 |
| 49 |

|
H | H |

|
H | 5.0 | 5.301 |
| 50 |

|
H | H |

|
H | 5.0 | 5.301 |
| 51 * |

|
H | H |

|
H | 2.0 | 5.699 |
| 52 |

|
H | H |

|
H | 16 | 4.796 |
| 53 * |

|
H | H |

|
H | 18 | 4.745 |
| 54 |

|
H | H |

|
H | 18 | 4.745 |
| 55 |

|
H | H |

|
H | 17 | 4.770 |
| 56 |

|
H | H |

|
H | 15 | 4.824 |
| 57 |

|
H | H |

|
H | 16 | 4.796 |
| 58 * |

|
H | H |

|
H | 3.0 | 5.523 |
| 59 |

|
H | H |

|
H | 17 | 4.770 |
| 60 |

|
H | H |

|
H | 2.0 | 5.699 |
| 61 |

|
H | H |

|
H | 18 | 5.699 |
| 62 |

|
H | H |

|
H | 16 | 4.796 |
| 63 |

|
H | H |

|
H | 17 | 4.770 |
| 64 |

|
H | H |

|
H | 5.0 | 5.301 |
| 65 |

|
H | H |

|
H | 15 | 4.824 |
| 66 |

|
H | H |

|
H | 1.9 | 5.721 |
| 67 |

|
H | H |

|
H | 2.0 | 5.699 |
| 68 |

|
H | H |

|
H | 2.1 | 5.678 |
| 69 |

|
H | H |

|
H | 1.4 | 5.854 |
| 70 |

|
H | H |

|
H | 1.7 | 5.770 |
| 71 |

|
H | H | OH | H | 1.6 | 5.796 |
| 72 |

|
H | H |

|
CH3 | 0.66 | 6.180 |
| 73 |

|
H | H |

|
CH3 | 0.41 | 6.387 |
| 74 * |

|
CH3 | H |

|
H | 0.79 | 6.102 |
| 75 * |

|
CH3 | H |

|
H | 1.2 | 5.921 |
| 76 |

|
H | H |

|
H | 4.0 | 5.398 |
| 77 |

|
H | H |

|
F | 0.45 | 6.347 |
| 78 * |

|
H | H |

|
F | 0.46 | 6.337 |
| 79 |

|
H | H |

|
F | 0.22 | 6.658 |
| 80 |

|
H | H |

|
F | 0.77 | 6.114 |
| 81 |

|
H | H |

|
F | 0.48 | 6.319 |
| 82 * |

|
H | H |

|
F | 0.35 | 6.456 |
| 83 |

|
H | H |

|
F | 0.59 | 6.229 |
| 84 * |

|
H | H |

|
F | 0.32 | 6.495 |
| 85 |

|
H | H |

|
CN | 1.0 | 6.000 |
| 86 |

|
H | H |

|
CN | 0.49 | 6.310 |
| 87 * |

|
H | H |

|
Cl | 0.25 | 6.602 |
| 88 * |

|
H | H |

|
Cl | 1.0 | 6.000 |
| 89 |

|
H | H |

|
Cl | 1.3 | 5.886 |
| 90 |

|
H | H |

|
Cl | 0.81 | 6.092 |
| 91 * |

|
H | H |

|
Cl | 0.62 | 6.208 |
| 92 |

|
H | H |

|
Cl | 0.44 | 6.357 |
| 93 |

|
H | H |

|
Cl | 0.55 | 6.260 |
| 94 |

|
H | H |

|
Cl | 0.85 | 6.071 |
| 95 |

|
H | H |

|
Cl | 0.91 | 6.041 |
| 96 |

|
H | H |

|
Cl | 1.0 | 6.000 |
| 97 |

|
H | H |

|
H | 0.9 | 6.046 |
| 98 |

|
H | H |

|
Cl | 2.0 | 5.699 |
| 99 |

|
H | H |

|
Cl | 0.45 | 6.347 |
| 100 |

|
H | H |

|
F | 0.35 | 6.456 |
| 101 * | H | H |

|
H |

|
189 | 3.724 |
| 102 * | H | H |

|
H |

|
308 | 3.511 |
| 103 * | H | H |

|
H |

|
461 | 3.336 |
| 104 * | H | H |

|
H |

|
110 | 3.959 |
| 105 | H | H |

|
H |

|
28 | 4.553 |
| 106 * | H | H |

|
H |

|
88 | 4.056 |
| 107 * | H | H | H |

|

|
379 | 3.421 |
| 108 | H | H | H |

|

|
15 | 4.824 |
| 109 | H | H | H |

|

|
32 | 4.495 |
| 110 | H | H | H |

|

|
24 | 4.620 |
| 111 | H | H | H |

|

|
71 | 4.149 |
| 112 | H | H | H |

|

|
59 | 4.229 |
| 113 | H | H | H |

|

|
103 | 3.987 |
| 114 * | H | H | H |

|

|
44 | 4.357 |
| 115 * | H | H | H |

|

|
127 | 3.896 |
| 116 * | H | H | H |

|

|
101 | 3.996 |
| 117 | H | H | H |

|

|
19 | 4.721 |
| 118 * | H | H | H |

|

|
22 | 4.658 |
| 119 | H | H | H |

|

|
16 | 4.796 |
| 120 * | H | H | H |

|

|
12 | 4.921 |
| 121 |

|
H | H |

|
H | 16 | 4.796 |
| 122 |

|
H | H |

|
H | 15 | 4.824 |
| 123 |

|
H | H |

|
H | 16 | 4.796 |
| 124 |

|
H | H |

|
H | 17 | 4.770 |
| 125 |

|
H | H |

|
H | 16 | 4.796 |
| 126 * |

|
H | H |

|
H | 16 | 4.796 |
| 127 * |

|
H | H |

|
H | 390 | 3.409 |
| 128 * |

|
H | H |

|
H | 15 | 4.824 |
| 129 |

|
H | H |

|
H | 0.4 | 6.398 |
| 130 |

|
H | H |

|
Cl | 0.90 | 6.046 |
| 131 |

|
H | H |

|
Cl | 1.0 | 6.000 |
| 132 |

|
H | H |

|
F | 0.34 | 6.585 |
* Test set.
3.2. Preparation of Protein
The crystal structure of BTK with high resolution was retrieved from the protein data bank (PDB ID: 5JRS) [20]. This crystal structure was prepared using a protein preparation module in Sybyl-X 2.0. Ligand and water molecules were removed. Furthermore, polar hydrogen atoms were added for investigating interactions between inhibitors and BTK.
3.3. Molecular Docking and Alignment
The molecular dockings of 1–132 were performed using Surflex-Dock (SFXC) module with default parameters, except that the maximum number of per molecular conformation was defined as 40 to ensure that the docked conformations in the BTKBTK-binding site were reasonable. The rational docked conformations of the compound in the protein-binding site were picked up from the clustered docking poses according to the principle of low energy and rational conformation [28]. The most potent compound, 79, with the rational conformation possessing the lowest energy, was chosen as the reference molecule. Rational conformations of the remaining inhibitors in the dataset based on the interactions with the BTK-enzyme-binding site were aligned on the common substructure of the reference compound (Figure 8). After the conformations were aligned in the BTKBTK-binding site, all selected conformations were conserved as a database file, which was used for 3D-QSAR study.
Figure 8.

The common scaffold of the dataset.
3.4. 3D-QSAR Analysis Studies
3D-QSAR analyses performed by the QSAR command bar of SYBYL X-2.0 (Tripos (DE), Inc., St. Louis, MO, USA) were carried out in the form of molecular spreadsheets to create CoMFA and CoMSIA fields from the database file acquired after molecular docking. The CoMFA [17] fields, including steric (S) and electrostatic (E) fields, were calculated under default settings with energy cutoff values of 30 kcal/mol. With the exception of the same fields in CoMFA, the CoMSIA [18] fields also containing hydrophobic (H) and hydrogen-bond donor (D) and acceptor (A) fields were derived using the same method as that of the CoMFA calculations. Both CoMFA and CoMSIA analyses were calculated in the standard settings with an attenuation factor α of 0.3. After 3D-QSAR analyses, the standard contour maps for both CoMFA and CoMSIA to visualize the results were developed using the field type StDev*Coeff.
3.5. Model Validation
All the developed CoMFA and CoMSIA models were checked for stability and robustness using the internal and external test set validations. Internal validation was carried out using a PLS [29] approach of cross-validation method to inspect the predictability of the dataset. The external test set containing 32 molecules not included in the model building was applied to verify the accuracy of the predictive abilities of the derived 3D-QSAR models. In the PLS approach, leave-one-out (LOO) method analysis generated the cross-validated q2 and the optimum number of components. The final CoMFA and CoMSIA models were developed using the obtained optimal number of components without cross-validation analysis. When the values of the coefficients fall between 1.0 and 0.5 [30], an accurate model is accepted. Furthermore, for better evaluation of the accuracy and robustness of the developed models, non-cross-validation analysis was employed to yield the conventional correlation coefficient r2 and the F-test value (F).
4. Conclusions
A 3D-QSAR study on carbazole inhibitors based on a common scaffold was conducted with the generation of rational docking conformations and CoMFA/CoMSIA models. The reasonable CoMFA (q2 = 0.761, r2 = 0.933) and CoMSIA (q2 = 0.891, r2 = 0.988) models displayed satisfactory correlations and predictive abilities. CoMFA and CoMSIA contour maps provided information (shown in Figure 9) indicating that structural optimization for improving activities can be predominantly considered by adding bulky negative electrostatic groups and hydrophilic groups at R1, by increasing hydrophilic groups at R4, and by raising H-bond donor and acceptor substituents at 1-position. Moreover, the predicted ability of 3D-QSAR models was validated for application in predicting the activities of newly designed compounds and further provided a valuable clue in the design of novel carbazole inhibitors for RA treatment.
Figure 9.
The structure–activity relationship (SAR) summarized based on our work.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (81202389).
Abbreviations
| BTK | Broton’s tyrosine kinase |
| RA | Rheumatoid Arthritis |
| BCR | B-cell receptor |
| NSAIDs | non-steroidal anti-inflammatory drug |
| SAARDs | slow acting anti-rheumatic drugs |
| DMARDs | disease-modifying anti rheumatic drugs |
| 3D-QSAR | three-dimensional quantitative structure–activity relationship |
| CoMFA | comparative molecular field analysis |
| CoMSIA | comparative molecular similarity indices analysis |
| PLS | partial least square |
| XLA | X-linked agammaglobulinemia |
| ALL | acute lymphoblastic leukemia |
| CML | chronic myeloid leukemia |
| CLL | chronic lymphocytic leukemia |
Author Contributions
Rui Li and Yongli Du conceived and designed the experiments; Rui Li performed the experiments; Rui Li and Zhipei Gao analyzed the data; Jingkang Shen contributed materials tools; Rui Li wrote the paper.
Conflicts of Interest
There are no conflicts of interest to declare.
References
- 1.Shaw M., Collins B.F., Ho L.A., Raghu G. Rheumatoid arthritis-associated lung disease. Eur. Respir. Rev. 2015;24:1–16. doi: 10.1183/09059180.00008014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brito Y., Glassberg M.K., Ascherman D.P. Rheumatoid Arthritis-Associated Interstitial Lung Disease: Current Concepts. Curr. Rheumatol. Rep. 2017;19:79. doi: 10.1007/s11926-017-0701-5. [DOI] [PubMed] [Google Scholar]
- 3.Smolen J.S., Landewe R., Breedveld F.C., Dougados M., Emery P., Gaujoux-Viala C., Gorter S., Knevel R., Nam J., Schoels M., et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs. Ann Rheum. Dis. 2010;69:964–975. doi: 10.1136/ard.2009.126532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma M.H., Cope A.P., Scott D.L. Safety of combination therapies in early rheumatoid arthritis: A systematic comparison between antirheumatic drugs and TNF inhibitors with methotrexate. Int. J. Clin. Rheumatol. 2010;5:547–554. doi: 10.2217/ijr.10.44. [DOI] [Google Scholar]
- 5.Takata M., Kurosaki T. A Role for Bruton’s Tyrosine Kinase in B Cell Antigen Receptor-mediated Activation of Phospholipase C-gamma 2. J. Exp. Med. 1996;184:31–40. doi: 10.1084/jem.184.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohamed A.J., Nore B.F., Christensson B., Smith C.I. Signalling of Bruton’s Tyrosine Kinase, Btk. Scand. J. Immunol. 1999;49:113–118. doi: 10.1046/j.1365-3083.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 7.Whang J.A., Chang B.Y. Bruton’s tyrosine kinase inhibitors for the treatment of rheumatoid arthritis. Drug Discov. Today. 2014;19:1200–1204. doi: 10.1016/j.drudis.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Pan Z., Scheerens H., Li S.J., Schultz B.E., Sprengeler P.A., Burrill L.C., Mendonca R.V., Sweeney M.D., Scott K.C., Grothaus P.G., et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. ChemMedChem. 2007;2:58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 9.Honigberg L.A., Smith A.M., Sirisawad M., Verner E., Loury D., Chang B., Li S., Pan Z., Thamm D.H., Miller R.A., et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc. Natl. Acad. Sci. USA. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J., Zhang M., Liu D. Acalabrutinib (ACP-196): A selective second-generation BTK inhibitor. J. Hematol. Oncol. 2016;9:21. doi: 10.1186/s13045-016-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshizawa T., Birkett J.T., Kawabata K. ONO-4059, A Novel Bruton’s Tyrosine Kinase (Btk) Inhibitor: Synergistic Activity in Combination with Chemotherapy in a ABC-DLBCL Cell Line. Blood. 2013;122:5151. [Google Scholar]
- 12.Evans E.K., Tester R., Aslanian S., Karp R., Sheets M., Labenski M.T., Witowski S.R., Lounsbury H., Chaturvedi P., Mazdiyasni H., et al. Inhibition of Btk with CC-292 provides early pharmacodynamic assessment of activity in mice and humans. J. Pharmacol. Exp. Ther. 2013;346:219–228. doi: 10.1124/jpet.113.203489. [DOI] [PubMed] [Google Scholar]
- 13.Park J.K., Byun J.Y., Park J.A., Kim Y.Y., Lee Y.J., Oh J.I., Jang S.Y., Kim Y.H., Song Y.W., Son J., et al. HM71224, a novel Bruton’s tyrosine kinase inhibitor, suppresses B cell and monocyte activation and ameliorates arthritis in a mouse model: A potential drug for rheumatoid arthritis. Arthritis Res. Ther. 2016;18:91. doi: 10.1186/s13075-016-0988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watterson S.H., De Lucca G.V., Shi Q., Langevine C.M., Liu Q., Batt D.G., Beaudoin Bertrand M., Gong H., Dai J., Yip S., et al. Discovery of 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide (BMS-986142): A Reversible Inhibitor of Bruton’s Tyrosine Kinase (BTK) Conformationally Constrained by Two Locked Atropisomers. J. Med. Chem. 2016;59:9173–9200. doi: 10.1021/acs.jmedchem.6b01088. [DOI] [PubMed] [Google Scholar]
- 15.Lee S.K., Xing J., Catlett I.M., Adamczyk R., Griffies A., Liu A., Murthy B., Nowak M. Safety, pharmacokinetics, and pharmacodynamics of BMS-986142, a novel reversible BTK inhibitor, in healthy participants. Eur. J. Clin. Pharmacol. 2017;73:689–698. doi: 10.1007/s00228-017-2226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillooly K.M., Pulicicchio C., Pattoli M.A., Cheng L., Skala S., Heimrich E.M., McIntyre K.W., Taylor T.L., Kukral D.W., Dudhgaonkar S., et al. Bruton’s tyrosine kinase inhibitor BMS-986142 in experimental models of rheumatoid arthritis enhances efficacy of agents representing clinical standard-of-care. PLoS ONE. 2017;12:e0181782. doi: 10.1371/journal.pone.0181782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cramer R.D., Patterson D.E., Bunce J.D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988;110:5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 18.Klebe G., Abraham U., Mietzner T. Molecular Similarity Indices in a Comparative Analysis (CoMSIA) of Drug Molecules to Correlate and Predict Their Biological Activity. J. Med. Chem. 1994;37:4130–4146. doi: 10.1021/jm00050a010. [DOI] [PubMed] [Google Scholar]
- 19.Li X., Zuo Y., Tang G., Wang Y., Zhou Y., Wang X., Guo T., Xia M., Ding N., Pan Z. Discovery of a series of 2,5-diaminopyrimidine covalent irreversible inhibitors of Bruton’s tyrosine kinase with in vivo antitumor activity. J. Med. Chem. 2014;57:5112–5128. doi: 10.1021/jm4017762. [DOI] [PubMed] [Google Scholar]
- 20.De Lucca G.V., Shi Q., Liu Q., Batt D.G., Beaudoin Bertrand M., Rampulla R., Mathur A., Discenza L., D’Arienzo C., Dai J., et al. Small Molecule Reversible Inhibitors of Bruton’s Tyrosine Kinase (BTK): Structure-Activity Relationships Leading to the Identification of 7-(2-Hydroxypropan-2-yl)-4-[2-methyl-3-(4-oxo-3,4-dihydroquinazolin-3-yl)phenyl]-9H-carbazole-1-carboxamide (BMS-935177) J. Med. Chem. 2016;59:7915–7935. doi: 10.1021/acs.jmedchem.6b00722. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q., Batt D.G., Lippy J.S., Surti N., Tebben A.J., Muckelbauer J.K., Chen L., An Y., Chang C., Pokross M., et al. Design and synthesis of carbazole carboxamides as promising inhibitors of Bruton’s tyrosine kinase (BTK) and Janus kinase 2 (JAK2) Bioorg. Med. Chem. Lett. 2015;25:4265–4269. doi: 10.1016/j.bmcl.2015.07.102. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q., Batt D.G., DeLucca G.V., Shi Q., Tebben A.J. Carbazole Carboxamide Compounds Useful as Kinase Inhibitors. 8,362,065. U.S. Patent. 2013 Jan 9;
- 23.Batt D.G., Bertrand M.B., DeLucca G., Galella M.A., Ko S.S., Langevine C.M., Liu Q., Shi Q., Srivastava A.S., Tino J.A., et al. Substituted Tetrahydrocarbazole and Carbazole Carboxamide Compounds. 9,334,290. U.S. Patent. 2016 May 10;
- 24.Clark M., Cramer R.D., Van Opdenbosch N. Validation of the General Purpose Tripos 5.2 Force Field. J. Comput. Chem. 1989;10:982–1012. doi: 10.1002/jcc.540100804. [DOI] [Google Scholar]
- 25.Cramer R.D., III, Patterson D.E., Bunce J.D. Recent advances in comparative molecular field analysis (CoMFA) Prog. Clin. Biol. Res. 1989;291:161–165. [PubMed] [Google Scholar]
- 26.Balasubramanian P.K., Balupuri A., Gadhe C.G., Cho S.J. 3D QSAR modeling study on 7-aminofuro [2,3-c] pyridine derivatives as TAK1 inhibitors using CoMFA and COMSIA. Med. Chem. Res. 2014;24:2347–2365. doi: 10.1007/s00044-014-1221-8. [DOI] [Google Scholar]
- 27.Cai B.Q., Jin H.X., Yan X.J., Zhu P., Hu G.X. 3D-QSAR and 3D-QSSR studies of thieno[2,3-d]pyrimidin-4-yl hydrazone analogues as CDK4 inhibitors by CoMFA analysis. Acta Pharmacol. Sin. 2014;35:151–160. doi: 10.1038/aps.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aalizadeh R., Pourbasheer E., Ganjali M.R. Analysis of B-Raf V600E inhibitors using 2D and 3D-QSAR, molecular docking and pharmacophore studies. Mol. Divers. 2015;19:915–930. doi: 10.1007/s11030-015-9626-y. [DOI] [PubMed] [Google Scholar]
- 29.Wold S., Ruhe A., Wold H., Dunn W.J. The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM J. Sci. Stat. Comput. 1984;5:735–743. doi: 10.1137/0905052. [DOI] [Google Scholar]
- 30.Wehrens R., Putter H., Buydens L.M. The bootstrap: A tutorial. Chemometr. Intell. Lab. Syst. 2000;54:35–52. doi: 10.1016/S0169-7439(00)00102-7. [DOI] [Google Scholar]










