Abstract
The present study aimed to investigate molecular mechanisms associated with liver cancer and provide a possible therapeutic target for the treatment of liver cancer. Liver cancer patients that were diagnosed and treated at the Central Hospital of China National Petroleum Corp. were included in the present study. microRNA (miR)-222 was predicted to target B-cell lymphoma-2 (Bcl-2) binding component 3 (BBC3, also known as p53 upregulated modulator of apoptosis) by a bioinformatics analysis with TargetScan, which was verified by a dual-luciferase reporter assay system. The correlations between BBC3 and miR-222 levels and the patients' characteristics were analyzed. Furthermore, reverse transcription-quantitative polymerase chain reaction was used to assess the mRNA levels of miRNA-222 in the HCC-LM3, MHCC97H and HepG2 cell lines. HepG2 cells were then transfected with miR-222 inhibitor or miR-negative control inhibitor. Cell proliferation, apoptosis, cell cycle, migration and invasion were evaluated by an MTT assay, flow cytometry, wound healing assay and Transwell assay, respectively. BBC3 was quantified by immunofluorescence and western blot analysis, and cyclin D1, Bcl-2 and caspase-3 levels were also evaluated by western blotting. miR-222 inhibitor obviously inhibited HepG2 cell proliferation, migration, invasion, BBC3 and cyclin D1 protein expression levels and enhanced HepG2 cell apoptosis as well as the protein levels of Bcl-2 and caspase-3. miR-222 level in tumors ≥5 cm (maximum) was significantly higher compared with tumors <5 cm (maximum) and was significantly higher in metastatic tumors compared with non-metastatic tumors, while BBC3 level showed the adverse changes. The results of the present study suggested that miR-222 inhibitor exerted anti-cancer effects against liver cancer cells, probably by targeting the 3′ untranslated region (UTR) of BBC3.
Keywords: miRNA-222, liver cancer, B-cell lymphoma-2 binding component 3, p53 upregulated modulator of apoptosis
Introduction
Liver cancer, the symptoms of which include a lump or pain on the right side below the rib cage, swelling of the abdomen, yellowish skin, and weight loss, is also known as hepatic cancer and starts in the liver (1). Primary liver cancer is the sixth most common cancer type (accounting for 6% of all cancers) worldwide (2). Therefore, it is urgently required to identify a novel therapeutic target for liver cancer and explore the possible associated molecular mechanisms.
B-cell lymphoma-2 (Bcl-2), encoded by BCL2 gene, is a major member of the Bcl-2 family, a group of proteins with the ability to induce or inhibit cell apoptosis in humans (3,4). However, since Bcl-2 is not a growth signal transducer, it is deemed as a crucial anti-apoptotic protein. Among all of the members of the Bcl-2-homology 3 (BH3)-only subgroup of the Bcl-2 family, Bcl-2 binding component 3 (BBC3), also known as tumor protein 53 (TP53) upregulated modulator of apoptosis (PUMA), a transcriptional target of TP53 (5,6), acts as an essential apoptosis inducer (7,8). BBC3 takes part in numerous pathological and physiological processes, for instance, cancer and bacterial or viral infections (9).
The first microRNA (miRNA/miR) was discovered in the early 1990s (10,11). miRNAs are a class of small non-coding RNA with a length of ~22 nucleotides, which function in RNA silencing and post-transcriptional gene regulation (12,13), and have a role in various biological processes (14–16). To date, >1,000 miRNAs encoded by the human genome have been identified (17). miRNA-based therapies are currently under investigation (18–20).
Mounting evidence demonstrates the involvement of miRNAs in liver cancer (21,22); however, the corresponding molecular mechanisms have remained to be fully elucidated. The present study aimed to explore this matter in order to provide novel approaches for the treatment and diagnosis/prognostication of liver cancer patients.
Materials and methods
Tissue samples
In the present study, liver cancer tissues and adjacent normal liver tissues that were ≥2 cm distant from tumor margins from 31 patients who underwent surgery at the Central Hospital of China National Petroleum Corp. (Langfang, China) were obtained. None of the patients received any other therapies, including chemotherapy and radiotherapy. The primary tumors from patients with and without the presence of metastasis were extracted. Tissues were quickly removed and immediately placed in liquid nitrogen. The present study was approved by the ethics committee of the Central Hospital of China National Petroleum Corp. (Langfang, China). Written informed consent for the use of the specimens for research purposes was provided by each patient prior to the surgery.
Cell culture and transfection
The human liver cancer cell lines HCC-LM3, SMCC7721 and HepG2, and 293 cells were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China) and cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both from GE Healthcare, Little Chalfont, UK) and 1% penicillin/streptomycin in an incubator with 5% CO2 at 37°C.
Hepatoblastoma-derived HepG2 cells (23) were randomly divided into 3 groups, i.e., the control group, miR-negative control (NC) inhibitor group and miR-222 inhibitor group. HepG2 cells were transfected with miR-222 inhibitor or miR-NC inhibitor (RiboBio, Guangzhou, China) for 48 h with Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in accordance with the manufacturer's instructions.
Luciferase activity assay
By using TargetScan (http://www.targetscan.org), BBC3 was predicted to be a target gene of miR-222; therefore, the recombinant reporter plasmids pmir-BBC3 wild-type (wt)-3′UTR and pmir-BBC3 mutant (mut)-3′UTR were constructed as followed: The 3′UTR of BBC3 was amplified from cDNA of 293 cells and cloned into pmir plasmid (Invitrogen; Thermo Fisher Scientific, Inc.) to construct pmir-BBC3 wt-3′UTR, while the pmir-BBC3 mut-3′UTR was obtained by Stratagene mutation kit (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA) based on the manufacture's protocol. Following incubation of HepG2 cells for 48 h when cells were allowed to attach after seeding, they were co-transfected with miR-222 inhibitor or miR-NC inhibitor and pmir-BBC3wt-3′UTR or pmir-BBC3mut-3′UTR by using Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h after transfection, the relative activity of luciferase in different groups was tested.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted and isolated from tissues or cultured HCC-LM3, SMCC7721 and HepG2 cells with a miRNeasy Mini Kit (Qiagen, Hilden, Germany). Complementary (c)DNA was first synthesized with an iScrip™ cDNA Synthesis Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). For the assessment of the miR-222 levels, cDNA was amplified with SYBR Premix Ex Taq™ (Takara Bio, Inc., Otsu, Japan) in a CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.). The conditions were as following: Denaturation at 95°C (5 min), followed by 40 cycles of denaturation at 95°C (10 sec) and annealing/extension at 60°C (30 sec). The relative content of miR-222 was calculated via the 2−ΔΔCq method (24). U6 was used as an internal control. Primers were as followed: miR-222 forward, GTGCAAGTGTAGATGCCGACAA and reverse, GTGCAAGTGTAGATGCCGACAA; U6 forward, CTCGCTTCGGCAGCACA and reverse, AACGCTTCACGAATTTGCGT.
Flow cytometric assays
The cell cycle distribution in the different groups was evaluated by flow cytometry. HepG2 cells were seeded into 96-well plates at the seeding density of 3×104/well and transfected with miR-222 inhibitor or miR-NC inhibitor. After 48 h of incubation, the cells were detached with 0.025% trypsin (Thermo Fisher Scientific, Inc.), washed with pre-cooled PBS and 70% cold ethanol, and finally fixed overnight at the temperature of 4°C. Subsequently, propidium iodide (PI) staining solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), RNaseA (cat no. R1253; Thermo Fisher Scientific, Inc.) and 0.2% Triton X-100 were added to HepG2 cells, followed by incubation at 4°C for 30 min in the dark. The cell suspension was then adjusted to the concentration of 1×106 cells/ml. The cellular DNA content was evaluated with a MoFlo XDP Cell Sorter (Beckman Coulter, Brea, CA, USA), and the percentage of cells at each phase was analyzed by FlowJo software version 7.6.3 (FlowJo LLC, Ashland, OR, USA).
To detect the apoptotic rate, the cells were seeded in 12-well plates at the seeding density of 3×105/well and cultured for 48 h. Thereafter, the cells were harvested with 0.025% trypsin (Thermo Fisher Scientific, Inc.) and collected. After washing with PBS three times, 5 µl fluorescein isothiocyanate-labeled Annexin V (FITC) and 5 µl PI was added into the cells and incubated in the dark for 15 min at 37°C. The cell apoptosis rate was analyzed by flow cytometry within 1 h.
MTT assay
HepG2 cell proliferation was measured by an MTT assay. HepG2 cells were seeded into 96-well plates at the density of 3×104 cells/well and cultured for 6, 12, 24 and 48 h, respectively. After the addition of MTT solution (20 µl, 5 mg/ml) to each well, HepG2 cells were further cultured at 37°C for 4 h. After the aspiration of culture medium from each well, 200 µl dimethylsulfoxide was added to fully dissolve the formazan crystals. A Synergy H1 Multi-Mode Reader (BioTek, Winooski, VT, USA) was used to read the absorbance of each well at the wavelength of 570 nm.
Wound healing assay
A wound healing assay was used for the detection of the cell migration ability. At 24 h after transfection of miR-222 inhibitor or miR-NC inhibitor, HepG2 cells were seeded into 6-well plates at 8×105 cells per well. After the attainment of 90% confluence, the surface of each well was scratched with a 20-µl pipette. After washing the wells with PBS for three times, they were further cultured at 37°C for 24 h and images were then captured.
Transwell invasion assay
The invasion ability of HepG2 cells was evaluated using Transwell chambers coated with Matrigel® (8 µm; BD Biosciences, Franklin Lakes, NJ, USA). The bottom chamber was filled with DMEM and 10% FBS. HepG2 cells (1×105 in 100 µl) were seeded in the upper wells with serum-free DMEM and incubated at 37°C. After 16 h, non-invading cells on the upper surface were removed, while cells invaded to the lower side of the polycarbonate membrane were fixed with ethanol for 30 min at room temperature and stained with crystal violet for 5 min at room temperature. The number of invaded cells in six independent fields of view was counted under a microscope.
Western blot analysis
HepG2 cells were lysed with radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) for the extraction of protein samples. Protein concentration was determined by the BCA method (Beyotime Institute of Biotechnology, Shanghai, China). Total protein (10 µg per lane) was separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). PVDF membranes were blocked with 5% bovine serum albumin (BSA; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) at room temperature for 1–2 h before the incubation with primary antibody to Bcl-2 (12789; 1:1,000; ProteinTech Group, Inc., Chicago, IL, USA), cyclin D1 (2978; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), BBC3 (sc-28226; 1:1,000, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), cleaved caspase-3 (9665; 1:1,000) and GAPDH (5174; 1:1,000) (both from Cell Signaling Technology, Inc.) at 4°C overnight, and then washed with TBST for 3 times for 10 min each. The PVDF membranes were treated with horseradish peroxidase-labeled goat anti-rabbit secondary antibody (ab97080; 1:5,000; Abcam, Cambridge, MA, USA) for 1 h at room temperature. Finally, protein bands were visualized by enhanced chemiluminescence (Pierce; Thermo Fisher Scientific, Inc.). GAPDH was used as an internal control.
Immunofluorescence
For immunofluorescence analysis, cells were incubated with primary and secondary antibody, respectively. Images were captured under a microscope. In brief, the protocol was as follows: HepG2 cells were seeded on glass slides in 24-well plates and cultured at 37°C for 24 h. After washing with PBS three times, HepG2 cells were treated with 4% paraformaldehyde for 30 min, and permeabilized/blocked with 0.1% Triton X-100 for 1 h at room temperature. Rabbit monoclonal BBC3 antibody (sc-28226; 1:1,000; Santa Cruz Biotechnology, Inc.) was used as primary antibody and incubated at 4°C overnight. Alexa Fluor 488 (A32731, 1:1,000, Invitrogen; Thermo Fisher Scientific, Inc.) was used as secondary antibody to detect fluorescence and incubated at room temperature for 2 h. The cell nuclei were stained by DAPI (Vector Laboratories, Ltd., Peterborough, UK) at room temperature for 2 h. Images were captured by Leica DM5000 B (Leica Microsystems, Buffalo Grove, IL, USA).
Statistical analysis
Statistical analyses were performed using SPSS 21.0 (IBM Corporation, Armonk, NY, USA). Values are expressed as the mean ± standard error of the mean. Data between two groups were compared with the Student's t-test, while data among 3 groups were compared by one-way analysis of variance followed by Newman-Keuls analysis. P<0.05 was considered to indicate a significant difference.
Results
miR-222 directly regulates the 3′UTR of the mRNA of BBC3
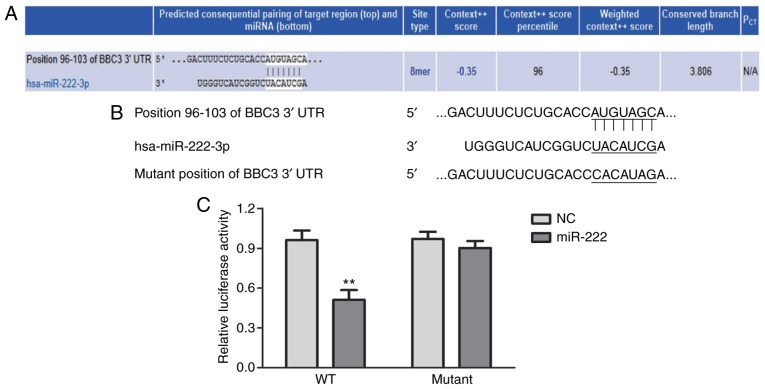
TargetScan was applied for the prediction of miRNAs that bind with the 3′UTR of the mRNA of BBC3. A miRNA that had a relatively high score was selected for the present study. As displayed in Fig. 1A, miR-222 was predicted to target the 3′UTR of the mRNA of BBC3. The predicted interaction between miR-222 and the 3′UTR of the mRNA of BBC3 was then verified by using a dual-luciferase reporter assay system. The position of the mutation introduced in the 3′UTR of the mRNA of BBC3 is presented in Fig. 1B. The relative luciferase activity in HepG2 cells that were transfected with miR-222 inhibitor and pmir-BBC3wt-3′UTR was obviously lower than that in HepG2 cells transfected with miR-222 inhibitor and pmir-BBC3mut-3′UTR (Fig. 1C). Thus, the 3′UTR of the mRNA of BBC3 was verified to be a direct target gene of miR-222.
Figure 1.
miR-222 targets the 3′UTR of the mRNA of BBC3. (A) miR-222 was predicted to target the mRNA of BBC3 3′UTR. (B) The mutation introduced in the 3′UTR of the mRNA of BBC3. (C) The relative luciferase activity in HepG2 cells that were transfected with miR-222 inhibitor and pmir-BBC3wt-3′UTR was obviously lower than that in HepG2 cells which were transfected with miR-222 inhibitor and pmir-BBC3mut-3′UTR. **P<0.01 miR-222 group vs. NC group. miR/miRNA, microRNA; BBC3, B-cell lymphoma 2 binding component 3; UTR, untranslated region; mut, mutant; wt, wild-type; NC, negative control; hsa, Homo sapiens.
miR-222 and BBC3 levels are associated with the tumor size and metastasis in liver cancer
The characteristics of 31 liver cancer patients, including sex, age, tumor size, stage and metastasis, are displayed in Table I. Regarding the patients' sex, age and tumor histological grade, no significant differences in miR-222 or BBC3 levels were identified between the respective sub-groups. As for the tumor size, the miR-222 levels in tumors with a maximum diameter of ≥5 cm (3.13±0.66) was significantly higher compared with that in tumors of <5 cm in maximum diameter (2.37±0.85; P=0.02); while significantly lower BBC3 levels were identified in tumors sized maximum ≥5 cm (2.29±0.36) than in those of <5 cm in maximum size (2.88±0.44; P=0.04). In addition, the miR-222 levels in metastatic tumors (3.27±0.34) were obviously higher than in non-metastatic tumors (2.49±0.63; P=0.04); however, BBC3 levels were downregulated in metastatic tumors (2.11±0.42) compared with those in non-metastatic tumors (2.99±0.49; P=0.01). Taken together, a negative correlation was identified between miR-222 and BBC3.
Table I.
Correlation between miR-222 or BBC3 and the characteristics of liver cancer patients.
| Factor | Cases (n) | miR-222 | P-value | BBC3 | P-value |
|---|---|---|---|---|---|
| Sex | |||||
| Male | 27 | 2.75±1.34 | 0.79 | 2.63±0.68 | 0.27 |
| Female | 4 | 2.95±1.12 | 2.67±0.47 | ||
| Age (years) | |||||
| <60 | 13 | 2.99±1.53 | 0.51 | 1.97±0.64 | 0.68 |
| ≥60 | 18 | 2.65±1.32 | 2.13±0.33 | ||
| Maximum tumor size (cm) | |||||
| ≥5 | 11 | 3.13±0.66 | 0.02 | 2.29±0.36 | 0.04 |
| <5 | 20 | 2.37±0.85 | 2.88±0.44 | ||
| Histological grade | |||||
| Well/moderate differentiation | 15 | 2.78±1.33 | 0.25 | 2.45±0.57 | 0.08 |
| Poor differentiation | 16 | 2.24±1.25 | 2.87±0.68 | ||
| Metastasis | |||||
| No | 28 | 2.49±0.63 | 0.04 | 2.99±0.49 | 0.01 |
| Yes | 3 | 3.27±0.34 | 2.11±0.42 |
Values are expressed as the mean ± standard error of the mean. BBC3, B-cell lymphoma-2 binding component 3; miR, microRNA.
miR-222 levels in HCC-LM3, SMCC7721 and HepG2 cell lines
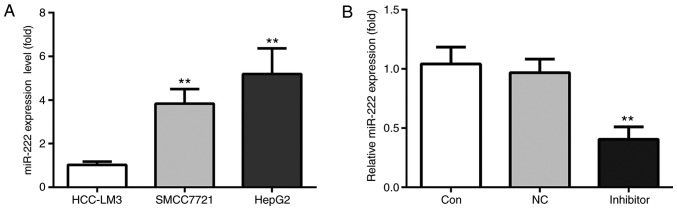
RT-qPCR was used to examine the miR-222 levels among 3 human liver cancer cell lines, including HCC-LM3, SMCC7721 and HepG2. The results indicated that HepG2 cells had the highest level of miR-222 among the 3 cell lines, which was not significantly different from that in SMCC7721 cells (Fig. 2A). Therefore, HepG2 cells were used in all of the subsequent experiments. First of all, HepG2 cells were randomly divided into 3 groups, namely the control group, the miR-NC inhibitor group and miR-222 inhibitor group. The results indicated no significant difference in miR-222 expression levels between the control group and the miR-NC inhibitor group, while miR-222 was notably decreased in the miR-222 inhibitor group (Fig. 2B). In conclusion, miR-222 inhibitor was successfully transfected into HepG2 cells.
Figure 2.
miR-222 expression is highest in the HepG2 cell line. (A) Among 3 human liver cancer cell lines, including HCC-LM3, SMCC7721 and HepG2, the latter had the highest level of miR-222. Therefore, HepG2 cells were used in the subsequent experiments. (B) No significant difference in the expression levels of miR-222 was detected between the control group and miR-NC inhibitor group, but it was notably decreased in the miR-222 inhibitor group. **P<0.01 miR-222 inhibitor group vs. NC group. miR, microRNA; NC, negative control.
miR-222 inhibitor induces apoptosis and decreases proliferation of HepG2 cells
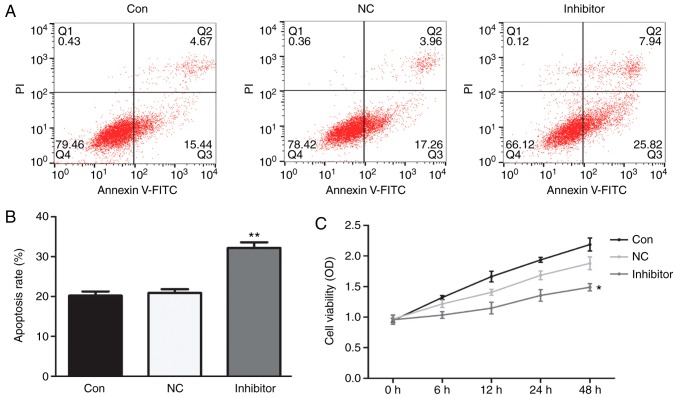
Cell apoptosis in the different groups was evaluated by flow cytometry. The results of the mean values of the apoptotic rate indicated that, compared with HepG2 cells in the control group (20.11%) and those transfected with miR-NC inhibitor (21.22%), the apoptotic rate was significantly higher in the group transfected with miR-222 inhibitor (33.76%) (Fig. 3A and B).
Figure 3.
miR-222 inhibitor increases the apoptotic rate of HepG2 cells and decreases their proliferation. No obvious changes in HepG2 cell apoptosis or proliferation were identified between the control and the miR-NC inhibitor group. (A and B) Compared with HepG2 cells in the control group and miR-NC inhibitor group, the apoptotic rate was significantly increased by treatment with miR-222 inhibitor, while (C) the proliferation ability was significantly reduced by miR-222 inhibitor. **P<0.01 miR-222 inhibitor group vs. NC group. miR, microRNA; NC, negative control; PI, propidium iodide; Con, control; FITC, fluorescein isothiocyanate; OD, optical density; Q, quadrant.
Changes in the proliferation of HepG2 cells in different groups were evaluated by an MTT assay. The results demonstrated that, compared with HepG2 cells in the control group (2.02%) and miR-NC inhibitor treatment group (1.75%), the proliferation ability was significantly reduced by treatment with miR-222 inhibitor (1.26%; Fig. 3C).
miR-222 inhibitor causes cell cycle arrest of HepG2 in G0/G1 phase
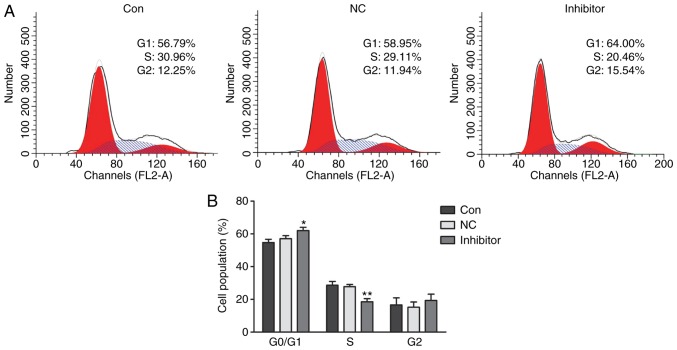
Regarding the cell cycle distribution of HepG2 cells, no obvious changes were observed between the control group and the miR-NC inhibitor group; furthermore, there was also no notable difference in the G2 phase population among the 3 groups. However, in the miR-222 inhibitor group, the G0/G1-phase population (64.00%) was significantly increase compared with that in the in control (56.79%) and miR-NC (58.95%) groups, and the S-phase population in the miR-222 inhibitor group (20.46%) was significantly decreased in comparison with that in the control group (30.96%) and miR-NC group (29.11%) (Fig. 4A and B).
Figure 4.
miR-222 inhibitor induces cell cycle arrest of HepG2 cells at G0/G1 phase. (A) Cell cycle distribution profiles and (B) quantified populations in each phase of the cell cycle are presented. No obvious changes in the cell populations in each phase were observed between the control group and the miR-NC inhibitor group. Furthermore, no notable difference in the number of cells in G2 phase was observed between the 3 groups. However, the G0/G1 phase population in the miR-222 inhibitor group (64.00%) was significantly increased compared with that in the control (56.79%) and miR-NC group (58.95%); and the S-phase population in the miR-222 inhibitor group (20.46%) was decreased in comparison with that in the control (30.96%) and miR-NC group (29.11%). *P<0.05, **P<0.01 miR-222 inhibitor group vs. NC group. miR, microRNA; NC, negative control.
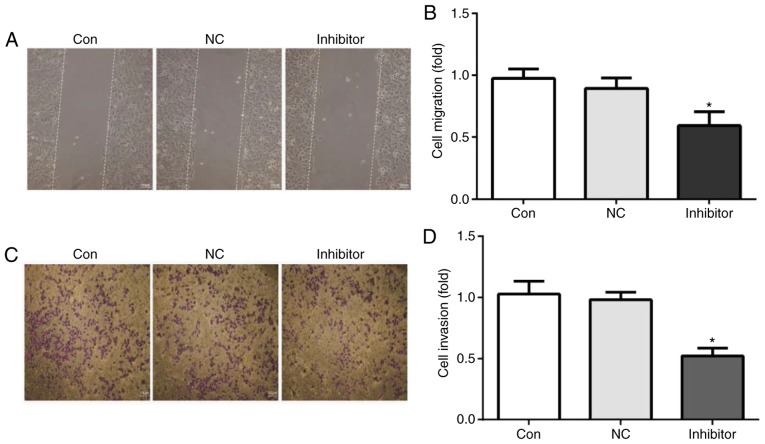
miR-222 inhibitor reduces HepG2 cell migration and invasion
The migration and invasion ability of HepG2 cells was assessed using a wound healing assay and Transwell chambers, respectively. The results indicated that, compared with HepG2 cells in the control group and the miR-NC inhibitor group, the cell migration ability (Fig. 5A and B) and cell invasion ability were significantly reduced by miR-222 inhibitor (Fig. 5C and D).
Figure 5.
miR-222 inhibitor impairs HepG2 cell migration and invasion. (A and B) The cell migration ability was tested by a wound healing assay and (C and D) the cell invasion ability of was assessed using Transwell chambers. The results indicated that, compared with HepG2 cells in the control and miR-NC inhibitor group, the cell migration and invasion ability were significantly reduced by miR-222 inhibitor (scale bar, 100 µm). miR, microRNA; NC, negative control; Con, control. *P<0.05 miR-222 inhibitor group vs. NC group. miR, microRNA; NC, negative control.
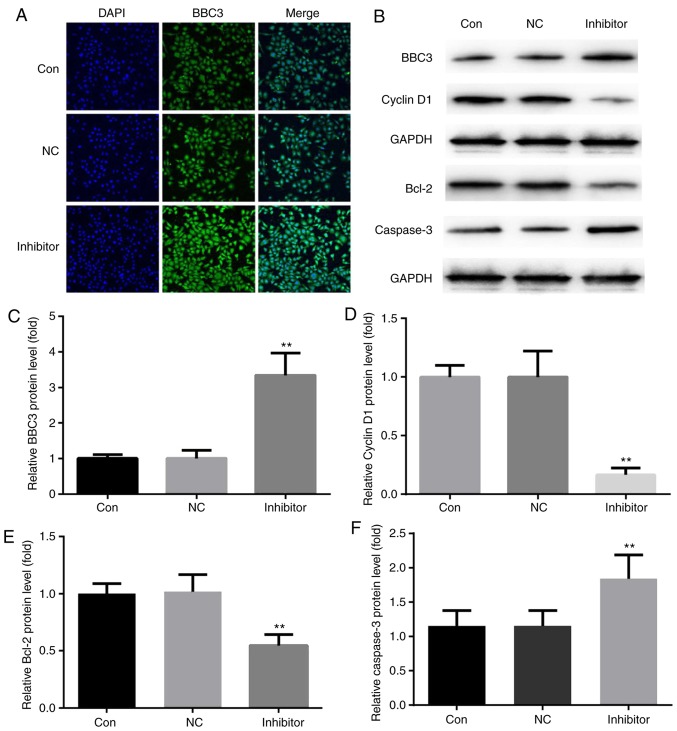
Effects of miR-222 inhibitor on Bcl-2/BBC3/cyclin D1/caspase-3 protein levels in HepG2 cells
Compared with the immunofluorescence of HepG2 cells in the control group and the miR-NC inhibitor group, miR-222 inhibitor significantly increased the immunofluorescence of BBC3 protein in HepG2 cells (Fig. 6A). Furthermore, western blot analysis indicated that compared with those in the control and miR-NC inhibitor groups, miR-222 inhibitor decreased the protein levels of Bcl-2/cyclin D1 and increased BBC3/cleaved caspase-3 protein levels in HepG2 cells (Fig. 6B–F).
Figure 6.
miR-222 inhibitor promotes the expression of BBC3 in HepG2 cells. (A) Compared with that in HepG2 cells in the control and miR-NC inhibitor groups, miR-222 inhibitor significantly increased the immunofluorescence intensity of BBC3 (magnification, ×400). (B) Western blot analysis indicated that compared with HepG2 cells in the control and miR-NC inhibitor groups, miR-222 inhibitor decreased Bcl-2 and cyclin D1 protein levels, increased the protein levels of BBC3 and cleaved caspase-3 in HepG2 cells. (C–F) The corresponding statistical data of BBC3, cyclin D1, Bcl-2, and cleaved caspase-3 were exhibited. Bcl-2, B-cell lymphoma 2; miR, microRNA; NC, negative control; Con, control; BBC3, Bcl-2 binding component 3. **P<0.01 miR222 inhibitor group vs. NC group.
Discussion
The present study indicated that miR-222 inhibitor obviously reduced the cell proliferation/migration/invasion/Bcl-2/cyclin D1 protein levels and promoted cell apoptosis as well as BBC3/caspase-3 protein levels, probably via binding with BBC3 in HepG2 cells. Taken together, the results of the present study suggested that miR-222 inhibitor reduced the progression of liver cancer cells by targeting the 3′UTR of the mRNA of BBC3, and providing a possible therapeutic target for the treatment of liver cancer.
Liver cancer starts in the liver (1). Primary liver cancer is the sixth most common cancer type (accounting for 6% of all cancers) worldwide (2). In order to improve the prognosis of liver cancer patients, it is required to identify novel therapeutic targets.
BBC3 promotes cell apoptosis and regulates pathophysiological processes during the development of cancer (6–8). The involvement of miRNAs has been reported in liver cancer, with indistinct molecular mechanisms (20,21). In the present study, TargetScan was used to predict miRNAs which recognize the 3′UTR of the mRNA of BBC3. Among all of the miRNAs that targeted the 3′UTR of the mRNA of BBC3, miR-222 had a relatively high score was selected for assessment in the present study. The relative luciferase activity in HepG2 cells transfected with miR-222 inhibitor and pmir-BBC3wt-3′UTR was identified to be obviously lower than that in cells transfected with miR-222 inhibitor and pmir-BBC3mut-3′UTR, which verified that the 3′UTR of the mRNA of BBC3 was indeed targeted by miR-222.
RT-qPCR was used to examine the levels of miR-222 mRNA in the HCC-LM3, SMCC7721 and HepG2 cell lines. In HepG2 cells, the level of miR-222 was highest among the 3 cell lines. HepG2 cells were therefore selected for the remaining experiments. The results suggested that miR-222 has an oncogenic function in liver cancer, which was in line with a previous study performed on HepG2 cells (25).
However, it has remained elusive whether miR-222 affects the behavior of liver cancer cells, e.g. HepG2, via BBC3. In the present study, HepG2 cells were randomly divided into 3 groups, including a control group, miR-NC group and miR-222 inhibitor group. An MTT assay, a wound healing assay, a Transwell invasion assay and flow cytometry were used to determine the roles of miR-222 in cell proliferation, migration, invasion, apoptosis and cell cycle, respectively. The results indicated that, compared with the control and miR-NC inhibitor groups, miR-222 inhibitor obviously inhibited the cell proliferation/migration/invasion and promoted the apoptosis of HepG2 cells, which suggested that miR-222 inhibitor has a role in ameliorating liver cancer. Furthermore, miR-222 inhibitor induced a block of HepG2 cells at G1/G0 phase. These results were consistent with a previous study, which reported that miR-222 inhibitor restrains prostate cancer cell migration and invasion, and promotes prostate cancer cell apoptosis (26,27). The corresponding molecules involved in the aforementioned processes remain to be elucidated.
The Bcl-2 family of proteins induces or represses cell apoptosis in humans (5,6). Bcl-2 itself is considered to have a crucial anti-apoptotic role. Caspase-3, encoded by the CASP3 gene, is activated in apoptotic cells through the extrinsic (death ligand) and intrinsic (mitochondrial) pathways (28,29); however, cleaved caspase-3 is active in normal and apoptotic cells (30). Cyclin D1, a protein encoded by the CCND1 gene in humans (31,32), is expressed in all adult tissues except cells derived from bone marrow stem cells (33,34). Overexpression of cyclin D1 has been associated with early cancer onset/progression (35), shorter overall survival and higher metastasis in patients (36,37). The protein levels of Bcl-2, BBC3, cyclin D1 and caspase-3 in 3 different groups were determined by western blot analysis. The results indicated that, compared with those in the control and miR-NC inhibitor groups, miR-222 inhibitor obviously inhibited Bcl-2/cyclin D1 protein levels and promoted BBC3/caspase-3 protein levels. Furthermore, miR-222 inhibitor promoted the immunofluorescence of BBC3.
Taken together, miR-222 was demonstrated to have a crucial role in the HepG2 cell line via targeting the 3′UTR of the mRNA of BBC3, and may be a therapeutic target for the treatment of liver cancer. HepG2 is a hepatoblastoma cell line, which is a rare childhood cancer; the results should be confirmed in other cell lines representing more common types of liver cancer, e.g. in SMCC7721, in future studies.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
ZL and CD designed the experiments. ZL, JS, BL and MZ performed the experiments, analyzed and interpreted the data. EX and CD were major contributors in writing the manuscript. The final version of the manuscript has been read and approved by all authors, and each author believes that the manuscript represents honest work.
Ethics approval and consent to participate
The present study was approved by the ethics committee of the Central Hospital of China National Petroleum Corp. (Langfang, China).
Consent for publication
Written informed consent for the use of the specimens for research purposes was provided by each patient prior to the surgery.
Competing interests
The authors declare that they have no competing interests regarding this study.
References
- 1.Adult Primary Liver Cancer Treatment (PDQ®)-Patient Version. NCI. 6 July 2016. Archived from the original on October 2016. Retrieved 29 September 2016. [Google Scholar]
- 2.World Cancer Report . World Health Organization. 6. Vol. 2014. 2014. p. 5. [Google Scholar]
- 3.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t (14;18) chromosome translocation. Science. 1984;226:1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 4.Cleary ML, Smith SD, Sklar J. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t (14;18) translocation. Cell. 1986;47:19–28. doi: 10.1016/0092-8674(86)90362-4. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/S1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 6.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/S1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 7.Chipuk JE, Green DR. PUMA cooperates with direct activator proteins to promote mitochondrial outer membrane permeabilization and apoptosis. Cell Cycle. 2009;8:2692–2696. doi: 10.4161/cc.8.17.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thakur VS, Ruhul Amin AR, Paul RK, Gupta K, Hastak K, Agarwal MK, Jackson MW, Wald DN, Mukhtar H, Agarwal ML. p53-Dependent p21-mediated growth arrest pre-empts and protects HCT116 cells from PUMA-mediated apoptosis induced by EGCG. Cancer Lett. 2010;296:225–232. doi: 10.1016/j.canlet.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl 1):S71–S83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V, Feinbaum A. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 11.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cel. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 15.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 16.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 17.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 18.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–S57. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Feng Y, Coukos G, Zhang L. Therapeutic microRNA strategies in human cancer. AAPS J. 2009;11:747–757. doi: 10.1208/s12248-009-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F. microRNA: Emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther. 2010;125:92–104. doi: 10.1016/j.pharmthera.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Gyugos M, Lendvai G, Kenessey I, Schlachter K, Halász J, Nagy P, Garami M, Jakab Z, Schaff Z, Kiss A. MicroRNA expression might predict prognosis of epithelial hepatoblastoma. Virchows Arch. 2014;464:419–427. doi: 10.1007/s00428-014-1549-y. [DOI] [PubMed] [Google Scholar]
- 22.von Frowein J, Pagel P, Kappler R, von Schweinitz D, Roscher A, Schmid I. MicroRNA-492 is processed from the keratin 19 gene and up-regulated in metastatic hepatoblastoma. Hepatology. 2011;53:833–842. doi: 10.1002/hep.24125. [DOI] [PubMed] [Google Scholar]
- 23.López-Terrada D, Cheung SW, Finegold MJ, Knowles BB. HepG2 is a hepatoblastoma-derived cell line. Hum Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Yang YF, Wang F, Xiao JJ, Song Y, Zhao YY, Cao Y, Bei YH, Yang CQ. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. Int J Clin Exp Med. 2014;7:893–902. [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Yang YM, Gan R, Zhao LX, Li W, Zhou HB, Wang XJ, Lu JX, Meng QH. Down-regulation of miR-221 and miR-222 restrain prostate cancer cell proliferation and migration that is partly mediated by activation of SIRT1. Plos One. 2014;9:e98833. doi: 10.1371/journal.pone.0098833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandhare-Dash J, Mantri CK, Gong Y, Chen Z, Dash C. XMRV accelerates cellular proliferation, transformational activity, and invasiveness of prostate cancer cells by downregulating p27 (Kip1) Prostate. 2012;72:886–897. doi: 10.1002/pros.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvesen GS. Caspases: Opening the boxes and interpreting the arrows. Cell Death Differ. 2002;9:3–5. doi: 10.1038/sj.cdd.4400963. [DOI] [PubMed] [Google Scholar]
- 29.Ghavami S, Hashemi M, Ande SR, Yeganeh B, Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ, Los M. Apoptosis and cancer: Mutations within caspase genes. J Med Genet. 2009;46:497–510. doi: 10.1136/jmg.2009.066944. [DOI] [PubMed] [Google Scholar]
- 30.Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 31.Motokura T, Bloom T, Kim HG, Jüppner H, Ruderman JV, Kronenberg HM, Arnold A. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature. 1991;350:512–515. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- 32.Lew DJ, Dulić V, Reed SI. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-W. [DOI] [PubMed] [Google Scholar]
- 33.Withers DA, Harvey RC, Faust JB, Melnyk O, Carey K, Meeker TC. Characterization of a candidate bcl-1 gene. Mol Cell Biol. 1991;11:4846–4853. doi: 10.1128/MCB.11.10.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inaba T, Matsushime H, Valentine M, Roussel MF, Sherr CJ, Look AT. Genomic organization, chromosomal localization, and independent expression of human cyclin D genes. Genomics. 1992;13:565–574. doi: 10.1016/0888-7543(92)90126-D. [DOI] [PubMed] [Google Scholar]
- 35.Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 36.Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: Perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–762. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- 37.Thomas GR, Nadiminti H, Regalado J. Molecular predictors of clinical outcome in patients with head and neck squamous cell carcinoma. Int J Exp Pathol. 2005;86:347–363. doi: 10.1111/j.0959-9673.2005.00447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.